Abstract
Spermatogenesis is regulated by a complex network of posttranslation modifications. Sumoylation (a modification by small ubiquitin-like modifiers, or SUMO proteins) was identified as an important cellular event in different cell types. SUMO proteins are highly expressed in the testis, and their role during spermatogenesis has begun to be elucidated. Given the important role of sumoylation in the regulation of mitosis and cancer progression in other tissues, the aim of the current study was to identify the targets of SUMO in proliferating mouse spermatogonia and human seminoma tissues and to initially examine the level of sumoylation in relation to the proliferative activity of the tissues. Using freshly purified spermatogonia and C18-4 spermatogonia cell line, mass spectrometry analysis identified several SUMO targets implicated into the proliferation of spermatogonia (such as heat shock protein 60 [HSP60] and prohibitin). Tissue array and western blot approaches showed that SUMO expression is a prominent feature of human seminomas and that the proliferative activity of the tumor tissues was positively correlated with the level of SUMO expression. Downregulation of sumoylation with si-RNA was not sufficient to significantly affect the proliferation of C18-4 spermatogonia; however, SUMO overexpression increased the proliferation rate of the cells. These data suggest that cells are more sensitive to an elevated level of SUMO, and that this situation may lead to an upregulated cellular proliferation and, possibly, cancer. Mass spectrometry analysis identified around a hundred SUMO targets in seminoma samples. Notably, many of the identified proteins (such as proliferating cell nuclear antigen [PCNA], DNA topoisomerase 2-alpha [Top2A], prohibitin, 14-3-3 protein, and others) were implicated in oncogenic transformation and cancer progression.
Keywords: proliferation, seminoma, spermatogonia, sumoylation
INTRODUCTION
Adult spermatogenesis consists of phases of stem cell and spermatogonial proliferation, meiosis of spermatocytes, and the postmeiotic maturation of spermatids (spermiogenesis). During the initial wave of spermatogenesis in mice, spermatogonia arise around day 4 postpartum (dpp) and undergo several rounds of proliferation and differentiation. Understanding of the molecular mechanisms that control the early steps of spermatogenesis is important as aberrant expression of signaling molecules may result in proliferating or differentiating defects.
Studies of sumoylation (a modification by small ubiquitin-like modifiers, or SUMO proteins) in male reproductive tissues were initiated almost 15 years ago. We, and others, have localized SUMO during mouse and human spermatogenesis. The localization pattern suggested diverse and potentially multiple roles of SUMO in testicular function and during spermatogenesis.1,2,3,4,5,6 Another study from our group has provided evidence that sumoylation is involved in the stress response in germ cells.1,2,3,4,5,6,7 We have also performed a broad-spectrum identification of SUMO targets in purified mouse spermatocytes and spermatids. Numerous proteins with important roles during spermatogenesis and meiosis have been identified.8,9 Notably, inhibition of sumoylation with the inhibitor Gingkolic acid (GA) arrested the G2/M1 transition in purified mouse spermatocytes in vitro.10 This inhibition was accompanied by significant changes in the activity of the two major kinases (polo-like kinase 1 [PLK1] and Aurora kinase B [AURKB]), which regulate meiotic prophase.10
Sumoylation regulates various aspects of mitosis in different cell types, which was implicated in the development and progression of cancer.11,12,13,14,15 Many important cell cycle regulators and oncogenes have been identified as sumoylation targets.12,16 Inactivation of sumoylation machinery in mouse resulted in early embryonic lethality with severe disruptions in mitosis, a finding that supports the indispensable role of sumoylation in cell cycle progression.17 However, targets and the role of sumoylation in proliferating spermatogonia remained to be elucidated. It is also unknown whether SUMO proteins are expressed in testicular cancerous tumors and if sumoylation plays a role in their initiation and progression.
As has been shown in previous studies from our and other laboratories, the identification of targets for sumoylation is a critical step toward understanding the cellular functions of SUMO.8,16,18,19,20,21,22 Given the important role of sumoylation in the regulation of mitosis in other tissues, the initial aim of the current study was to identify the targets of sumoylation in mouse spermatogonia. To achieve this aim, freshly purified mouse spermatogona and a stem-like C18-4 spermatogonia cell line were used for a mass-spectromic identification of sumoylated proteins. To obtain insights into a possible role of sumoylation in cell cycle control in human germ cell tumors, we have also identified SUMO targets in cancerous cells from seminoma patients and initially examined the level of sumoylation in relation to the proliferative activity of the tissues.
MATERIALS AND METHODS
Differentiating plating procedure
Differentiating plating procedure was used to separate mouse Sertoli cells from spermatognia using pubertal testes. Sertoli cells attach to Matrigel-coated dishes, but spermatogonia remain suspended in the medium. For the attachment of Sertoli cells, culture flasks were coated with Matrigel (BD Biosciences, San Jose, CA, USA). Briefly, Matrigel was thawed in a 4°C refrigerator overnight. Each milliliter of Matrigel was diluted with 7 ml of cold Dulbecco's Modified Eagle completed media (DMEM) basal media (Life Technologies, Carlsbad, CA, USA), and the solution was added to T-75 flasks to cover the whole surface of the flask. The flasks were incubated overnight, and the excess Matrigel solution was then removed. The flasks were washed with the medium.
C57BL/6NCrl mice were purchased from Charles River (Kingston, NY, USA). The Animal Committee of Albert Einstein College of Medicine (Bronx, NY, USA), approved all animal protocols. For the isolation of spermatogonia, 15–18 pubertal mice (7–8 dpp) were sacrificed, and their testes were isolated, decapsulated, and enzymatically digested, first with collagenase (1 mg ml−1) and DNase I (1 μg ml−1) for 4 min to remove Leydig cells, and then with collagenase (1 mg ml−1), trypsin (0.5 mg ml−1), hyaluronidase (1.5 mg ml−1), and DNase I (1 μg ml−1) together for 8 min. The enzymes were obtained from Sigma-Aldrich (St. Louis, MO, USA). Both the digestions were performed with constant shaking at 150 rpm in a 34°C water bath (C76 water bath shaker, New Brunswick Scientific, Edison, NJ, USA). The cells were filtered through a 70-μm filter, counted, centrifuged at 300g at 4°C for 7 min, and re-suspended in a prewarmed DMEM (Sigma-Aldrich) at an approximate concentration of 0.5–1 × 107 cells per ml. Each milliliter of the cell suspension was added per one T75 flask coated with Matrigel. The flasks were incubated for about 4 h or overnight at 34°C. After the incubation, the nonadhesive spermatogonia were collected, and the flasks were washed several times with gentle agitation to release spermatogoia from Sertoli cells. The Sertoli cells were then removed from the flask by brief trypsinization. The purity of the isolation was confirmed using germ- and Sertoli-specific markers (anti-germ cell-specific antigen antibody [TRA98],23 and GATA-Binding Factor 4 [GATA4],24 respectively). Both antibodies were from Abcam (Cambridge, MA, USA). Nuclei of all cells were counterstained by 4´,6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich). The percentage of the fraction purity was calculated by dividing the number of cells positive for the specific marker by the total number of DAPI-positive cells from several microscopic fields.
C18-4 spermatogonia cell line
The C18-4 cell line was generously provided by Dr. Marie-Claude Hofmann (MD Anderson Cancer Center, Huston, TX, USA). This cell line was established from type A spermatogonia obtained from testes of 6-day-old mice.25 The C18-4 cells express various markers for proliferating spermatogonia, and spermatogonial stem cells (SSCs), including germ cell nuclear antigen 1 (GCNA1), mouse ortholog of VASA, deleted in azoospermia-like (DAZL), proliferating cell nuclear antigen (PCNA), octamer-binding transcription factor 4 (OCT-4), glial cell line-derived neurotrophic factor family receptor alpha-1 (GFRA1), rearranged during transfection (RET), and promyelocytic leukemia zinc finger (PLZF).26 This cell line was used in several studies as a model for spermatogonia stem cells.26,27,28,29 The C18-4 cell line was grown in DMEM media with 5% fetal bovine serum (FBS; 16140-071; Thermo Fisher, Waltham, MA, USA), 5% bovine growth serum (SH30541.03, Thermo Fisher), 1% penicillin/streptomycin (15140-122, Thermo Fisher), and 0.5% Fungizone (15290-018, Thermo Fisher) at 37°C with 5% CO2.
Human samples and tissue arrays
Human testicular cancer arrays were purchased from Biomax Inc. (Rockville, MD, USA). Overall, about twenty semimona cases were analyzed using the tissue arrays.
Frozen tissues from one normal male and four seminoma patients were purchased from Cybrdi Inc. (Rockville, MD, USA). The normal sample was obtained from a 25-year-old male (confirmed as having normal spermatogenesis); the seminoma samples were from patients at age of 50 (two patients), 60, and 68 years. No information about the stage of seminoma was available for the frozen samples. All frozen samples were used for the preparation of the protein lysates and western blot analysis (below). For the mass spectrometry (MS) identification, a portion of the sample from the 50-year-old seminoma patient was used. Before the analysis, a small piece of the sample was chopped to prepare a cell suspension. The cells were attached to poly-lysine-coated microscopic slides, fixed with 1% paraformaldehyde (PFH; Sigma-Aldrich), and stained with anti-SUMO antibody (Abcam) to confirm the SUMO expression.
Tissues were homogenized in the whole cell extraction buffer (produced from the Millipore kit) supplemented with N-ethylmaleimide (NEM; desumoylation inhibitor, Sigma-Aldrich) at a final concentration of 20 mmol l−1. The lysate was collected after a centrifugation at high speed at 4°C for 20 min and used for western blot or immunoprecipitation (IP).
Immunoprecipitation and mass spectrometry
IP using a nondenatured lysate and SUMO1-agarose conjugate (sc-5308 AC, Santa Cruz Biotechnology, Dallas, TX, USA) followed by gel electrophoresis and western blot was performed as described in detail in our recent publication.8 Agarose resin without an antibody was used as a negative control. Gel fixation and staining followed by MS and data analysis was performed with the assistance of the Laboratory for Macromolecular Analysis and Proteomics at the Albert Einstein College of Medicine of Yeshiva University as described by our group.8 The identified proteins were divided into functional groups based on a literature search.8
The percentage of targets in each group was calculated by dividing the number of targets in this group by the total number of the identified targets.
Immunohistochemistry and immunofluorescence
Immunohistochemistry (IHC) and immunofluorescent (IF) analyses were performed as described in our previous studies.8,10,30 A rabbit polyclonal antibody against SUMO2/3 (ab3742), a rabbit monoclonal against SUMO1 (ab32058), and a rat monoclonal against TRA98 (ab82527) antibodies were purchased from Abcam and used at 1:100 dilution for IHC and IF. A rabbit polyclonal antibody against PCNA (Abcam, ab29) was used at 1:1000 dilution. A mouse monoclonal antibody against GFRA1 (E-11) was obtained from Santa Cruz Biotechnology (sc-271546) and used at 1:50 dilution.8,10,30
Real-time PCR
Reverse transcription (RT)-quantitative polymerase chain reaction (qPCR) was performed as described by our laboratory.30 The primer sequences for GFRA1 and RET were used as previously described.31
Overexpression and downregulation of SUMO, western blot, and statistical analysis
UBC9 (the SUMO-conjugating enzyme) and control siRNAs were purchased from Santa Cruz Biotechnology (sc-36773 and sc-36869). 0.5 × 106 cells were seeded onto separate 10-cm tissue culture dishes and grown overnight at 5% CO2 and 37°C. Eighty pmol of siRNA was transfected into each dish of cells using Lipofectamine® RNAiMAX Transfection Reagent (Thermo Fisher) at a ratio of 0.3 μl of reagent per pmol of siRNA. The transfection procedure followed the manufacturer's instructions. The cells were subjected to 6 h of transfection followed by a 48-h recovery period prior to analysis.
The cells were transfected with control vector (pCMV-SPORT6) or SUMO2-overexpressing plasmid (Open biosystem [currently Thermo Fisher]) using Lipofectamine 3000 (Thermo Fisher). The cells were collected 48 h after the transfection for western blot assay.
Whole-cell protein lysates were prepared as previously described, using the whole-cell extraction kit and protease inhibitor from Millipore (2910, Sigma-Aldrich) complemented with 2.5 mg ml−1 of NEM (a de-sumoylation inhibitor; E3876-100G, Sigma-Aldrich) according to the manufacturer's instructions. Protein concentrations were determined via bicinchoninic acid (BCA) protein assay (23225, Thermo Fisher) using bovine serum albumin (BSA) as the standard. Gel electrophoresis was performed under reducing conditions using NuPAGE 4%–12% gradient bis-tris polyacrylamide gels (Thermo Fisher) and MOPS running buffer (Thermo Fisher). Protein electrophoresis (and transfer) was performed using the Invitrogen XCell SureLock Mini-Cell electrophoresis system (Thermo Fisher) at a constant voltage (200 V). After electrophoresis, the proteins were transferred to a nitrocellulose membrane (Novex nitrocellulose membrane, 0.45-μm pore size [Thermo Fisher]) using NuPAGE transfer buffer (Thermo Fisher). The membrane was first blocked with 2% membrane-blocking agent (RPN2125V, GE Healthcare UK Limited, Little Chalfont, UK) in phosphate-buffered saline (PBS) +0.02% (v/v) Tween 20 (00-3005, Life Technologies) for 1 h at room temperature. The membrane was then incubated with primary antibodies in PBS containing 2% BSA and 0.1% sodium azide for either 2 h at room temperature or overnight at 4°C. Rabbit polyclonal anti-SUMO1 (Abcam, ab32058) and anti-SUMO2/3 (Abcam, ab3742) antibodies were used in a 1:500 dilution; a rabbit polyclonal antibody against PCNA (Abcam, ab29) was used at 1:1000 dilution. Equal loading was ensured with monoclonal anti-β-actin (sc-1615, Santa Cruz) or monoclonal anti-β-tubulin (1:2000; Invitrogen) antibody in a 1:1000 dilution. Following three washes with PBS-T, the membrane was further incubated with secondary antibodies that were diluted to 1:5000 in PBS-T for 1 h at room temperature. The secondary antibodies used in this study included the following: anti-rabbit IgG horseradish peroxidase (HRP) linked (NA934V, GE Healthcare UK Limited) and goat anti-mouse IgG (H + L) HRP (AP308P, EMD Millipore Corporation, Sigma Aldrich). Western blot detection was performed using Luminata™ Forte (Sigma-Aldrich), in accordance with the manufacturer's instructions. Quantitative densitometry analyses were performed using the Quantity One software (Bio-Rad Laboratories, Hercules, CA, USA), and the density values were normalized to actin or tubulin. In each experiment, controls (untreated samples) were considered as 1, and other samples were normalized to the controls. Each experiment was repeated three times. To calculate the difference between the samples, Student's paired t-test was used. P < 0.05 was considered statistically significant.
RESULTS
Identification of sumoylated targets in mouse spermatogonia
We and others have previously described the localization pattern of SUMO in testicular cells. All germ and somatic cells were found to be positive for SUMO.6,32 Furthermore, SUMO-related genes express in a stage-specific manner during mouse spermatogenesis.33 SUMO-positive spermatogonia are found at the edge of seminiferous tubules and can be clearly distinguished from the Sertoli cells by their nuclear morphology (Figure 1a). In a similar manner, all cells in the human testis seem to be positive for SUMO, although the expression level of the sumoylated proteins may vary in a cell- and stage-specific manner (Figure 1b). In order to understand the regulation of spermatogenesis by sumoylation, it is important to know what proteins are being modified by SUMO in a cell-specific manner. While sumoylated targets in mouse spermatocytes and spermatids have been identified by our group,8 the sumoylome of spermatogonia has not been studied. Furthermore, as our previously published data implicated sumoylation in the regulation of meiosis, whether SUMO proteins regulate mitosis of germ cells is currently not known. Therefore, we have used spermatogonia from pubertal mouse testes for mass spectrometry identification of sumoylated targets. Spermatogonia-enriched fraction was obtained using a differentiating plating procedure and mouse testes from day 7–8 postpartum. While Sertoli cells attach to the Mitrogel-coated dishes, spermatogonia remain suspended in the medium. Sertoli cells are then detached from the flask using light trypsinization.
Figure 1.
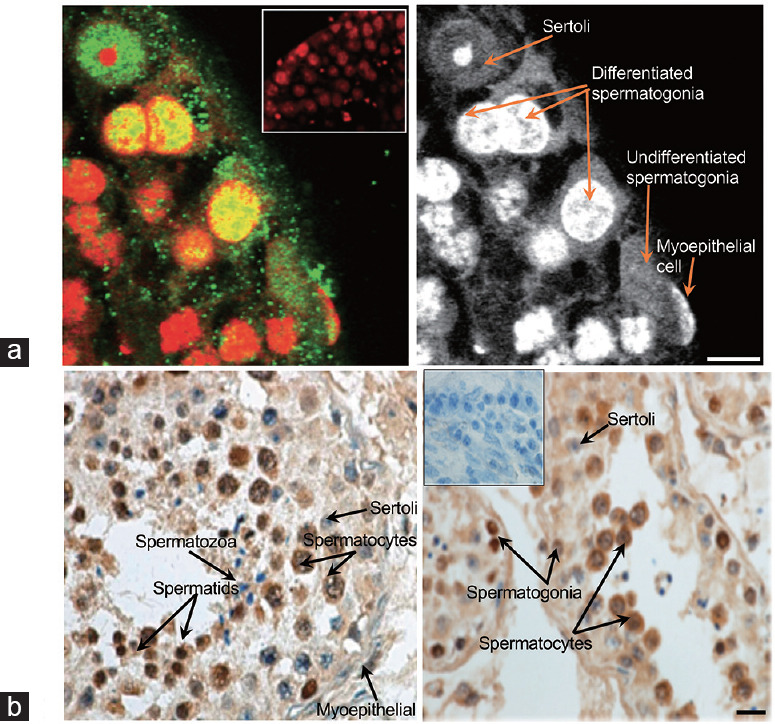
(a) Spermatogonia at the edge of seminiferous tubules are positive for SUMO (green; nuclei are counterstained by PI [red]). The PI-only staining is provided to demonstrate the nuclear morphology of different cells. A negative control of seminiferous tubules (where the primary antibody was omitted) is shown as an inset. Scale bar = 10 μm. (b) SUMO (brown color) is expressed in normal human testis. Different cell types are indicated by the arrows. A negative control of seminiferous tubules (where the primary antibody was omitted) is shown as an insert. Scale bar = 10 μm. SUMO: small ubiquitinlike modifier; PI: propidium iodide.
The purity of the isolation was confirmed using germ- and Sertoli-specific markers (Tra 98 [an antibody against anti-germ cell-specific antigen],23 and GATA4,24 respectively). Nuclei of all cells were counterstained by DAPI. The percentage of the fraction purity was calculated by dividing the number of cells positive for the specific marker by the total number of DAPI-positive cells from several microscopic fields. Representative images of the results are shown in Figure 2. Almost all spermatogonia are positive for Tra 98 and Sertoli for GATA4. Only few cells that are positive for DAPI but negative for the markers are seen in both fractions. Separation of spermatogonia from Sertoli cells resulted in our hands in the fraction purity being over 80% for spermatogonia and over 90% for Sertoli cells.
Figure 2.
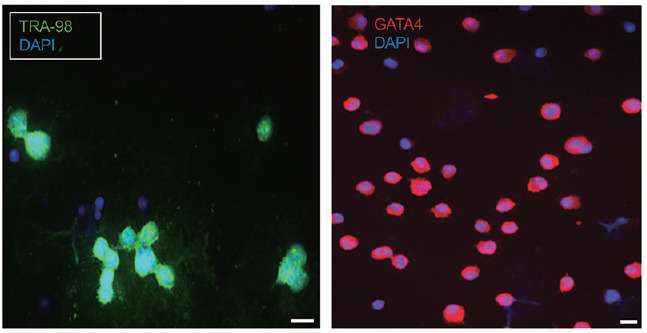
Separation of spermatogonia from Sertoli cells using differentiating plating. Fraction purity was confirmed using germ- and Sertoli-specific markers, TRA-98 and GATA4, respectively. Nuclei of all cells were counterstained by DAPI. Only few cells are positive for DAPI but negative for the marker can be seen in both fractions. The percentage of the purity was calculated by dividing the number of cells positive for the specific marker by the number of all cells from several microscopic fields. Scale bars = 10 μm. TRA-98: anti-germ cell-specific antigen antibody; GATA4: GATA-binding factor 4; DAPI: 4´,6-diamidino-2-phenylindole.
Given a limited number of spermatogonia that can be obtained from pubertal testes, proteomic studies in spermatogonia are challenging. Nevertheless, an IP using anti-SUMO antibody followed by a mass spectrometry analysis resulted in the identification of several SUMO targets uniquely in the antibody fractions but not in the negative control. The identified targets included proteins regulating transcription, translation, cytoskeleton functions, and stress response (Supplementary Table 1). Importantly, the major target of SUMO, Ran-GAP, was also identified, supporting the specificity of the identification. Interestingly, one of the identified targets, HSP60, was specifically localized to proliferating spermatogonia in rodent and human testes.34,35 Notably, in human biopsies, the number of HSP60-positive spermatogonia decreased with the loss of spermatogenic function and was close to zero in the testis with a spermatogenic arrest before entrance into meiosis.35 Another notable SUMO target in spermatogonia is prohibitin which was implicated in the suppression of proliferation in different cell types.36 This finding is consistent with the fact that only quiescent and not proliferating rodent spermatogonia expressed prohibitin.37 Interestingly, prohibitin can be ubiqitinated,38 but its possible sumoylation in mouse and human germ cells remained to be confirmed. Given the critical role of SUMO in the regulation of mitosis, sumoylation of HSP60, prohibitin, and other targets in spermatogonia should be further explored.
Supplementary Table 1.
Identification of sumoylated proteins in mouse spermatogonia
| Identified sumoylated targets | Accession number | MW | Number of peptides | Function |
|---|---|---|---|---|
| Annexin A2 | gi|6996913 | 39 kDa | 3 | Membrane |
| Elongation factor 1-delta | gi|13124192 [4] | 31 kDa | 5 | Translation |
| Disulfide-isomerase A3 precursor | gi|112293264 [4] | 57 kDa | 2 | ER chaperon |
| 60 kDa heat shock protein, mitoch | gi|183396771 [2] | 61 kDa | 2 | Proliferation |
| Ubiquitin-60S ribosomal protein L40 | gi|568962710 [2] | 17 kDa | 2 | Translation/degrad |
| ATP synthase subunit beta, mitoch | gi|31980648 (+5) | 56 kDa | 3 | Enzyme |
| Immunoglobulin heavy chain | gi|1794157 | 50 kDa | 4 | Immune resp. |
| RAN GTPase activating protein 1 | gi|148672614 (+2) | 73 kDa | 3 | Nucl/cyt. transp |
| Citrate synthase | gi|13385942 | 52 kDa | 2 | Enzyme |
| Prohibitin | gi|6679299 [3] | 30 kDa | 2 | Proliferation |
| THO complex subunit 4 | gi|6755763 | 27 kDa | 2 | Transcription |
| Junction plakoglobin | gi|28395018 (+1) | 82 kDa | 4 | Cytoskelet |
| Laminin subunit beta-1 | gi|114326497 (+5) | 202 kDa | 3 | Cytoskelet |
| Laminin, gamma 1 | gi|148707494 [4] | 165 kDa | 7 | Cytoskelet |
| Nidogen-1 precursor | gi|171543883 (+2) | 137 kDa | 3 | Cytoskelet |
Protein accession number, MW, the number of unique peptides (number of peptides), and function is indicated for each protein. Proteins of specific interest are indicated in bold. MW: molecular weight
Identification of sumoylated targets in C18-4 stem-like spermatogonia cell line
To obtain some insights into proteins regulated by sumoylation in stem-like spermatogonia and supplement the information obtained from the analysis of spermatogonia, we used a C18-4 stem-like spermatogonia cell line. This cell line is derived from type A spermatogonia and expresses multiple stem cell markers including GCNA1, VASA, DAZL, PCNA, OCT-4, GFRA1, RET, and PLZF.26 This cell line was used in several studies as a model of spermatogonia stem cells.26,27,28,29 We have confirmed the expression of GFRA1a and RET in C18-4 spermatogonia (Figure 3a and 3b).
Figure 3.
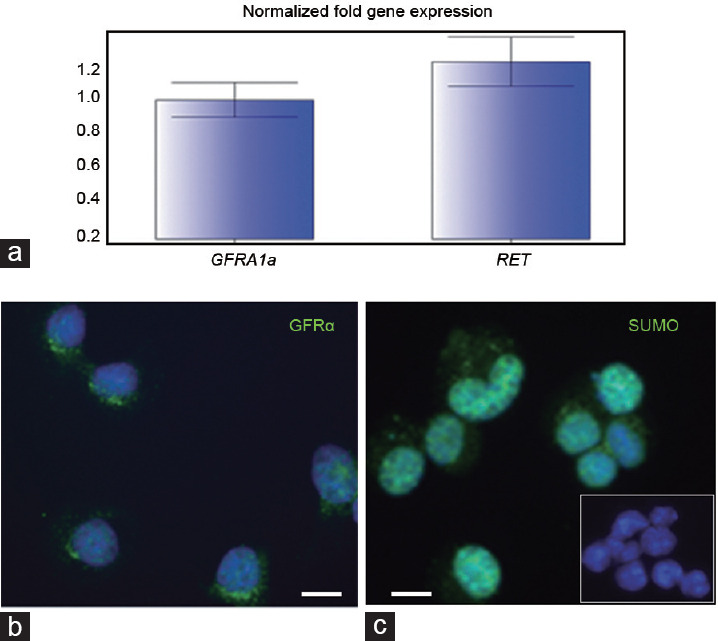
C18-4 spermatogonia express GFRA1a and RET. (a) Real-time RTPCR analysis of GFRA1a and RET expression levels; qPCR was performed in triplicate for each sample; the values were normalized to the actin content of each sample. (b) GFRA1a protein expression (green) in C18-4 spermatogonia. (c) Expression pattern of SUMO (green) in C18-4 cells; a negative control of 18-4 cells (where the primary antibody was omitted) is shown as an insert. Scale bars = 10 μm. GFRA1a: glial cell line-derived neurotrophic factor family receptor alpha-1; RET: rearranged during transfection; PCNA: proliferating cell nuclear antigen; SUMO: small ubiquitin-like modifiers; RT-PCR: reverse transcription-polymerase chain reaction; qPCR: quantitative polymerase chain reaction.
Similar to mouse spermatogonia, the expression pattern of SUMO in C18-4 cells was mostly nuclear (Figure 3c vs Figure 1a).
Mass spectrometry analysis identified 42 targets in C18-4 spermatogonia (Supplementary Table 2). These identified proteins were subdivided into several groups. The largest group (33%) of the SUMO targets included proteins involved in transcription, translation, and splicing. Other functional categories included cytoskeletal proteins (27%), membrane-associated and vesicle trafficking proteins (21%), proteins that mediate nuclear–cytoplasmic transport (6%), involved in ubiquitination (6%), and stress-related proteins/chaperones (6%). This functional distribution was similar to the one found in mouse spermatocytes and spermatids, with some SUMO targets being the same in all germ cell fractions (such as importin beta, heat shock 70 kDa protein 4, ubiquitin, heterogeneous nuclear ribonucleoproteins A2/B1, and several ribosomal proteins) and others being specific for the C18-4 fraction.8 Similar to the results in spermatogonia, prohibitin (a regulator of cellular proliferation) was identified as a SUMO target in the C18-4 fractions. Other notable targets of SUMO included calpirin (whose absence in other cell types resulted in defects in cellular proliferation39), heat shock 70 kDa protein 4 (which is required for mouse spermatogenesis), and importins (implicated in the regulation of male germ cell development40,41). Similar to the results in mouse spermatocytes and spermatids, several enzymes involved in ubiquitination were also identified as SUMO targets, supporting a close crosstalk between ubiquitination and sumoylation. The major target of sumoylation Ran GAP was identified again, confirming the specificity of the identification. Further studies should address the role of sumoylation in the regulation of the identified proteins and other posttranslational modifications.
Supplementary Table 2.
Identification of sumoylated proteins in mouse C18-4 spermatogonia cell line
| Transcription, translation, cell cycle | Accession number | MW | Number of peptides C18 | spg |
|---|---|---|---|---|
| p100 co-activator | gi|6009521 [4] | 99 kDa | 4 | |
| Caprin-1 | gi|162329566 (+1) | 77 kDa | 2 | |
| 14-3-3 eta | gi|1526541 [13] | 28 kDa | 7 | |
| Prohibitin-2 | gi|126723336 [2] | 33 kDa | 2 | X |
| Heterogeneous nuclear ribonucleoproteins A2/B1 | gi|109134362 [5] | 36 kDa | 2 | |
| mKIAA0120 protein | gi|50510369 | 24 kDa | 3 | |
| Nucleoside diphosphate kinase B | gi|6679078 [4] | 17 kDa | 2 | |
| ADP-ribosylation factor 3 | gi|4502203 [2] | 21 kDa | 5 | |
| 60S ribosomal protein L13-like | gi|149251177 [3] | 24 kDa | 2 | |
| 40S ribosomal protein S9 | gi|14141193 | 23 kDa | 3 | |
| 60S ribosomal protein L10a | gi|1709972 [2] | 25 kDa | 2 | |
| Nuclear-cytoplasmic transport | ||||
| RAN GTPase activating protein 1 | gi|148672614 (+2) | 73 kDa | 2 | X |
| Importin beta | gi|1669535 [5] | 97 kDa | 4 | |
| Cytoskeleton | ||||
| Plec1 protein | gi|71534062 [3] | 94 kDa | 20 | |
| AHNAK nucleoprotein isoform 1 | gi|61743961 | 604 kDa | 3 | |
| Arpc4 (Actin-related protein 4) protein | gi|33244031 (+2) | 20 kDa | 2 | |
| Ras GTPase-activating-like protein IQGAP2 | gi|118344444 [2] | 181 kDa | 12 | |
| Cytoskeleton-associated protein 4 | gi|62526118 | 64 kDa | 3 | |
| Transgelin | gi|6755714 | 23 kDa | 4 | |
| Cytoplasmic dynein 1 heavy chain 1 | gi|134288917 | 532 kDa | 8 | |
| Profilin-1 | gi|6755040 (+1) | 15 kDa | 2 | |
| Ezrin | gi|50881 [2] | 69 kDa | 6 | |
| Ubiqituination , proteasomal degradation | ||||
| Ubiquitin-like modifier-activating enzyme 1 | gi|209862989 [2] | 118 kDa | 2 | |
| Ubiquitin-60S ribosomal protein L40-like | gi|568962710 [2] | 17 kDa | 2 | X |
| Murine valosin-containing protein | gi|55217 (+6) | 89 kDa | 6 | |
| Membrane-associated, vesicle trafficing | ||||
| Annexin A 1 | gi|70912321 [2] | 39 kDa | 4 | X |
| Ras-related protein Rab-2A | gi|10946940 | 24 kDa | 2 | |
| Ras-related protein Rab-7a | gi|34147513 | 23 kDa | 4 | X |
| Ras-related protein Rab-1A isoform 1 | gi|4758988 [8] | 23 kDa | 3 | |
| Coatomer protein complex, subunit gamma | gi|148666818 [4] | 98 kDa | 2 | |
| Coatomer protein complex subunit alpha | gi|148707095 [6] | 138 kDa | 2 | |
| Chaperones/stress-related proteins | ||||
| Hypoxia up-regulated protein 1 | gi|157951706 [4] | 111 kDa | 4 | |
| Heat shock 70 kDa protein 4 | gi|112293266 [4] | 94 kDa | 3 | |
| Enzymes | ||||
| L-lactate dehydrogenase B chain | gi|6678674 | 37 kDa | 3 | |
| Phosphoglycerate mutase 1 | gi|114326546 [2] | 29 kDa | 4 | |
| GDP dissociation inhibitor 2 | gi|148700276 [4] | 53 kDa | 2 | |
| Protein disulfide-isomerase | gi|112293264 [4] | 57 kDa | 6 | X |
| Triosephosphate isomerase | gi|3287374 [4] | 27 kDa | 7 | |
| ATP synthase subunit beta | gi|31980648 (+5) | 56 kDa | 3 | X |
| Fatty acid synthas | gi|148702861 [2] | 274 kDa | 0 | |
| Immune response | ||||
| Immunoglobulin heavy chain | gi|1794157 | 50 kDa | 3 |
Functional group, protein accession number, MW, and the number of unique peptides (number of peptides) is indicated for each protein. Proteins that were also identified as SUMO targets in mouse spermatogonia (spg) are indicated by “X.” Proteins of specific interest are indicated in bold. GDP: guanosine diphosphate; mw: molecular weight
Analysis of sumoylation in human seminoma samples
SUMO proteins are abundantly expressed in mouse and human testes,6,32 however, their expression in testicular cancer tissues has not been characterized. Testicular cancer is the most common cancer in men aged 20–39 years. The incidence of testicular cancer has almost doubled since the 1930s and continues to increase, although effective treatments have led to a decline in mortality.42,43 Common testicular germ cell tumors include seminomas, embryonal carcinomas, yolk sac tumors, and teratomas.44 Seminoma tumors are the most common testicular germ cell tumors, accounting for nearly 50% of them. Seminomas are suggested to originate from embryonic germ cells that failed to differentiate normally.45 However, the exact mechanism of carcinogenesis is not well understood.44 To examine a possible role of sumoylation in germ cell cancer, we initially looked into the expression of SUMO in samples obtained from patients with seminomous tumors. Using a tissue array approach together with anti-SUMO antibody, we found that SUMO expression is a prominent feature of human seminomas. However, the intensity of the immunostaining varied to some degree between the samples (Figure 4). Using an anti-PCNA (marker of cell proliferation) antibody on the same tissue arrays, we noticed that the proliferative activity of the tumor tissues was often positively correlated with the level of SUMO expression (Figure 4). Interestingly, both PCNA and SUMO staining intensity varied among the samples corresponded to the stage 1 (T1) of the diseases (samples A2, A9, and B8). These data suggest that this stage might be further subdivided into several substages based on the proliferation activity of the tissue. A western blot using frozen tissues from a normal and four seminoma cases confirmed the positive correlation between overall SUMO expression and the level of PCNA (Figure 5).
Figure 4.
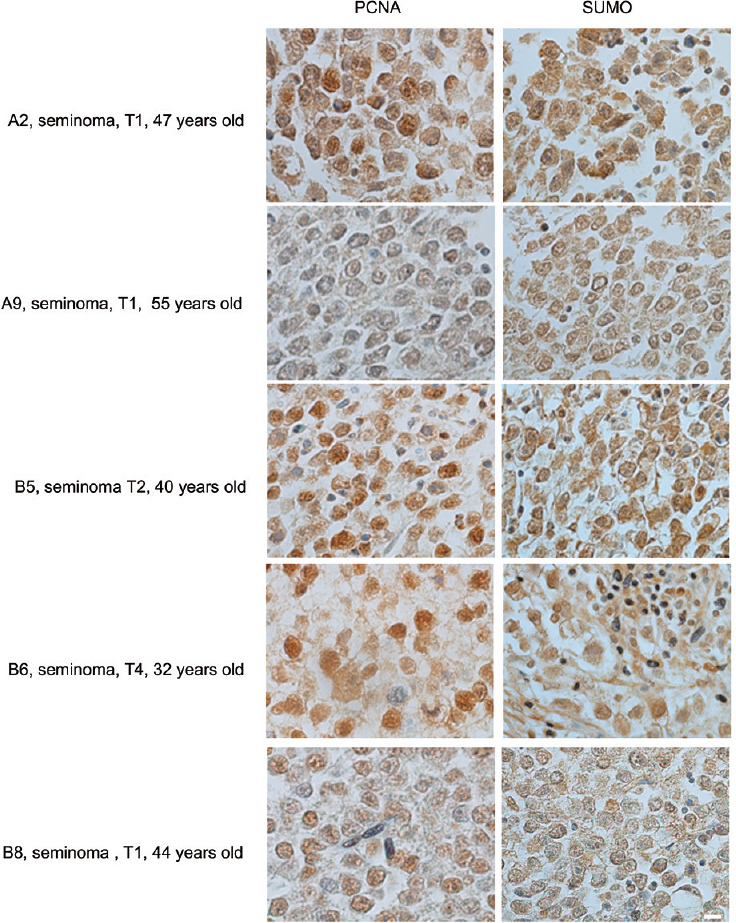
Correlation between PCNA and SUMO expression in human seminomas. Several seminoma samples from a tissue array are shown side by side after immunostaining with either PCNA- or SUMO1-antibody. The age of the patients and the stage of the disease are indicated (provided by the company). The parameters used for the diagnosis were as follows: T1 – tumor invades submucosa; T2 – tumor invades muscularis propria; T3 – tumor invades through muscularis propria into subserosa or into nonperitonealized pericolic or perirectal tissues; T4 – tumor directly invades other organs or structures and/or perforate visceral peritoneum. Scale bar = 10 μm. PCNA: proliferating cell nuclear antigen; SUMO: small ubiquitin-like modifiers.
Figure 5.
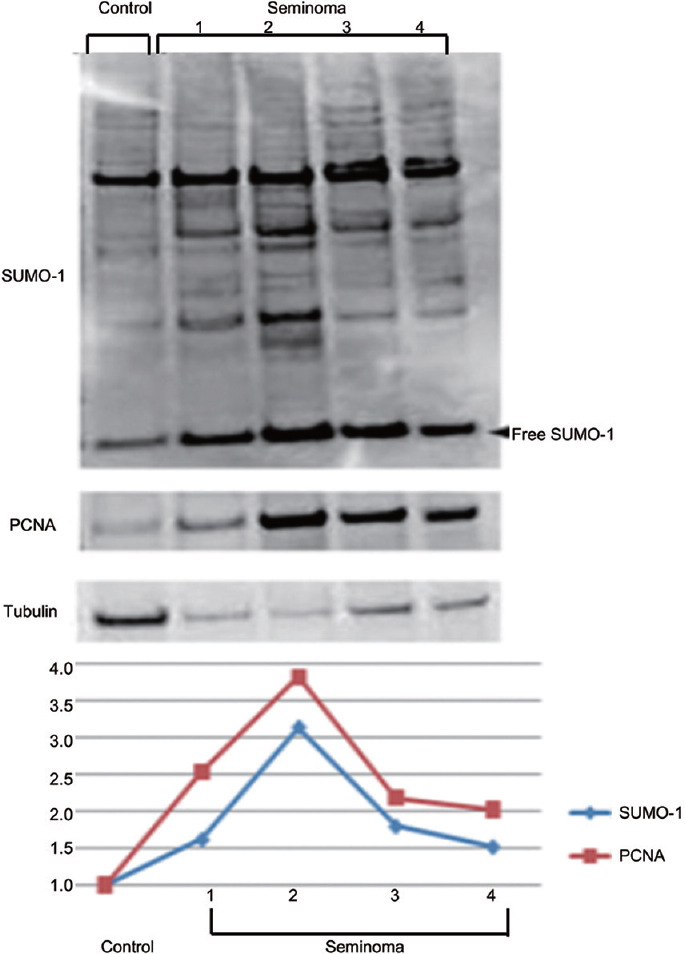
Positive correlation between the expression level of SUMO and PCNA in seminoma cells. Western blot analysis using cell lysates from a normal human sample (control) and samples from four patients with seminoma showed positive correlation between overall SUMO expression and the level of PCNA. Densitometric analysis of the western blot is shown. Both total SUMO and PCNA density values were normalized to the tubulin content of each sample. The results were normalized to control which was considered as 1. PCNA: proliferating cell nuclear antigen; SUMO: small ubiquitin-like modifiers.
In order to better understand how sumoylation regulates cell proliferation and other processes in cancerous tissues, we performed an immunoprecipitation with an anti-SUMO antibody followed by a mass spectrometry identification of SUMO targets in seminoma cells. Before the analysis, a small portion of the seminoma sample was used to prepare slides and confirm the SUMO expression. SUMO was detected in the nucleus and cytoplasm of the cells; this pattern was similar to that obtained using the tissue microarray analysis (Supplementary Figure 1 (153.7KB, tif) ). The mass spectrometry analysis identified 91 proteins that were subdivided into several groups according to their previously published functions (Supplementary Table 3). The largest group (with about 27% of the SUMO targets) included proteins involved in RNA/DNA binding, transcription, splicing, and translation. Notably, several proteins in this group were implicated in the regulation of cell replication and cancerogenesis (Supplementary Table 3). It was interesting to identify PCNA as one of the SUMO targets because, as discussed above, there is a correlation between the expression levels of these two proteins (Figure 6). Notably, it has been shown that PCNA can be modified by SUMO at the replication fork, and that impairment of this modification facilitates formation of double-strand DNA breaks.46 Therefore, aberrant sumoylation of PCNA can contribute to aberrant recombination events and development of cancer.
Supplementary Table 3.
Identification of sumoylated proteins in human seminoma
| RNA/DNA binding, transcription, translation | Accession number | MW | Number of peptides seminoma | Mouse testis | ||
|---|---|---|---|---|---|---|
| Cytes/tids | spg | C18 | ||||
| Heterogeneous nuclear ribonucleoproteins A2/B1 | gi|14043072 (+1) | 37 kDa | 9 | X | X | |
| Heterogeneous nuclear ribonucleoprotein A3 | gi|34740329 [4] | 40 kDa | 7 | X | ||
| Heterogeneous nuclear ribonucleoprotein K | gi|119583080 (+11) | 51 kDa | 3 | |||
| Heterogeneous nuclear ribonucleoprotein M | gi|119589327 (+6) | 78 kDa | 5 | |||
| Heterogeneous nuclear ribonucleoprotein H2 | gi|9624998 | 49 kDa | 2 | X | ||
| Heterogeneous nuclear ribonucleoprotein -E1 | gi|460771 (+1) | 38 kDa | 2 | |||
| Heterogeneous nuclear ribonucleoprotein -F protein | gi|16876910 [3] | 46 kDa | 3 | |||
| Heterogeneous nuclear ribonucleoproteins C1/C2 | gi|117189975 [14] | 34 kDa | 3 | |||
| Small nuclear ribonucleoprotein Sm D2 | gi|237649049 (+1) | 12 kDa | 2 | X | ||
| Polyadenylate-binding protein 1 | gi|41386798 | 71 kDa | 15 | |||
| Tat binding protein 1, TBP-1=transcriptional activator | gi|263098 (+6) | 49 kDa | 2 | |||
| Elongation factor 1-delta isoform 2 | gi|25453472 [3] | 31 kDa | 6 | X | X | |
| Elongation factor 2 | gi|4503483 | 95 kDa | 10 | X | ||
| Elongation factor 1 gamma | gi|119594432 (+2) | 48 kDa | 4 | |||
| Fus-like protein, partial | gi|1040970 (+7) | 53 kDa | 4 | X | ||
| Ewing sarcoma breakpoint region 1 | gi|119580187 (+9) | 47 kDa | 4 | |||
| Eukaryotic initiation factor 4A-I | gi|4503529 [4] | 46 kDa | 7 | |||
| Transcription activator SMARCA4/BRG1 | gi|192807314 [12] | 182 kDa | 4 | X | ||
| AT-rich interactive domain-containing protein 1A | gi|21264565 [2] | 242 kDa | 4 | X | ||
| Rna Polymerase II | gi|353251633 [3] | 163 kDa | 7 | X | ||
| splicing factor proline/glutamine-rich | gi|119627826 [4] | 66 kDa | 5 | X | ||
| DNA topoisomerase 2-alpha isoform X1 | gi|530412779 | 173 kDa | 4 | X | ||
| TAR DNA binding protein, TDP43 | gi|119592079 [9] | 34 kDa | 2 | X | ||
| Far upstream element-binding protein 2 | gi|154355000 (+4) | 73 kDa | 2 | X | ||
| FUSE binding protein 1 | gi|119626762 (+8) | 69 kDa | ||||
| Guanine nucleotide binding protein (G protein) | gi|119574084 | 40 kDa | 3 | |||
| Vigilin isoform a | gi|4885409 (+3) | 141 kDa | 5 | |||
| ATP-dependent RNA helicase DDX5 | gi|197692465 [5] | 69 kDa | ||||
| Nuclear receptor coactivator 6 isoform 1 | gi|32307128 (+8) | 219 kDa | 3 | |||
| MCM4 minichromosome maintenance deficient 4 | gi|119607091 (+6) | 96 kDa | 2 | |||
| Nucleolin | gi|119591368 (+5) | 59 kDa | 2 | |||
| Proliferating cell nuclear antigen | gi|4505641 (+2) | 29 kDa | 2 | |||
| ADP-ribosylation factor 3 isoform X2 | gi|578823704 | 16 kDa | 2 | X | ||
| 14-3-3 protein zeta/delta | gi|4507953 [6] | 28 kDa | 10 | X | ||
| Nm23 Human Nucleoside Diphosphate Kinase B | gi|1421609 [8] | 17 kDa | 4 | X | ||
| Prohibitin isoform 1 | gi|4505773 (+2) | 30 kDa | 4 | X | X | |
| Prohibitin-2 isoform 3 | gi|390608669 | 29 kDa | 2 | X | X | |
| Ribosomal protein S4, X-linked | gi|119592221 [6] | 43 kDa | 3 | |||
| Ribosomal protein S16 | gi|119577297 (+1) | 16 kDa | 3 | |||
| Ribosomal protein S3 | gi|15718687 (+3) | 27 kDa | 4 | |||
| Ras-related protein Rab-1B | gi|13569962 [2] | 22 kDa | 2 | X | ||
| 60S ribosomal protein L12 | gi|4506597 | 18 kDa | 3 | |||
| 40S ribosomal protein S7 | gi|4506741 | 22 kDa | 3 | |||
| Ribosomal protein L7 | gi|307388 | 29 kDa | 4 | |||
| 60S ribosomal protein L27 | gi|4506623 (+1) | 16 kDa | 2 | |||
| Ribosomal protein L7a | gi|119608472 | 33 kDa | 3 | |||
| Ribosomal protein SA | gi|119584991 (+5) | 20 kDa | 4 | |||
| Ribosomal protein L23a | gi|119571516 (+3) | 22 kDa | 2 | X | ||
| Ribosomal protein S10 | gi|119624187 (+3) | 20 kDa | 3 | |||
| 40S ribosomal protein S9 | gi|14141193 [2] | 23 kDa | 3 | X | ||
| Ribosomal protein L21 variant | gi|62087780 | 19 kDa | 2 | X | ||
| Ribosomal protein, large, P0 | gi|12654583 (+4) | 34 kDa | 4 | |||
| 40S ribosomal protein S20 isoform 1 | gi|226246671 (+1) | 16 kDa | 3 | |||
| 60S ribosomal protein L11 isoform 2 | gi|315221152 (+1) | 20 kDa | 2 | x | ||
| Ribosomal protein S19 | gi|119577478 (+2) | 17 kDa | 3 | x | ||
| Similar to ribosomal protein L23 | gi|13097600 (+2) | 14 kDa | 4 | x | ||
| 60S ribosomal protein L31 isoform 2 | gi|148746199 (+6) | 15 kDa | 3 | |||
| 40S ribosomal protein S13 | gi|4506685 | 17 kDa | 3 | |||
| 60S ribosomal protein L26 | gi|4506621 (+1) | 17 kDa | 4 | x | ||
| 60S ribosomal protein L10a | gi|4506621 (+1) | 17 kDa | 4 | x | x | |
| Ribosomal protein S15a ribosomal protein S3A 40S ribosomal protein S14 | gi|12804561 (+5) gi|119625394[8] gi|5032051 | 15 kDa 28 kDa 16 kDa | 3 2 | x | ||
| 60S ribosomal protein L24 | gi|4506619 | 18 kDa | 2 | |||
| 40S ribosomal protein S2-like isoform 5 | gi|114647215 [3] | 31 kDa | 2 | |||
| Ribosomal protein S6 | gi|337514 | 29 kDa | 2 | |||
| Ribosomal protein S26 | gi|119617283 (+5) | 12 kDa | 2 | x | ||
| Ribosomal protein S5 | gi|119592989 (+2) | 22 kDa | 2 | |||
| Ribosomal protein L30 | gi|119612175 [2] | 10 kDa | 2 | |||
| Cytoskeleton | ||||||
| Plectin isoform 1a | gi|41322923 (+1) | 516 kDa | 13 | X | ||
| Actin-related protein 2/3 complex subunit 4 | gi|5031595 (+3) | 20 kDa | 18 | X | ||
| Actin-related protein 3 isoform 2 | gi|460417294) | 42 kDa | 2 | |||
| Myosin light polypeptide | gi|113812151 (+6) | 16 kDa | 2 | X | ||
| Myosin-9 | gi|12667788 | 227 kDa | 22 | |||
| Cofilin-1 | gi|5031635 | 19 kDa | 3 | |||
| Spectrin beta chain, non-erythrocytic 1 | gi|112382250 (+8) | 275 kDa | 4 | |||
| Spectrin, alpha, non-erythrocytic 1 | gi|119608216 | 286 kDa | 2 | |||
| Transgelin 2, isoform CRA_e | gi|119573148 (+4) | 22 kDa | 16 | X | ||
| Talin-2 | gi|344179032 [6] | 167 kDa | 10 | X | ||
| Asap3 , ankyrin repeat and PH domain 3 | gi|297787706 (+7) | 55 kDa | 2 | |||
| Catenin delta-1 isoform 1A | gi|146231938 (+21) | 104 kDa | 2 | |||
| Filamin-A isoform 1 | gi|116063573 [7] | 280 kDa | 2 | |||
| Filamin-B isoform 2 | gi|105990514 [18] | 278 kDa | 2 | X | ||
| Lamin A/C | gi|119573381 (+13) | 78 kDa | 8 | |||
| Lamin B1 | gi|15126742 (+1) | 66 kDa | 12 | X | X | |
| IQ motif containing GTPase activating protein 1 | gi|141797011 [3] | 189 kDa | 2 | |||
| Dynein, cytoplasmic 1, heavy chain 1 | gi|119602165 (+5) | 526 kDa | 2 | X | ||
| Vinculin | gi|119574932 [6] | 109 kDa | 3 | X | ||
| Triple functional domain (PTPRF interacting) | gi|119628449 (+3) | 268 kDa | 2 | |||
| Alpha-centractin isoform 2 | gi|114632563 (+6) | 37 kDa | 2 | |||
| Porin 31HM | gi|238427 (+3) | 31 kDa | 5 | |||
| Ubiqituination , proteasomal degradation | 10 | |||||
| Valosin-containing protein | gi|111305821 (+1) | 89 kDa | 4 | X | ||
| Ubiquitin B | gi|119624910 | 13 kDa | 3 | X | X | |
| Ubiquitin associated Protein (AD-012 protein) | gi|7688705 | 61 kDa | 6 | |||
| Ubiquitin associated protein 2-like | gi|13111995 (+17) | 115 kDa | 6 | X | ||
| 26S proteasome subunit p97 | gi|1060888 (+11) | 100 kDa | 2 | X | ||
| Cullin-associated NEDD8-dissociated protein 1 | gi|16758920 | 136 kDa | 3 | |||
| Membrane-associated, vesicle trafficing | 5 | |||||
| GPI-anchored membrane protein 1 | gi|119588582 [2] | 77 kDa | 4 | X | X | |
| annexin A1 annexin V chloride intracellular channel protein 1 | gi|4502101 gi|342350777[2] gi|14251209 [8] | 39 kDa 36 kDa 27 kDa | 4 3 | X | X | |
| Clathrin heavy chain 1 isoform 1 | gi|4758012 [6] | 192 kDa | 17 | |||
| ras-related protein Rab-2A isoform b | gi|336391093) | 21 kDa | 3 | X | ||
| Ras-related protein Rab-7a | gi|34147513 | 23 kDa | 3 | X | X | |
| ras GTPase-activating protein-binding protein 1G3BP | gi|49168554 [3] | 52 kDa | 4 | X | ||
| rab11 In Complex With Rab11-Fip2 | gi|114794171 (+11) | 19 kDa | 2 | |||
| ras-related protein Rab-14 | gi|18390323 | 24 kDa | 2 | X | ||
| rho GDP-dissociation inhibitor 1 isoform b | gi|297374785 (+3) | 18 kDa | 2 | X | ||
| Chaperones/stress-related proteins | 10 | |||||
| Heat shock 70kDa protein 4 | gi|119582699 (+3) | 88 kDa | 4 | X | X | |
| Heat shock protein gp96 precursor | gi|15010550 (+2) | 90 kDa | 5 | |||
| Heat shock protein 47, member 1 | gi|119595384 (+4) | 41 kDa | 2 | |||
| Chaperonin containing TCP1, subunit 4 (delta) | gi|119620393 (+2) | 58 kDa | 3 | |||
| Chaperonin containing TCP1, subunit 3 (gamma) | gi|14124984 (+7) | 60 kDa | 3 | |||
| Chaperonin containing TCP1 subunit 8KIAA0002 | gi|1136741 (+3) | 59 kDa | 2 | |||
| Hypoxia up-regulated protein 1 precursor | gi|5453832 [4] | 111 kDa | 2 | X | ||
| Peroxiredoxin 1, isoform CRA_b | gi|119627382 (+5) | 21 kDa | 4 | |||
| t-complex, isoform CRA b | gi|119568002 (+3) | 35 kDa | 2 | X | ||
| Enzymes | ||||||
| ATP synthase, H+transporting | gi|119630208 (+3) | 17 kDa | 2 | |||
| ATP synthase subunit beta, mitochondrial precursor | gi|32189394 (+2) | 57 kDa | 3 | X | X | X |
| ADP/ADT translocator protein | gi|339920 [2] | 33 kDa | 10 | |||
| Triosephosphate isomerase | gi|441670320 | 23 kDa | 6 | X | X | |
| Phosphate carrier protein, mitochondrial isoform b | gi|4505775 (+5) | 40 kDa | 3 | X | ||
| Malate dehydrogenase, mitochondrial isoform 1 | gi|21735621 (+5) | 36 kDa | 6 | |||
| Beta-enolase isoform X1 | gi|530410063 | 48 kDa | 2 | |||
| 6-phosphogluconate dehydrogenase | gi|40068518 [4] | 53 kDa | 2 | |||
| Human glutamate oxaloacetate transaminase 1 | gi|255918055 (+3) | 48 kDa | 2 | |||
| GAPDH-2 like | gi|2282013 (+5) | 45 kDa | 2 | |||
| Aconitase precursor | gi|3600098 | 65 kDa | 5 | |||
| Arginyl-tRNA synthetase | gi|119581911 (+6) | 75 kDa | 3 | |||
| Glutamine-fructose-6-phosphate transaminase 1 | gi|119620257 (+4) | 65 kDa | 2 | |||
| Acyl-CoA thioesterase 7 | gi|119591933 (+11) | 30 kDa | 2 | |||
| Multifunctional protein CAD | gi|1228049 (+7) | 243 kDa | 3 | |||
| Nuclear-cytoplasmic transport | ||||||
| Karyopherin (importin) beta 1 | gi|119615215 (+3) | 94 kDa | 4 | X | X | |
| Ran GTPase activating protein 1 | gi|119580824 (+6) | 72 kDa | 3 | X | X | X |
| Exportin-2 isoform 1 | gi|29029559 (+5) | 110 kDa | 2 | X | ||
| Immune response | ||||||
| Chain A, Cyclophilin B | gi|1310882 (+5) | 20 kDa | 2 | |||
| Chain B, Traf6 Chain A, The 25 Kda subunit of human cleavage facto | gi|238538004 (+5) gi|168177225 | 26 kDa | 2 | |||
| Expressed only in cancer tissues | ||||||
| TRK-fused gene/anaplastic large cell lymphoma kinas | gi|20269390 (+1) | 89 kDa | 2 | |||
Protein accession number, MW, and the number of unique peptides identified for each protein (number of peptides) is indicated. Proteins that were also identified as SUMO targets in mouse spermatocytes and/or spermatids, spermatogonia, or C18-4 cells (cytes/tids, spg, C18-4 respectively) are indicated by “X.” Proteins of specific interest are indicated in bold. FUSE: far upstream element; MW: molecular weight
Figure 6.
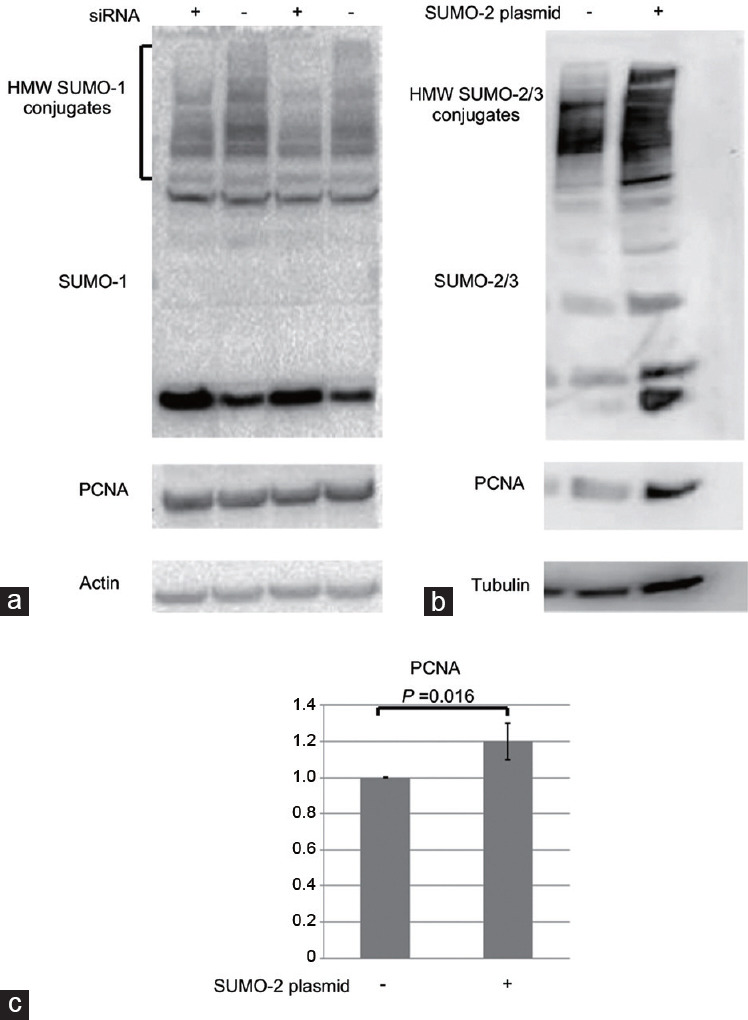
(a) The cells were subjected to 6 h of transfection with UBC9 (the sumo-conjugating enzyme; [+]) and control si-RNAs (-) followed by a 48-h recovery period prior to western blot assays. The membranes were blotted with anti-SUMO-1, anti-PCNA, or anti-actin antibodies. (b) Cells were transfected with SUMO2-overexpressing plasmid (+) or control vector (-). Cells were collected 48 h after the transfection for western blot assays. The membranes were blotted with anti-SUMO2/3, anti-PCNA, or anti-tubulin antibodies. A representative image for each experiment is shown. (c) For three different experiments of over-expression, both total SUMO and PCNA density values were normalized to the tubulin or actin content of each sample. The results were normalized to control which was considered as 1. The P value is shown for the statistically significant results. PCNA: proliferating cell nuclear antigen; SUMO: small ubiquitin-like modifiers.
Both AT-rich interactive domain-containing protein 1A and Brahma-related gene-1 (BRG1)/SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily a, member 4(SMARCA4; (Supplementary Table 3) act as tumor suppressors and are subunits of the SWI/SNF chromatin remodeling complex, which is silenced in many cancer types.47,48 Notably, BRG1/SMARCA4 was also identified by our group as a SUMO target in mouse spermatocytes, supporting its possible regulation by sumoylation.8
Topoisomerase 2-alpha (Top2A) is another identified SUMO target in the seminoma fraction (Supplementary Table 3). Type 2 topoisomerases play an important role in cohesion and separation of sister chromatids during mitosis and meiosis. Blocking SUMO modification of Top2 in mitotic cells prevented the dissociation of sister chromatids at the metaphase–anaphase transition, supporting the idea that SUMO conjugation is crucial for proper chromosome segregation.49,50 We have previously confirmed sumoylation of Top2A in mouse testicular cells.7 The role of Top2A sumoylation in human seminomas should be addressed in the follow-up studies.
TAR DNA-binding protein 43 (TDP43), a DNA/RNA binding protein, was also identified as a SUMO target (Supplementary Table 3). TDP43 was recently implicated in cancer progression as its loss inhibited the progression of triple-negative breast cancer.51 TDP43 was also identified by our group among SUMO targets in mouse germ cells,8 supporting its possible sumoylation.
The 14-3-3 beta is also a notable SUMO target in seminomous tumors as it has gained importance in the initiation and progression of several types of cancer including seminoma.52 Compared with healthy testes, where 14-3-3 beta expression was very low, malignant seminoma cells revealed a high expression of 14-3-3 beta, suggesting an oncogenic potential of the protein.53 Interestingly, 14-3-3 protein was found in the complex with Cyclin-A1 (CcnA1) and SUMO in mouse testes, supporting its possible sumoylation in germ cells.54
Other proteins implicated in cancerogenesis included Ewing sarcoma breakpoint region 1, vigilin, Nm23 Human Nucleoside Diphosphate Kinase B, and prohibitins (Supplementary Table 3).36,55,56,57 Prohibitin was also identified as a SUMO target in freshly isolated and C18-4 permatogonia, supporting its possible regulation by sumoylation. Prohibitins regulate the proliferation of different cell types (including embryonic stem cells), and were implicated in cancerogenesis in different tissues.36,58 Given the suggested origin of seminomas from embryonic cells, prohibitin is an interesting SUMO target to study further.
Many ribosomal proteins were also identified as SUMO targets in seminoma fraction. Interestingly, while ribosomal proteins play important role in protein translation, recent studies revealed that they may have additional extra-ribosomal functions in the regulation of apoptosis, cell cycle arrest, cell proliferation, neoplastic transformation, and cell migration and invasion.59
Cytoskeletal proteins represented 16% of SUMO targets in seminoma cells (Supplementary Table 3). Several of the identified targets (e.g., transgelin, myosin light polypeptide, talin, and catenin delta) were implicated in the initiation and progression of cancer.60,61,62,63
Stress-related and heat shock proteins represented 7% of sumoylated targets in seminomas. Some of these proteins were shown to be upregulated in different tumors and associated with the tumor invasiveness. For example, hypoxia up-regulated 1 (HYOU1) protein was overexpressed in breast cancer tumors and peroxiredoxin 1 (PRDX1) regulated the progression and metastasis of breast, esophageal, and lung cancers.64,65
Membrane-associated, vesicle trafficking, and endoplasmic reticulum (ER) proteins represented 8% of sumoylated proteins (Supplementary Table 3). Some members of this group, such as ras-related in brain 2A and 11 (Rab2A and Rab 11), were shown to promote tumor cell migration and invasion by regulating various cell signaling pathways.66,67
Similar to the results in mouse germ cells, several proteins involved in ubiquitination, including ubiquitin-activating and -conjugating enzymes, ubiquitin-associated proteins, and proteasome subunits, were identified as SUMO targets (Supplementary Table 3). This finding supports a possible evolutionary-conserved interplay between sumoylation and ubiquitination at several levels.8 During cancerogenesis, proteins regulating ubiqitination control cell cycle and DNA repair processes. For example, ubiquitin-associated protein 2 is overexpressed in several cancers, and its silencing inhibits proliferation of cancer cells.68 In a similar manner, an elevated expression of valosin-containing protein (p97) is associated with poor prognosis of prostate cancer.69
Proteins that mediate nuclear–cytoplasmic transport comprised 2% of seminoma cell sumoylome (Supplementary Table 3). Once again, Ran GTPase activating protein 1 (RanGAP1) was identified among the SUMO targets, supporting the specificity of the identification. Other proteins in this group included importin beta and exportin 2, which are known to interact with RanGAP1.70 Metabolic enzymes represented 16% of SUMO targets. Interestingly, numerous targets identified in human seminomas were also identified by our group as SUMO targets in mouse germ cells8 and in C18-4 cells (marked by X in Supplementary Table 3).
Upregulation and downregulation of sumoylation in C18-4 spermatogonia
To test whether changes in the level of sumoylated proteins affect the proliferation of C18-4 cell, a downregulation and overexpression of SUMO was performed using UBC9 (the SUMO conjugating enzyme) si-RNA and SUMO-2 plasmid, respectively. The experiments were followed by western blot with the use of anti-SUMO and anti-PCNA antibodies. Equal loading was ensured with anti-β-actin or anti-β-tubulin antibody (Figure 6a and 6b). The density values were normalized to actin or tubulin. In each experiment, controls (untreated samples) were considered as 1, and other samples were normalized to the controls. Using PCNA as a proliferative marker, the results of the experiments revealed that downregulation of sumoylation using si-RNA was not sufficient to significantly affect the level of cellular proliferation (Figure 6a). However, the overexpression of SUMO statistically significantly increased the proliferation rate of the cells (P = 0.016; n = 3; Figure 6b and 6c).
DISCUSSION
Sumoylation was implicated in the regulation of mitotic progression and cancerogenesis, but its role in the proliferation of normal and cancer male germ cells has not been characterized. SUMO proteins are highly expressed in spermatogonia where they may regulate various cellular processes. Mass spectrometry analysis of freshly purified mouse spermatogonia identified several SUMO targets implicated in the control of spermatogonia proliferation, such as HSP60 and prohibitin. Possible sumoylation of these proteins and the consequences of inhibition or overexpression of sumoylation in spermatogonia on the activity of these targets should be further addressed using both in vitro and in vivo approaches. Interestingly, only five targets were identified in both stem-like C18-4 and differentiating spermatogonia, suggesting a dynamic nature of sumoylation during spermatogonia differentiation. However, given the limited number of spermatogonia obtained from the pubertal testes, sumoylation of some targets could be below the detection limit of the analysis. Nevertheless, specific proteins of interest identified in the C18-4 fraction can now be studied in spermatogonia stem cells and differentiated spermatogonia using more sensitive techniques (such as a co-immunoprecipitation [co-IP] analysis with the use of antibodies).
Our analysis has also identified around 100 SUMO targets in human seminoma. To the best of our knowledge, this is the first study to identify the seminoma sumoylome and also the largest one to characterize the seminoma proteome. These data provide important insights into the types of proteins expressed in seminoma cells and their possible regulation by sumoylation. Several important targets implicated in the initiation and progression of cancer (such as PCNA, Top2A, 14-3-3 beta, prohitin, and others) have been identified. Interestingly, 44 out of 91 targets identified in human seminoma cells were also identified by our group as SUMO targets in mouse germ cells.8 These data support a significant conservation between sumoylome of mouse and human germ cells.
An interesting finding was that the proliferation activity of cancerous tissues correlated with the overall sumoylation level in several seminoma samples. These results are in line with several other studies in normal and cancer tissues, demonstrating a possible link between cellular proliferation by sumoylation.11,12,71 Whether sumoylation contributes or just correlates with the high proliferation rate in male germ cells in vivo, remains to be determined. While downregulation by si-RNA was not sufficient to significantly affect C18-4 proliferation, SUMO overexpression increased their proliferation rate. These data suggest that cells are more sensitive to an elevated level of SUMO, and that this situation may lead to upregulated cellular proliferation and, possibly, cancer. Further studies using overexpression of sumolylation in stem cells or its inactivation in cancerous cells, together with studies of specific SUMO targets identified in the current study, should provide a better understanding of the SUMO-mediated molecular control of events in normal and cancerous male germ cells.
AUTHOR CONTRIBUTIONS
TS, RL, BL, and SK carried out study design, execution, and analysis of data. MV was involved in study design, data analysis, and manuscript writing and revision. All authors read and approved the final manuscript.
COMPETING INTERESTS
All authors declare no competing interests.
Expression of SUMO proteins in frozen seminoma samples. A small piece from the frozen seminoma sample, used for the mass spectrometry analysis, was cut with a blade to prepare a cell suspension and slides. Cells were stained with anti-SUMO antibody to confirm SUMO expression. SUMO immunostaining is shown in green, nuclear staining by DAPI in blue. Most of the cells show SUMO expression.
ACKNOWLEDGMENTS
This study was supported by the NIH, NICHD, and Academic Research Enhancement Award 1R15HD067944-01A1 (MV, PI). Undergraduate student research was supported by Selma and Jacques H. Mitrani Foundation and by Stern College for Women, Yeshiva University, New York, NY, USA. We would like to thank the Laboratory for Macromolecular Analysis and Proteomics (LMAP) at Albert Einstein College of Medicine, Bronx, NY, USA, for their assistance in proteomic analyses.
Supplementary Information is linked to the online version of the paper on the Asian Journal of Andrology website.
REFERENCES
- 1.Brown PW, Hwang K, Schlegel PN, Morris PL. Small ubiquitin-related modifier (SUMO)-1, SUMO-2/3 and SUMOylation are involved with centromeric heterochromatin of chromosomes 9 and 1 and proteins of the synaptonemal complex during meiosis in men. Hum Reprod. 2008;23:2850–7. doi: 10.1093/humrep/den300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Metzler-Guillemain C, Depetris D, Luciani JJ, Mignon-Ravix C, Mitchell MJ, et al. In human pachytene spermatocytes, SUMO protein is restricted to the constitutive heterochromatin. Chromosome Res. 2008;16:761–82. doi: 10.1007/s10577-008-1225-7. [DOI] [PubMed] [Google Scholar]
- 3.Rogers RS, Inselman A, Handel MA, Matunis MJ. SUMO modified proteins localize to the XY body of pachytene spermatocytes. Chromosoma. 2004;113:233–43. doi: 10.1007/s00412-004-0311-7. [DOI] [PubMed] [Google Scholar]
- 4.Vigodner M. Sumoylation precedes accumulation of phosphorylated H2AX on sex chromosomes during their meiotic inactivation. Chromosome Res. 2009;17:37–45. doi: 10.1007/s10577-008-9006-x. [DOI] [PubMed] [Google Scholar]
- 5.Vigodner M, Ishikawa T, Schlegel PN, Morris PL. SUMO-1, human male germ cell development, and the androgen receptor in the testis of men with normal and abnormal spermatogenesis. Am J Physiol Endocrinol Metab. 2006;290:E1022–33. doi: 10.1152/ajpendo.00527.2005. [DOI] [PubMed] [Google Scholar]
- 6.Vigodner M, Morris PL. Testicular expression of small ubiquitin-related modifier-1 (SUMO-1) supports multiple roles in spermatogenesis: silencing of sex chromosomes in spermatocytes, spermatid microtubule nucleation, and nuclear reshaping. Dev Biol. 2005;282:480–92. doi: 10.1016/j.ydbio.2005.03.034. [DOI] [PubMed] [Google Scholar]
- 7.Shrivastava V, Pekar M, Grosser E, Im J, Vigodner M. SUMO proteins are involved in the stress response during spermatogenesis and are localized to DNA double-strand breaks in germ cells. Reproduction. 2010;139:999–1010. doi: 10.1530/REP-09-0492. [DOI] [PubMed] [Google Scholar]
- 8.Xiao Y, Pollack D, Andrusier M, Levy A, Callaway M, et al. Identification of cell-specific targets of sumoylation during mouse spermatogenesis. Reproduction. 2016;151:149–66. doi: 10.1530/REP-15-0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiao Y, Pollack D, Nieves E, Winchell A, Callaway M, et al. Can your protein be sumoylated? A quick summary and important tips to study SUMO-modified proteins. Anal Biochem. 2015;477:95–7. doi: 10.1016/j.ab.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiao Y, Lucas B, Molcho E, Vigodner M. Cross-talk between sumoylation and phosphorylation in mouse spermatocytes. Biochem Biophys Res Commun. 2017;487:640–5. doi: 10.1016/j.bbrc.2017.04.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alarcon-Vargas D, Ronai Z. SUMO in cancer – wrestlers wanted. Cancer Biol Ther. 2002;1:237–42. doi: 10.4161/cbt.74. [DOI] [PubMed] [Google Scholar]
- 12.Baek SH. A novel link between SUMO modification and cancer metastasis. Cell Cycle. 2006;5:1492–5. doi: 10.4161/cc.5.14.3008. [DOI] [PubMed] [Google Scholar]
- 13.Guo WH, Yuan LH, Xiao ZH, Liu D, Zhang JX. Overexpression of SUMO-1 in hepatocellular carcinoma: a latent target for diagnosis and therapy of hepatoma. J Cancer Res Clin Oncol. 2011;137:533–41. doi: 10.1007/s00432-010-0920-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morris JR, Boutell C, Keppler M, Densham R, Weekes D, et al. The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature. 2009;462:886–90. doi: 10.1038/nature08593. [DOI] [PubMed] [Google Scholar]
- 15.Mo YY, Moschos SJ. Targeting Ubc9 for cancer therapy. Expert Opin Ther Targets. 2005;9:1203–16. doi: 10.1517/14728222.9.6.1203. [DOI] [PubMed] [Google Scholar]
- 16.Ganesan AK, Kho Y, Kim SC, Chen Y, Zhao Y, et al. Broad spectrum identification of SUMO substrates in melanoma cells. Proteomics. 2007;7:2216–21. doi: 10.1002/pmic.200600971. [DOI] [PubMed] [Google Scholar]
- 17.Nacerddine K, Lehembre F, Bhaumik M, Artus J, Cohen-Tannoudji M, et al. The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev Cell. 2005;9:769–79. doi: 10.1016/j.devcel.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 18.Sarge KD, Park-Sarge OK. Detection of proteins sumoylated in vivo and in vitro. Methods Mol Biol. 2009;590:265–77. doi: 10.1007/978-1-60327-378-7_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sarge KD, Park-Sarge OK. Sumoylation and human disease pathogenesis. Trends Biochem Sci. 2009;34:200–5. doi: 10.1016/j.tibs.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andersen JS, Matic I, Vertegaal AC. Identification of SUMO target proteins by quantitative proteomics. Methods Mol Biol. 2009;497:19–31. doi: 10.1007/978-1-59745-566-4_2. [DOI] [PubMed] [Google Scholar]
- 21.Zhao C, Huo R, Wang FQ, Lin M, Zhou ZM, et al. Identification of several proteins involved in regulation of sperm motility by proteomic analysis. Fertil Steril. 2007;87:436–8. doi: 10.1016/j.fertnstert.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 22.Vigodner M, Shrivastava V, Gutstein LE, Schneider J, Nieves E, et al. Localization and identification of sumoylated proteins in human sperm: excessive sumoylation is a marker of defective spermatozoa. Hum Reprod. 2013;28:210–23. doi: 10.1093/humrep/des317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanaka H, Pereira LA, Nozaki M, Tsuchida J, Sawada K, et al. A germ cell-specific nuclear antigen recognized by a monoclonal antibody raised against mouse testicular germ cells. Int J Androl. 1997;20:361–6. doi: 10.1046/j.1365-2605.1998.00080.x. [DOI] [PubMed] [Google Scholar]
- 24.Chen SR, Tang JX, Cheng JM, Li J, Jin C, et al. Loss of Gata4 in Sertoli cells impairs the spermatogonial stem cell niche and causes germ cell exhaustion by attenuating chemokine signaling. Oncotarget. 2015;6:37012–27. doi: 10.18632/oncotarget.6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hofmann MC, Braydich-Stolle L, Dettin L, Johnson E, Dym M. Immortalization of mouse germ line stem cells. Stem Cells. 2005;23:200–10. doi: 10.1634/stemcells.2003-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He Z, Jiang J, Kokkinaki M, Golestaneh N, Hofmann MC, et al. Gdnf upregulates c-Fos transcription via the Ras/Erk1/2 pathway to promote mouse spermatogonial stem cell proliferation. Stem Cells. 2008;26:266–78. doi: 10.1634/stemcells.2007-0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He Z, Jiang J, Kokkinaki M, Dym M. Nodal signaling via an autocrine pathway promotes proliferation of mouse spermatogonial stem/progenitor cells through Smad2/3 and Oct-4 activation. Stem Cells. 2009;27:2580–90. doi: 10.1002/stem.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lucas BE, Fields C, Joshi N, Hofmann MC. Mono-(2-ethylhexyl)-phthalate (MEHP) affects ERK-dependent GDNF signalling in mouse stem-progenitor spermatogonia. Toxicology. 2012;299:10–9. doi: 10.1016/j.tox.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garcia TX, Parekh P, Gandhi P, Sinha K, Hofmann MC. The NOTCH ligand JAG1 regulates GDNF expression in Sertoli cells. Stem Cells Dev. 2017;26:585–98. doi: 10.1089/scd.2016.0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pollack D, Xiao Y, Shrivasatava V, Levy A, Andrusier M, et al. CDK14 expression is down-regulated by cigarette smoke in vivo and in vitro . Toxicol Lett. 2015;234:120–30. doi: 10.1016/j.toxlet.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suzuki H, Ahn HW, Chu T, Bowden W, Gassei K, et al. SOHLH1 and SOHLH2 coordinate spermatogonial differentiation. Dev Biol. 2012;361:301–12. doi: 10.1016/j.ydbio.2011.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vigodner M. Roles of small ubiquitin-related modifiers in male reproductive function. Int Rev Cell Mol Biol. 2011;288:227–59. doi: 10.1016/B978-0-12-386041-5.00006-6. [DOI] [PubMed] [Google Scholar]
- 33.La Salle S, Sun F, Zhang XD, Matunis MJ, Handel MA. Developmental control of sumoylation pathway proteins in mouse male germ cells. Dev Biol. 2008;321:227–37. doi: 10.1016/j.ydbio.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paranko J, Seitz J, Meinhardt A. Developmental expression of heat shock protein 60 (HSP60) in the rat testis and ovary. Differentiation. 1996;60:159–67. doi: 10.1046/j.1432-0436.1996.6030159.x. [DOI] [PubMed] [Google Scholar]
- 35.Werner A, Meinhardt A, Seitz J, Bergmann M. Distribution of heat-shock protein 60 immunoreactivity in testes of infertile men. Cell Tissue Res. 1997;288:539–44. doi: 10.1007/s004410050839. [DOI] [PubMed] [Google Scholar]
- 36.Koushyar S, Jiang WG, Dart DA. Unveiling the potential of prohibitin in cancer. Cancer Lett. 2015;369:316–22. doi: 10.1016/j.canlet.2015.09.012. [DOI] [PubMed] [Google Scholar]
- 37.Choongkittaworn NM, Kim KH, Danner DB, Griswold MD. Expression of prohibitin in rat seminiferous epithelium. Biol Reprod. 1993;49:300–10. doi: 10.1095/biolreprod49.2.300. [DOI] [PubMed] [Google Scholar]
- 38.Xu YR, Fan YS, Yang WX. Mitochondrial prohibitin and its ubiquitination during spermatogenesis of the swimming crab Charybdis japonica. Gene. 2017;627:137–48. doi: 10.1016/j.gene.2017.06.025. [DOI] [PubMed] [Google Scholar]
- 39.Wang B, David MD, Schrader JW. Absence of caprin-1 results in defects in cellular proliferation. J Immunol. 2005;175:4274–82. doi: 10.4049/jimmunol.175.7.4274. [DOI] [PubMed] [Google Scholar]
- 40.Held T, Barakat AZ, Mohamed BA, Paprotta I, Meinhardt A, et al. Heat-shock protein HSPA4 is required for progression of spermatogenesis. Reproduction. 2011;142:133–44. doi: 10.1530/REP-11-0023. [DOI] [PubMed] [Google Scholar]
- 41.Loveland KL, Major AT, Butler R, Young JC, Jans DA, et al. Putting things in place for fertilization: discovering roles for importin proteins in cell fate and spermatogenesis. Asian J Androl. 2015;17:537–44. doi: 10.4103/1008-682X.154310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sharpe RM, Irvine DS. How strong is the evidence of a link between environmental chemicals and adverse effects on human reproductive health? BMJ. 2004;328:447–51. doi: 10.1136/bmj.328.7437.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walschaerts M, Muller A, Auger J, Bujan L, Guerin JF, et al. Environmental, occupational and familial risks for testicular cancer: a hospital-based case-control study. Int J Androl. 2007;30:222–9. doi: 10.1111/j.1365-2605.2007.00805.x. [DOI] [PubMed] [Google Scholar]
- 44.Bahrami A, Ro JY, Ayala AG. An overview of testicular germ cell tumors. Arch Pathol Lab Med. 2007;131:1267–80. doi: 10.5858/2007-131-1267-AOOTGC. [DOI] [PubMed] [Google Scholar]
- 45.Rajpert-de Meyts E, Hoei-Hansen CE. From gonocytes to testicular cancer: the role of impaired gonadal development. Ann N Y Acad Sci. 2007;1120:168–80. doi: 10.1196/annals.1411.013. [DOI] [PubMed] [Google Scholar]
- 46.Gali H, Juhasz S, Morocz M, Hajdu I, Fatyol K, et al. Role of SUMO modification of human PCNA at stalled replication fork. Nucleic Acids Res. 2012;40:6049–59. doi: 10.1093/nar/gks256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Q, Yan HB, Wang J, Cui SJ, Wang XQ, et al. Chromatin remodeling gene AT-rich interactive domain-containing protein 1A suppresses gastric cancer cell proliferation by targeting PIK3CA and PDK1. Oncotarget. 2016;7:46127–41. doi: 10.18632/oncotarget.10060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ramos P, Karnezis AN, Hendricks WP, Wang Y, Tembe W, et al. Loss of the tumor suppressor SMARCA4 in small cell carcinoma of the ovary, hypercalcemic type (SCCOHT) Rare Dis. 2014;2:e967148. doi: 10.4161/2167549X.2014.967148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee MT, Bachant J. SUMO modification of DNA topoisomerase II: trying to get a CENse of it all. DNA Repair (Amst) 2009;8:557–68. doi: 10.1016/j.dnarep.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Azuma Y, Arnaoutov A, Dasso M. SUMO-2/3 regulates topoisomerase II in mitosis. J Cell Biol. 2003;163:477–87. doi: 10.1083/jcb.200304088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ke H, Zhao L, Zhang H, Feng X, Xu H, et al. Loss of TDP43 inhibits progression of triple-negative breast cancer in coordination with SRSF3. Proc Natl Acad Sci U S A. 2018;115:E3426–35. doi: 10.1073/pnas.1714573115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Freeman AK, Morrison DK. 14-3-3 Proteins: diverse functions in cell proliferation and cancer progression. Semin Cell Dev Biol. 2011;22:681–7. doi: 10.1016/j.semcdb.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Graf M, Brobeil A, Sturm K, Steger K, Wimmer M. 14-3-3 beta in the healthy and diseased male reproductive system. Hum Reprod. 2011;26:59–66. doi: 10.1093/humrep/deq319. [DOI] [PubMed] [Google Scholar]
- 54.Panigrahi SK, Manterola M, Wolgemuth DJ. Meiotic failure in cyclin A1-deficient mouse spermatocytes triggers apoptosis through intrinsic and extrinsic signaling pathways and 14-3-3 proteins. PLoS One. 2017;12:e0173926. doi: 10.1371/journal.pone.0173926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Paronetto MP. Ewing sarcoma protein: a key player in human cancer. Int J Cell Biol. 2013;2013:642853. doi: 10.1155/2013/642853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang WL, Wei L, Huang WQ, Li R, Shen WY, et al. Vigilin is overexpressed in hepatocellular carcinoma and is required for HCC cell proliferation and tumor growth. Oncol Rep. 2014;31:2328–34. doi: 10.3892/or.2014.3111. [DOI] [PubMed] [Google Scholar]
- 57.Hartsough MT, Steeg PS. Nm23/nucleoside diphosphate kinase in human cancers. J Bioenerg Biomembr. 2000;32:301–8. doi: 10.1023/a:1005597231776. [DOI] [PubMed] [Google Scholar]
- 58.Kowno M, Watanabe-Susaki K, Ishimine H, Komazaki S, Enomoto K, et al. Prohibitin 2 regulates the proliferation and lineage-specific differentiation of mouse embryonic stem cells in mitochondria. PLoS One. 2014;9:e81552. doi: 10.1371/journal.pone.0081552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu X, Xiong X, Sun Y. The role of ribosomal proteins in the regulation of cell proliferation, tumorigenesis, and genomic integrity. Sci China Life Sci. 2016;59:656–72. doi: 10.1007/s11427-016-0018-0. [DOI] [PubMed] [Google Scholar]
- 60.Zhou HM, Fang YY, Weinberger PM, Ding LL, Cowell JK, et al. Transgelin increases metastatic potential of colorectal cancer cells in vivo and alters expression of genes involved in cell motility. BMC Cancer. 2016;16:55. doi: 10.1186/s12885-016-2105-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Khuon S, Liang L, Dettman RW, Sporn PH, Wysolmerski RB, et al. Myosin light chain kinase mediates transcellular intravasation of breast cancer cells through the underlying endothelial cells: a three-dimensional FRET study. J Cell Sci. 2010;123:431–40. doi: 10.1242/jcs.053793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fang KP, Dai W, Ren YH, Xu YC, Zhang SM, et al. Both Talin-1 and Talin-2 correlate with malignancy potential of the human hepatocellular carcinoma MHCC-97 L cell. BMC Cancer. 2016;16:45. doi: 10.1186/s12885-016-2076-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang H, Dai SD, Zhang D, Liu D, Zhang FY, et al. Delta-catenin promotes the proliferation and invasion of colorectal cancer cells by binding to E-cadherin in a competitive manner with p120 catenin. Target Oncol. 2014;9:53–61. doi: 10.1007/s11523-013-0269-6. [DOI] [PubMed] [Google Scholar]
- 64.Stojadinovic A, Hooke JA, Shriver CD, Nissan A, Kovatich AJ, et al. HYOU1/Orp150 expression in breast cancer. Med Sci Monit. 2007;13:BR231–9. [PubMed] [Google Scholar]
- 65.Ding C, Fan X, Wu G. Peroxiredoxin 1 – an antioxidant enzyme in cancer. J Cell Mol Med. 2017;21:193–202. doi: 10.1111/jcmm.12955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Luo ML, Gong C, Chen CH, Hu H, Huang P, et al. The Rab2A GTPase promotes breast cancer stem cells and tumorigenesis via Erk signaling activation. Cell Rep. 2015;11:111–24. doi: 10.1016/j.celrep.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bhuin T, Roy JK. Rab11 in disease progression. Int J Mol Cell Med. 2015;4:1–8. [PMC free article] [PubMed] [Google Scholar]
- 68.Wang W, Zhang M, Peng Y, He J. Ubiquitin Associated Protein 2-like (UBAP2L) overexpression in patients with hepatocellular carcinoma and its clinical significance. Med Sci Monit. 2017;23:4779–88. doi: 10.12659/MSM.907071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tsujimoto Y, Tomita Y, Hoshida Y, Kono T, Oka T, et al. Elevated expression of valosin-containing protein (p97) is associated with poor prognosis of prostate cancer. Clin Cancer Res. 2004;10:3007–12. doi: 10.1158/1078-0432.ccr-03-0191. [DOI] [PubMed] [Google Scholar]
- 70.Roscioli E, Di Francesco L, Bolognesi A, Giubettini M, Orlando S, et al. Importin-beta negatively regulates multiple aspects of mitosis including RANGAP1 recruitment to kinetochores. J Cell Biol. 2012;196:435–50. doi: 10.1083/jcb.201109104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jin L, Shen K, Chen T, Yu W, Zhang H. SUMO-1 gene silencing inhibits proliferation and promotes apoptosis of human gastric cancer SGC-7901 cells. Cell Physiol Biochem. 2017;41:987–98. doi: 10.1159/000460836. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression of SUMO proteins in frozen seminoma samples. A small piece from the frozen seminoma sample, used for the mass spectrometry analysis, was cut with a blade to prepare a cell suspension and slides. Cells were stained with anti-SUMO antibody to confirm SUMO expression. SUMO immunostaining is shown in green, nuclear staining by DAPI in blue. Most of the cells show SUMO expression.


