Abstract
Keloids are a skin fibroproliferative condition characterized by the hyperproliferation of fibroblasts and the excessive deposition of extracellular matrix (ECM) components. Previous studies have determined that Caveolin-1 controlled hyperresponsiveness to mechanical stimuli through Runt-related transcription factor 2 (Runx2) activation in keloids. However, the molecular mechanism of Runx2 regulating the pathological progression of keloids has not been elucidated. Gene Ontology and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis revealed that most of the differentially expressed genes (DEGs), including Runx2, were significantly enriched in the biological processes ‘Positive regulation of cell proliferation’, in the cellular components ‘Extracellular matrix’, in the molecular functions ‘Extracellular matrix structural constituents’ and in the KEGG ‘PI3K-Akt signaling pathway’. The aim of the present study was to investigate the expression levels of the Runx2 in human keloid tissues and primary human keloid fibroblasts (HKFs), and to determine the underlying molecular mechanisms involved in the fibrotic roles of Runx2 in keloid formation. Runx2 expression levels were analyzed in patient keloid tissues and HKFs using western blotting, reverse transcription-quantitative PCR (RT-qPCR) and immunofluorescence microscopy. Primary HKFs were transfected with a small interfering RNA (si) specifically targeting Runx2 (si-Runx2). Subsequently, Cell Counting Kit-8, wound healing and Transwell assays, flow cytometry, RT-qPCR and western blotting were applied to evaluate the proliferation, migration, apoptosis, ECM deposition and PI3K/AKT signaling pathway of HKFs, respectively. In addition, western blotting was also used to determine the expression levels of phosphorylated AKT and PI3K in HKFs. The results revealed that Runx2 expression levels were upregulated in keloid tissues and primary HKFs compared with the normal skin tissues and human normal fibroblasts. Following the transfection with si-Runx2, the proliferative and migratory abilities of HKFs were significantly reduced and the apoptotic rate was increased. The expression levels of type I, type III collagen, fibronectin, and α-smooth muscle actin were downregulated in si-Runx2-transfected cells, which was hypothesized to occur through following the downregulation of the phosphorylation levels of PI3K and AKT. In conclusion, the findings of the present study indicated that Runx2 silencing in HKFs might significantly inhibit the cell proliferation, migration and the expression levels of ECM-related proteins, and promote apoptosis via suppressing the PI3K/AKT signaling pathway. Thus, Runx2 siRNA treatment may reverse the pathological phenotype of keloids through the inhibition of PI3K/AKT signaling in patients.
Keywords: runt-related transcription factor 2, keloid fibroblasts, small interfering RNA, PI3K/AKT signaling
Introduction
Keloids are a skin fibroproliferative condition usually accompanied by the loss of the sense of touch, pain and itching, amongst other physical and psychosocial symptoms (including restricted joint movement, anxiety, depression, low self-esteem) (1). Keloids usually occur secondary to the abnormal healing of skin wounds, and grow in an invasive manner, extending beyond the original injured tissue area (2). Histopathological examinations have revealed that compared with normal skin, keloids are characterized by the hyperproliferation of fibroblasts, an increased proportion of myofibroblasts and the excessive deposition of extracellular matrix (ECM) components, such as collagen I and collagen III (3). The increased synthesis of ECM collagen is considered to be related to the overactivation of keloid fibroblasts. The proliferation and migration of fibroblasts have been identified as essential for keloid formation. To some extent, keloids exhibit invasive and immortal growth, similar to tumor tissues (4). Based on the RNA-Seq analysis of keloid and normal skin samples, previous studies indicated that changes in the expression levels of pro-fibrotic genes are found to serve a significant role in keloid formation (5,6). Although various therapeutic modalities are used to treat keloids, such as local surgical resection combined with radiotherapy, laser treatments, cryotherapy and chemotherapy, the clinical effects remain unsatisfactory (7). This is because these methods have the potential to cause relapse, leading to contour deformity caused by keloids repeatedly pulling on the skin and dysfunction caused by keloids around the joint, seriously affecting the physical and mental health, and social life of patients (8). These issues remain a major challenge for keloid treatment in the field of plastic surgery. Therefore, determining the molecular mechanisms involved in the genesis of keloids is essential for developing novel and effective therapeutic strategies.
The emerging development of high-throughput sequencing has enabled the identification of hub genes and dysfunctional biological pathways involved in the pathological process of numerous types of disease. Gene Ontology (GO) functional term and Kyoto Encyclopedia of Genes and Genomes (KEGG) signaling pathway enrichment analyses are common methods used in bioinformatics analysis for annotating genes and identifying underlying biological pathways (9). For example, Liu et al (10) applied weighted gene co-expression network analysis to identify the main functions of hub genes and keloid-related pathways. The results of this study revealed that the expression change of some hub genes (such as DKK3 and MMP3) have been verified by immunohistochemical staining. In addition, Fagone et al (11) used functional term and signaling pathway enrichment analyses to verify that macrophage migratory inhibitory factor (MIF) receptors were variably expressed in CD4+ T cells and central nervous lesions. Similarly, Presti et al (12) also used The Cancer Genome Atlas data to determine the accurate and underlying biological mechanism in glioblastoma multiforme, and further confirmed that MIF and its receptors were associated with glioblastoma multiforme progression and maintenance. Runt-related transcription factors (Runx) are a family of three genes, which have been discovered to serve an important role in cell migration, proliferation and apoptosis (13). Runx2 is a member of the Runx family of transcription factors containing the runt DNA-binding domain (14). Runx2 has been revealed to be essential for osteoblastic differentiation and skeletal morphogenesis, as well as tumor formation and progression (15). In addition, Runx2 contributed to the profibrotic function and promoted the progression of lung fibrosis, suggesting that targeting Runx2 may be of therapeutic value for curing fibrosis-related diseases (14). Numerous specific molecular pathways have been suggested to serve a role in liver fibrosis, mainly related to chronic viral hepatitis B or C (HBV or HCV) (16). For example, Fagone et al (17) reported that Runx2 was identified as a hub gene that participated in the progression of hepatic fibrosis, while the knockdown of Runx2 expression levels could ameliorate liver cirrhosis. Similarly, Chen et al (18) revealed that the protein expression levels of Runx2 were upregulated in aortic valve fibrosis, indicating that the AMPK-activated protein kinase α/Runx2 pathway may participate in high-fat diet-induced fibrosis in aortic valves. A previous study also demonstrated that Runx2 was differentially expressed in alveolar epithelial cells and fibroblasts in pulmonary fibrosis. In fact, the cell-specific targeting of Runx2 signaling pathways was suggested as a therapeutic approach for idiopathic pulmonary fibrosis (14). Runx2 expression levels were also found to be crucial in autoimmune diseases. For instance, Tchetina et al (19) discovered that Runx2 expression levels were upregulated in the peripheral blood and may predict an improved response of patients with rheumatoid arthritis to methotrexate treatment, indicating the potential of Runx2 as a prognostic marker. However, to the best of our knowledge, the role of Runx2 in keloid pathogenesis remains unknown. Therefore, the present study aimed to investigate the expression levels of Runx2 in human keloid fibroblasts (HKFs) and to determine its role in keloid formation.
Materials and methods
Patient samples
Keloid tissues and normal skin tissues were obtained from 9 patients (age range, 13–36, mean, 25.4 years; 5 women and 4 men) with keloids between September 2018 and May 2020 at Tongji Hospital of Huazhong University of Science and Technology (Wuhan, China). The patients recruited in the present study had not been pretreated for keloid for ≥3 months and the patients keloids were in the active stage. The diagnosis of keloid was based on clinical appearance, symptoms, persistence for >1 year and extension beyond the original margins. The nature of the samples was confirmed by pathological examination. The present study was approved by the ethics committee of Tongji Hospital of Huazhong University of Science and Technology. Written informed consent was obtained from all participants.
Bioinformatics analysis
Genome-wide microarray data for HKFs (from nine different patients with keloids) and human dermal fibroblasts (HDFs, from four different control patients) (dataset no. GSE44270) (20) were downloaded from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo). GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r) was used to identify the differentially expressed genes (DEGs, |fold change| >1.5 and P<0.05) between HKFs and HDFs in the GSE44270 dataset (21). DEGs were then subjected to functional term enrichment analysis using Gene Ontology (GO) analysis and signaling pathway enrichment analysis using Kyoto Encyclopedia for Genes and Genomes (KEGG) analysis on the Database of Annotation, Visualization and Integrated Discovery (DAVID; http://david.abcc.ncifcrf.gov). In addition, a protein-protein interaction (PPI) network of the DEGs was constructed using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database (http://string-db.org) and a threshold combined score of ≥0.4 was set as the cut-off. Cytoscape software (https://cytoscape.org/index.html; version 3.6.1) was used to visualize the PPI network. The plug-in MCODE (https://cytoscape.org/index.html; version 3.6.1) in Cytoscape was used to analyze the genes in the PPI network and to identify hub genes based on the node degree value (degree cut-off ≥2, node score cut-off ≥0.2, K-core ≥2, and max depth = 100).
Primary cell culture and small interfering RNA (siRNA/si) transfection
HKFs were isolated from keloid tissues (n=3) with high Runx2 expression level and HDFs were isolated from normal skin tissues (n=3) with low Runx2 expression level, respectively, using the collagenase digestion method. Briefly, the adipose tissues and epidermis were removed from the samples. Subsequently, specimens were cut into 1×1 mm sections and digested with collagenase (0.5 mg/ml) and trypsin (0.2 mg/ml) for 6 h at 37°C. Following centrifugation at 300 × g at 37°C. for 5 min, the supernatant was discarded and the precipitate retained. Fibroblasts were cultured in DMEM/F12 medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.), 100 IU/ml penicillin and 100 IU/ml streptomycin, and maintained at 37°C with 5% CO2. HDFs and HKFs were used in the present study following 3–5 cell passages (22).
siRNAs targeting Runx2 (si-Runx2; 5′-CAGAAGAATGGTACAAATCCAAG-3′) and a negative control (NC) siRNA (si-NC; 5′-TTCTCCGAACGTGTCACGTdTdT-3′) were constructed by Guangzhou RiboBio Co., Ltd. HKFs at a density of ~2×106/well were cultured to 50% confluence in 6-well plates and transfected with 20 µmol/ml of si-Runx2 or si-NC using 10 µl Lipofectamine® 3000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) in Opti-MEM medium (Gibco; Thermo Fisher Scientific, Inc.) at 37°C for 24 h, according to the manufacturer's protocol. Following 24 h of transfection, HKFs were harvested for further experiments.
Fluorescence and light microscopy
The expression and distribution of Runx2, collagen I, collagen III, and fibronectin from human keloid tissues and normal skin tissues were analyzed using fluorescence microscopy (magnification, ×200). Meanwhile, the expression and distribution of vimentin in HKFs and HDFs was also analyzed using fluorescence microscopy (magnification, ×400). The morphology of fibroblasts (included HKFs and HDFs) was observed under the light microscope (magnification, ×40). Briefly, fresh tissues collected in the operating rooms were immediately fixed with 4% paraformaldehyde at 37°C for 24 h and embedded in paraffin after dehydration with an ascending alcohol series (23). Sections were cut at 5 µm. Following paraffinization, the tissues were rehydrated using a descending alcohol series. Sections were washed with xylene at 37°C, deparaffinized, blocked with 5% bovine serum albumin (Sangon Biotech Co., Ltd.) for 30 min at 37°C and incubated overnight at 4°C with the specific primary antibodies (anti-Runx2 1:1,000, cat. no. 55725-1-AP; anti-collagen I 1:1,000, cat. no. 14695-1-AP; anti-collagen III 1:1,000, cat. no. 14737-1-AP; anti-fibronectin 1:1,000, cat. no. 49225-1-AP; anti-vimentin, 1:1,000, cat. no. 82132-1-AP, all from ProteinTech Group, Inc.). Following the primary antibody incubation, the slides were incubated for 1 h at 37°C with the corresponding specific secondary antibody (HRP-conjugated Affinipure Goat Anti-Rabbit IgG(H+L), 1:2,500, cat. no. SA00001-2; HRP-conjugated Affinipure Goat Anti-Mouse IgG(H+L), 1:2,500, cat. no. SA00001-1; both from ProteinTech Group, Inc.), and staining with the DAB solution (Dako; Agilent Technologies, Inc.). Nuclei were stained with DAPI for 45 min at 37°C.
Western blotting
HKFs at a density of approximately 2×106/well were cultured to 50% confluence in 6-well plates and transfected with si-Runx2 or si-NC at 37°C for 24 h. Total protein was extracted from cells using RIPA lysis buffer (Boster Biological Technology). Total protein was quantified using a BCA protein assay kit and a total of 50 µg proteins loaded per lane were separated via 12% SDS-PAGE. The separated proteins were subsequently transferred onto polyvinylidene fluoride membranes (24) and blocked with 5% blocking buffer (Invitrogen; Thermo Fisher Scientific, Inc.) in TBS-0.1% Tween-20 (TBST) for 2 h at 37°C. The membranes were then incubated with the following primary antibodies overnight at 4°C: Anti-Runx2 (1:1,000, cat. no. 55725-1-AP), anti-α-smooth muscle actin (SMA, 1:1,000, cat. no. 55135-1-AP), anti-collagen I (1:1,000, cat. no. 14695-1-AP), anti-collagen III (1:1,000, cat. no. 14737-1-AP), anti-fibronectin (1:1,000, cat. no. 49225-1-AP), anti-phosphorylated (p)-AKT (1:500, cat. no. 66444-1-Ig), anti-AKT (1:500, cat. no. 60203-2-Ig), anti-p-PI3K (1:500, cat. no. 56321-1-Ig), anti-PI3K (1:1,000, cat. no. 48631-2-Ig) and anti-GAPDH (1:2,500, cat. no. 63714-1-AP; all from ProteinTech Group, Inc.). Following the primary antibody incubation, the membranes were washed three times with TBST and incubated with the secondary antibody [HRP-conjugated Affinipure Goat Anti-Rabbit IgG(H+L), 1:2,500, cat. no. SA00001-2; HRP-conjugated Affinipure Goat Anti-Mouse IgG(H+L), 1:2,500, cat. no. SA00001-1; both from ProteinTech Group, Inc.], and staining with the DAB solution (Dako; Agilent Technologies, Inc.) for 2 h at 37°C. Protein bands were visualized using an ECL detection kit (Beyotime Institute of Biotechnology) and then the signal intensity of proteins were quantified using ImageJ software (version 1.8.0; National Institutes of Health). All experiments were performed in triplicate.
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from HKFs using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). Total RNA was spectrophotometrically quantified at an absorbance of 260 nm by a spectrophotometer (Genova; Jenway) and reverse transcribed into cDNA using a PrimeScript RT reagent kit (Takara Bio, Inc.) at 85°C for 5 sec, 37°C for 10 min and 4°C for 15 min. qPCR was subsequently performed (one initial cycle at 95°C for 30 sec, followed by 40 cycles at 95°C for 5 sec and at 60°C for 30 sec.) using a Power SYBR Green PCR Master mix (Takara Bio, Inc.) on a Real-Time Thermal cycler (Bio-Rad Laboratories, Inc.). The following primers were used for the qPCR: Fibronectin forward, 5′-CGGTGGCTGTCAGTCAAAG-3′ and reverse, 5′-AAACCTCGGCTTCCTCCATAA-3′; Runx2 forward, 5′-ACGAGGCAAGAGTTTCACCT-3′ and reverse, 5′-TGTCTGTGCCTTCTTGGTTC-3′; GAPDH forward, 5′-ACCACAGTCATGCCATCAC-3′ and reverse, 5′-TCCACCACCCTGTTGCTGTA-3′; collagen I forward, 5′-GGGCAAGACAGTGATTGAATA-3′ and reverse, 5′-ACGTCFAAGCCGAATTCCT-3′; collagen III forward, 5′-AGGTCCTGCGGGTAACACT-3′ and reverse, 5′-ACTTTCACCCTTGACACCCTG-3′; and α-SMA forward, 5′-CTGTTCCAGCCATCCTTCAT-3′ and reverse, 5′-CCGTGATCTCCTTCTGCATT-3′. The expression levels were quantified using the 2−∆∆Cq method and normalized to GAPDH expression levels (25).
Transwell migration assay
The migratory ability of HKFs from different groups was evaluated using a Transwell assay. Briefly, 1×104 HKFs/well in 200 µl serum-free DMEM/F12 Glucose medium were seeded into the upper compartments of the Transwell chambers (Corning, Inc.). The lower compartments were filled with basic medium supplemented with 20% FBS as a chemoattractant. The cells were incubated at 37°C with 5% CO2 to allow the migration through the porous membrane. Following 24 h of incubation, the cells on the upper surface of the membrane were gently removed with a cotton swab. The migratory cells in the lower chamber were fixed with 4% paraformaldehyde for 30 min, stained with 0.1% crystal violet for 10 min at 37°C and gently rinsed 3 times with PBS. The stained cells were counted in 3 randomly selected fields under a light microscope (magnification, ×200) (26).
Wound healing assay
A wound healing assay was used to analyze the migratory ability of HKFs. Briefly, the fibroblasts were seeded into 6-well culture plates at a density of 1×106 cells/well and transfected with si-Runx2 or si-NC for 24 h at 37°C. A scratch was created using a sterile 200-µl pipette tip in the cell monolayer upon the fibroblasts reaching 95–100% confluence. The wells were rinsed with PBS and incubated in serum-free DMEM/F12 Glucose medium at 37°C with 5% CO2. The migration of the cells in the different groups into the wound area was observed under a light microscope (magnification, ×100) at 0, 12 and 24 h. ImageJ software (version 1.8.0; National Institutes of Health) was used for analysis.
Proliferation assay
The proliferative ability of cells was determined using a Cell Counting Kit-8 (CCK-8) assay (Dojindo Molecular Technologies, Inc.). Following 24 h of transfection at 37°C, HKFs were seeded into 96-well culture plates at a density of 5×104 cells/well in 100 µl DMEM/F12 Glucose medium supplemented with 10% FBS and cultured for 24 h at 37°C. Following the incubation, 10 µl CCK-8 reagent was added to each well and incubated for 1 h at 37°C. The absorbance at 450 nm was measured using a microplate reader.
Flow cytometric analysis of early and late apoptosis
The apoptotic rate of HKFs was analyzed using an Annexin V-FITC/PI apoptosis detection kit (BD Biosciences). Briefly, HKFs were seeded into 6-well plates at a density of 1×106 cells/well and incubated for 12 h at 37°C. Cells were transfected with si-Runx2 or si-NC for 24 h at 37°C, subsequently harvested (300 × g; 5 min at 37°C), washed with PBS and resuspended with 200 µl the Annexin V-PI binding buffer at a final density of 2×106 cells/flow tube. The cell suspension was mixed with 5 µl Annexin V-FITC and 5 µl propidium iodide (PI) solution and incubated in the dark for 15 min at 4°C. The apoptotic cells from each sample were detected using a fluorescence-activated cell sorting flow cytometer (BD Biosciences) and analyzed using FlowJo software (version 10.2; FlowJo LLC). The total number of cells was assessed using quadrant statistical method (upper right quadrant-advanced stage apoptosis cell percentage; lower right quadrant-prophase apoptosis cell percentage) (27).
Statistical analysis
Statistical analysis was performed using GraphPad Prism version 8.0.1 software (GraphPad Software, Inc.) and all data are presented as the mean ± SD. Statistical differences between two groups were analyzed using an unpaired two-tailed Student's t-test, whereas differences between ≥3 groups were analyzed using a one-way ANOVA followed by a Tukey's post hoc test for multiple comparisons. The primary fibroblasts (including HKFs and HDFs) used in the experiment were derived from 3 different individuals. P<0.05 was considered to indicate a statistically significant difference.
Results
Identification of differentially expressed mRNAs in keloid fibroblasts
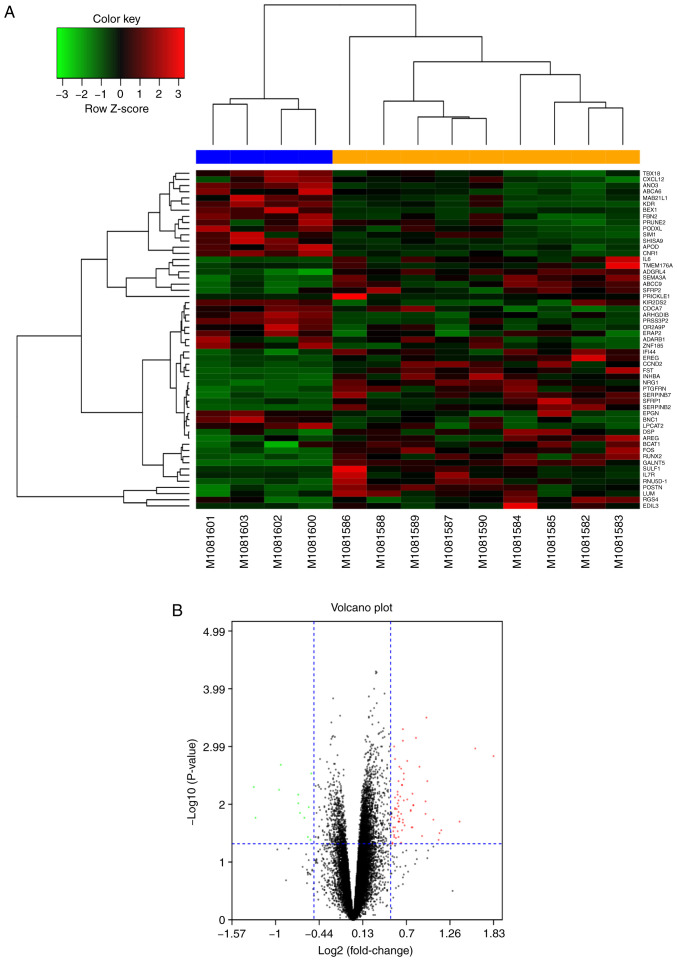
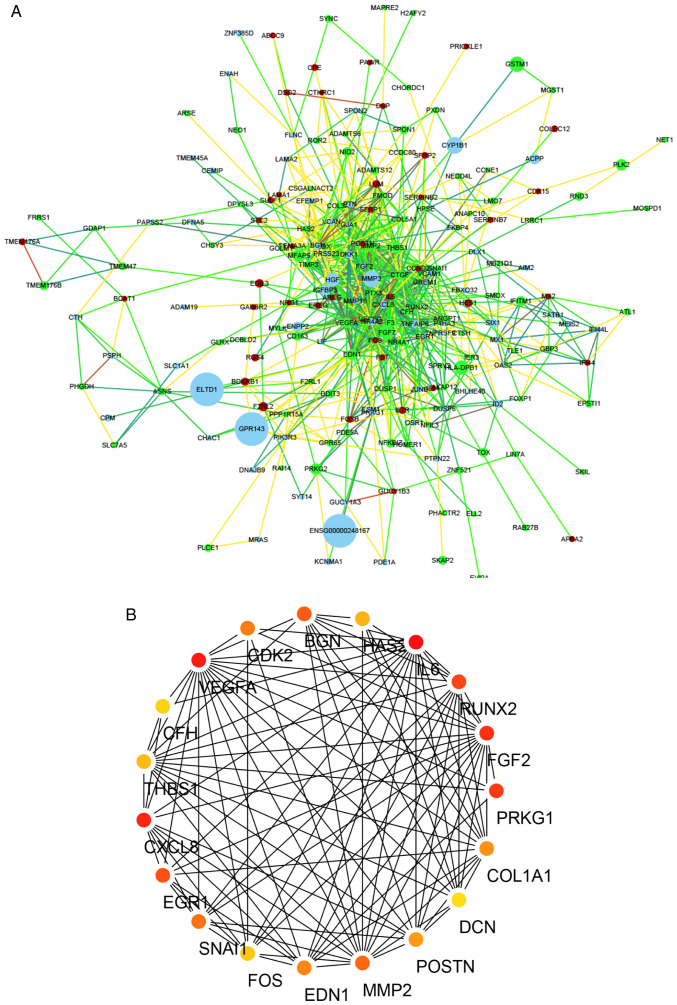
The microarray dataset GSE44270 was downloaded from the GEO database. This dataset comprised nine samples of HKFs (GSM1081582-GSM1081590) and four samples of HDFs (GSM1081600-GSM1081603). GEO2R was used to identify the DEGs in the GSE44270 dataset. The 60 most DEGs (30 upregulated, including Runx2, FOS, AREG and BCAT1 and 30 downregulated, including TBX18, ANO3, SIM1 and APOD) were represented in hierarchical clustering maps (Fig. 1A) and volcano maps (Fig. 1B). A PPI network was generated using the STRING online tool and the result revealed that MMP3, VEGF, Runx2 and CTGF were the most prominent proteins and had the most prominent interactions in the PPI network (Fig. 2A). The hub genes (network-centric genes, including Runx2) from the PPI network were identified and visualized using the plug-in MCODE in Cytoscape (Fig. 2B). These results indicated that Runx2 may serve an important regulatory role in the pathological process of keloids.
Figure 1.
Heat and volcano map of DEGs. (A) Heat map of DEGs identified using a threshold of P<0.05. The color key represents fold change. (B) Volcano map of all DEGs. The red dots represent genes that are upregulated in keloids and the green dots represent downregulated genes. DEGs, differentially expressed genes.
Figure 2.
DEGs PPI network complex. (A) PPI network of DEGs was generated using the Search Tool for the Retrieval of Interacting Genes/Proteins database. The gradual node color represented the fold change (HKFs vs. HDFs) of DEGs in GSE44270 dataset. The redder the color of the node, the greater the fold change of the gene. Conversely, the greener the color of the node, the smaller the fold change of the gene. The gradient color line represented the degree of connection between each node (the green line represented the closest connection, the red line represented the most distant connection) The gradual node volume represented the P-value of each node, and the larger the P-value, the larger the volume of the node. (B) The hub genes (network-centric genes; including PRKG1, FGF2, Runx2, IL6, HAS2, BGN, CDK2, VEGFA, CFH, THBS1, CXCL8, EGR1, SNAI1, FOS, EDN1, MMP2, POSTN, DCN, and COL1A1) from the PPI network were identified and visualized using the plug-in MCODE in Cytoscape. The different colors represented different degree value and the redder the color, the greater the degree value of the node. DEGs, differentially expressed genes; PPI, protein-protein interaction; HKFs, primary human keloid fibroblasts; HDFs, human dermal fibroblasts; PRKG1, protein kinase cGMP-dependent 1; FGF2, fibroblast growth factor 2; Runx2, RUNX family transcription factor 2; IL6, interleukin 6; HAS2, hyaluronan synthase 2; BGN, biglycan; CDK2, cyclin dependent kinase 2; VEGFA, vascular endothelial growth factor A; CFH, complement factor H; THBS1, thrombospondin 1; CXCL8, C-X-C motif chemokine ligand 8; EGR1, early growth response 1; SNAI1, snail family transcriptional repressor 1; FOS, Fos proto-oncogene, AP-1 transcription factor subuni; EDN1, endothelin 1; MMP2, matrix metallopeptidase 2; POSTN, periostin; DCN, decorin; COL1A1, collagen type I α 1 chain.
Functional term and signaling pathway enrichment analyses
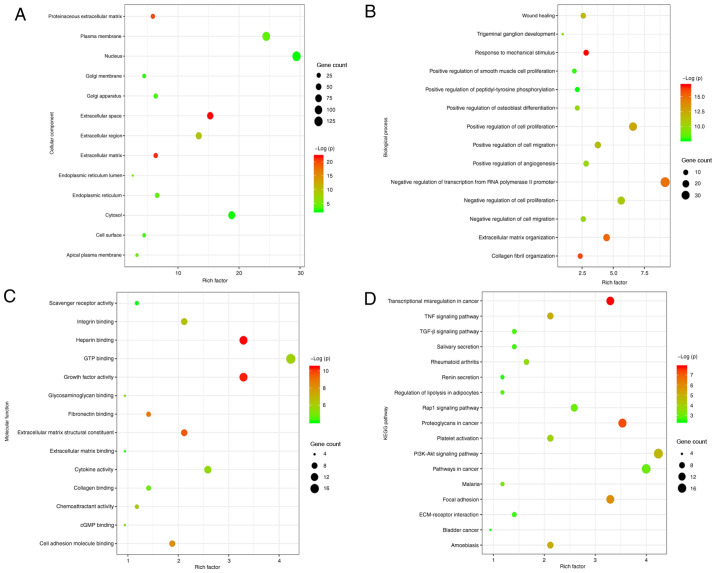
GO consists of three structured ontologies that describe gene function terms based on their associated biological processes (BP), cellular components (CC) and molecular functions (MF) (28). Meanwhile, the KEGG database is a database applied to search genomes, promising drug candidates and biological pathways (29). GO functional term and KEGG signaling pathway enrichment analyses are widely applied to investigate pathogenetic pathways and novel therapeutic approaches for the treatment of several pathological conditions, including autoimmune, neoplastic, psychiatric and neurodegenerative disorders (11,30). DAVID was used to analyze the BPs, CCs and MFs, and the associated signaling pathways of the DEGs (Fig. 3). A total of 427 upregulated genes associated with keloids were screened for further functional term and signaling pathway enrichment analyses. CC analyses indicated that the DEGs closely associated with the pathological progression of keloids were strongly enriched in the ‘Extracellular matrix’, ‘Extracellular space’ and ‘Extracellular region’ (Fig. 3A). BP analyses identified that the genes closely associated with the pathological progression of keloids were mainly enriched in ‘Extracellular matrix organization’, ‘Positive regulation of cell proliferation’ and ‘Positive regulation of cell migration’ (Fig. 3B). MF analyses revealed that the DEGs closely associated with the pathological progression of keloids were enriched in ‘Extracellular matrix structural constituents’, ‘Fibronectin binding’, ‘Growth factor activity’ and ‘Collagen binding’ (Fig. 3C). KEGG analyses revealed that the genes closely associated with the pathological progression of keloids were mainly enriched in the ‘PI3K-Akt signaling pathway’, ‘TGF-β signaling pathway’, ‘ECM-receptor interaction’ and ‘Focal adhesion’ (Fig. 3D). Overall, GO and KEGG analysis revealed that most of the DEGs closely associated with the pathological progression of keloids (including Runx2) were significantly enriched in the BP ‘Positive regulation of cell proliferation’, in the CC ‘Extracellular matrix’, in the MF ‘Extracellular matrix structural constituents’ and in the KEGG ‘PI3K-Akt signaling pathway’.
Figure 3.
GO functional term and KEGG signaling pathway enrichment analyses of DEGs associated with the pathological process of keloids. GO functional term enrichment analysis of (A) Cellular component, (B) Biological process and (C) Molecular function. (D) KEGG signaling pathway enrichment analysis. The y-axes represent the GO or KEGG terms and the x-axes represent the enrichment factor of each term. GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Expression levels of Runx2 in keloid tissues and HKFs
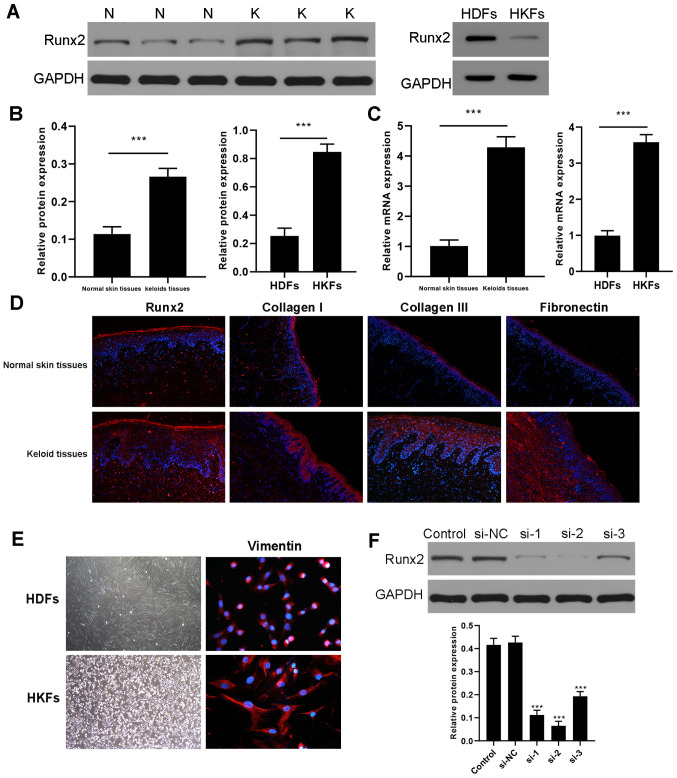
The expression levels of Runx2 in keloid tissues and HKFs compared with normal skin tissues and HDFs were analyzed. The protein expression levels of Runx2 were determined by western blotting and the results indicated that Runx2 expression levels were significantly upregulated in human keloid tissues and HFKs compared with normal skin tissues and HDFs, respectively (Figs. 4A and B and S1). Consistent results were obtained in the mRNA expression levels using RT-qPCR (Fig. 4C). Immunofluorescence subsequently revealed that the fluorescence intensities of Runx2, collagen I, collagen III and fibronectin in keloid tissues were markedly higher compared with the normal skin tissues (Fig. 4D). Primary fibroblasts (including HKFs and HDFs) appeared as long fusiform cells under the light microscope (magnification, ×40) and immunofluorescence imaging of vimentin expression was performed using fluorescence microscope (magnification, ×400; Fig. 4E). The results of the immunofluorescence experiments revealed that mesenchymal-derived skin fibroblasts (including HKFs and HDFs) could express vimentin. To investigate the biological function of Runx2 in HKFs, Runx2 was knocked down by transfection with si-Runx2. The result of western blotting demonstrated that Runx2 expression was markedly decreased after si-Runx2 transfection compared to the si-NC transfected HKFs (Fig. 4F). Compared with si-1 and si-3, si-2 had the most significant silencing efficiency and then was selected for subsequent experiments.
Figure 4.
Upregulation of Runx2 expression levels in keloid tissues and HKFs. (A) Expression levels of Runx2 were analyzed in keloid and normal tissue and HDFs and HKFs using western blotting. (B) Semi-quantification of the expression levels from part (A). ***P<0.001 keloid tissues vs. normal skin tissues (n=3); ***P<0.001 HKFs vs. HDFs (n=3). (C) mRNA expression levels of Runx2 were analyzed using reverse transcription-quantitative PCR. ***P<0.001 keloid tissues vs. normal skin tissues (n=3); ***P<0.001 HKFs vs. HDFs (n=3). (D) Immunofluorescence was used to determine that the expression of Runx2, collagen I, collagen III and fibronectin in the keloid tissues was upregulated compared with normal skin tissues (magnification, ×200). (E) Left panel: Morphology of fibroblasts under the light microscope (magnification, ×40). Primary fibroblasts (including HKFs and HDFs) appeared as long fusiform cells. Right panel: immunofluorescence imaging of vimentin expression (red field, magnification, ×400). (F) Western blotting revealed a significantly downregulation in the expression levels of Runx2 in HKFs transfected with si-Runx2 (si-1, si-2 and si-3) compared with si-NC. The silencing effect of si-2 was much stronger than those of si-1 and si-3 in HKFs. ***P<0.001 vs. si-NC (N=3). Data are presented as the mean ± SD of 3 independent experiments. N, normal skin tissue; K, keloid tissue; HKFs, human keloid fibroblasts; HDFs, human dermal fibroblasts; si, small interfering RNA; NC, negative control; Runx2, Runt-related transcription factor 2.
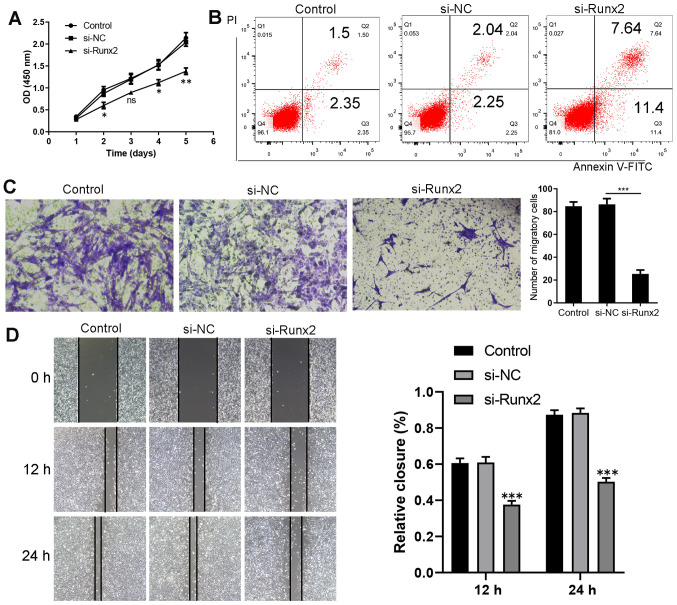
Regulation of proliferation, migration and apoptosis of HKFs following Runx2 knockdown
The effect of Runx2 on HKF proliferation was analyzed using CCK-8 assays. As a consequence of Runx2 knockdown by the transfection with si-Runx2, HKF proliferation was significantly suppressed compared with the si-NC group (Fig. 5A). Using Annexin V//PI staining, the percentage of apoptotic cells was markedly increased after Runx2 expression was knocked down (Fig. 5B). The population of migratory HKFs was significantly decreased following the knockdown of Runx2 compared with the si-NC group (Fig. 5C). A wound healing assay demonstrated that the cell number in the wound of the si-Runx2 group was significantly decreased compared with the si-NC group at 12 and 24 h (Fig. 5D). Taken together, these results suggested that the knockdown of Runx2 in HKFs may inhibit cell proliferation and migration.
Figure 5.
Runx2 knockdown inhibits the migration and proliferation, and promotes the apoptosis of HKFs. (A) Cell Counting Kit-8 assay revealed that the proliferation of HKFs was inhibited following Runx2 knockdown. *P<0.05, **P<0.01 vs. si-NC (n=3). (B) Promotion of apoptosis following the transfection of HKFs with si-Runx2, as shown by flow cytometry using an Annexin V-FITC/PI assay. The lower left quadrant (Annexin V-/PI-) represents live cells, the lower right quadrant (Annexin V+/PI-) represents early apoptotic stage cells, the upper right quadrant (Annexin V+/PI+) represents late apoptotic stage or necrotic cells. (C) ***P<0.001 si-Runx2 vs. si-NC (n=3). (D) Evaluation of cell migration using a wound healing assay (magnification, ×100). 12 h ***P<0.001 si-Runx2 vs. si-NC (n=3); 24 h ***P<0.001 si-Runx2 vs. si-NC (n=3). Data are presented as the mean ± SD of three independent experiments. HKFs, human keloid fibroblasts; si, small interfering RNA; NC, negative control; Runx2, Runt-related transcription factor 2; PI, propidium iodide; OD, optical density.
Downregulation of ECM protein expression levels due to Runx2 knockdown in HKFs
Keloids are characterized by the presence of large numbers of myofibroblasts and the excessive deposition of ECM components, such as collagen I and collagen III (31). The transfection of HKFs with si-Runx2 resulted in the significant downregulation of α-SMA, collagen I, collagen III and fibronectin expression levels at both the mRNA and protein level compared with the si-NC group (Fig. 6A and B).
Figure 6.
Regulation of ECM deposition through the PI3K/AKT signaling pathway. (A) Reverse transcription-quantitative PCR was used to demonstrate that the expression levels of α-SMA, collagen I, collagen III and fibronectin were downregulated in HKFs following transfection. α-SMA, ***P<0.001 si-Runx2 vs. si-NC (n=3); collagen I, ***P<0.001si-Runx2 vs. si-NC (n=3); collagen III, **P<0.01 si-Runx2 vs. si-NC (n=3); fibronectin, **P<0.01 si-Runx2 vs. si-NC (n=3). (B) Western blotting was used to demonstrate that the expression levels of α-SMA, collagen I, collagen III and fibronectin were downregulated in HKFs following transfection. α-SMA, ***P<0.001 si-Runx2 vs. si-NC (n=3); collagen I, ***P<0.001 si-Runx2 vs. si-NC (n=3); collagen III, ***P<0.001 si-Runx2 vs. si-NC (n=3); fibronectin, **P<0.01 vs. si-NC (n=3). (C) Expression levels of AKT, p-AKT, PI3K and p-PI3K were analyzed using western blotting. Statistical analysis was based on the ratio between the levels of phosphorylated protein and total protein. Following normalization to GAPDH, no difference was observed in the PI3K and AKT total protein expression levels in the si-Runx2 group compared with the si-NC group. ***P<0.001, p-AKT vs. AKT (n=3);***P<0.001, p-PI3K vs. PI3K (n=3). The relative protein expression levels were semi-quantified using ImageJ software. Data are presented as the mean ± SD of 3 independent experiments. α-SMA, α-smooth muscle actin; si, small interfering RNA; NC, negative control; Runx2, Runt-related transcription factor 2; p-, phosphorylated.
Knockdown of Runx2 suppresses the phosphorylation of AKT and PI3K in HKFs
KEGG signaling pathway enrichment analyses revealed that the genes that served an important role in the pathology of keloids were mainly enriched in the PI3K/AKT signaling pathway (Fig. 3D). Therefore, to investigate the effect of Runx2 knockdown on the PI3K/AKT signaling pathway, the expression levels of AKT, p-AKT, PI3K and p-PI3K were analyzed using western blotting. The results revealed that the expression levels of p-AKT and p-PI3K were significantly downregulated in HKFs following Runx2 knockdown compared with the si-NC group (Fig. 6C).
Discussion
Keloids are a type of benign mass that occur in different shapes, which are characterized by a reddish protuberance of the skin and a tough texture (32). Patients with keloids often experience pain and itching as the main symptoms and often suffer from infections due to scratching or physical friction (33). Although keloids are benign masses and do not metastasize, they sometimes seriously impact the confidence of an individual with their appearance, which subsequently affects the life quality (8). However, there is currently no effective treatment for improving keloids prognosis and reducing recurrence (34). At present, it is known that genetics, gene mutation, inflammatory cytokines and autoimmunity are associated with keloid formation, but the specific pathogenesis remains unclear (35). Therefore, further investigations to determine the process of keloid formation using HKFs and to identify effective treatments have become necessary for plastic surgery clinicians and researchers. HKFs are the main effector cells involved in the development of keloids. The proliferation and migration of HKFs was discovered to be closely related to the formation of keloids (36). Compared with normal skin tissues, HKFs were reported to secrete more ECM components, such as collagen I, collagen III and mucin. Simultaneously, HKFs can also secrete related cytokines, including TGF-β and platelet-derived growth factor to promote keloid hyperplasia (37). Therefore, an in-depth understanding of the molecular mechanisms that regulate HKF function may provide novel therapeutic strategies for keloids.
In the present study, GO and KEGG pathway analysis were applied to investigate the possible biological functions and potential mechanisms of DEGs in keloid formation. GO analysis revealed that most of the DEGs (including Runx2) were significantly enriched in the BP ‘Positive regulation of cell proliferation’, in the CC ‘Extracellular matrix’, in the MF ‘Extracellular matrix structural constituents’. Previous studies have confirmed that the proliferation of fibroblasts and the excessive deposition of ECM served an indispensable role in the pathological process of keloids (3,31,36), which is consistent with the GO enrichment analysis. KEGG analysis of these DEGs (including Runx2) proposed that the ‘PI3K-Akt signaling pathway’, ‘TGF-β signaling pathway’, ‘ECM-receptor interaction’ and ‘Focal adhesion’ might serve a pathological role in keloid formation. The abovementioned findings revealed that the identified hub genes and pathways using GO and KEGG analysis could significantly enrich our understanding of the development of keloids and provide the potential mechanisms in keloid formation.
Runx2 has been discovered to be closely associated with the differentiation of mesenchymal cells to osteoblast lineage cells and is essential for cell proliferation and ECM formation (38). Mümmler et al (14) reported that Runx2 expression levels were upregulated in ATII cells isolated from fibrotic mouse lungs, and Runx2 knockdown using siRNA decreased the migratory ability of A549 cells. Herreño et al (15) demonstrated that Runx2 promoted epithelial-mesenchymal transition and increased the migratory capacity in lung adenocarcinoma cells. In the present study, the results revealed that the expression levels of Runx2 in HKFs and keloid tissues were significantly upregulated compared with HDFs and normal skin tissues. Runx2 knockdown with siRNA inhibited HKF proliferation, migration, and the deposition of the ECM (including collagen I, collagen III, and fibronectin). Furthermore, the apoptotic cell population in HKFs was markedly increased following Runx2 knockdown. Consequently, this may cause HKFs to lose their highly proliferative and migratory nature, and the excessive deposition of the ECM, which may thereby result in the delayed progression of keloids, indicating the possibility of Runx2 as a potential target for the treatment of keloids.
Previously, it was demonstrated that the PI3K/AKT signaling pathway was involved in the regulation of cell proliferation, differentiation, apoptosis and glucose transport (39). In the current study, KEGG signaling pathway enrichment analysis revealed that the DEGs in keloids were mainly enriched in the PI3K/AKT signaling pathway. It was also discovered that the phosphorylation levels of PI3K and AKT significantly decreased following the transfection with si-Runx2, which indicated that the PI3K/AKT signaling pathway was involved in HFK functions.
The protein mTOR consists of the serine/threonine kinase TOR and at ≥5 other proteins. mTOR participates in several signaling transduction networks, especially the PI3K/AKT signaling pathway, to regulate cell cycle progression, cell survival, proliferation, invasion and apoptosis (40). Research has revealed that mTOR is a key component of the PI3K/AKT/mTOR signaling pathway, and it is considered as an attractive therapeutic target in breast cancer (41). In addition, mTOR was reportedly involved in regulating the pathogenesis of autoimmune diseases and viral infections, such as human immunodeficiency virus (HIV) and SARS-CoV-2 (42). By analyzing the clinical data and measuring the plasma levels of p-PI3K, p-AKT and p-mTOR using ELISAs, Ge et al (43) found that the PI3K/AKT/mTOR signaling pathway was closely associated with the pathogenesis of lupus nephritis. In addition, to determine possible novel therapeutic targets, Fagone et al (44) applied the anti-signature perturbation analysis to predict mTOR as a potential target to delay the progression of Covid-19 through analyzing transcriptomic profiles of primary human lung epithelium following SARS-CoV-2 infection. Nicoletti et al (45) also identified a relationship between dysregulated mTOR activation and the pathogenesis of HIV, which suggested that mTOR might be a multifunctional therapeutic target in HIV infection. Interestingly, Tu et al (46) discovered that CUDC-907, which is a PI3K/Akt/mTOR pathway inhibitor, could significantly inhibit the proliferation, migration, invasion and ECM deposition of HKFs. Therefore, based on these aforementioned findings, it was hypothesized that inhibiting the PI3K/AKT/mTOR signaling pathway may reverse the pathological phenotype of HKFs.
Gene therapy is a recently developed and novel approach for treatment, which has demonstrated tremendous potential (47). For example, the feasibility of gene therapy applications of a retroviral vector expressing LAMB3 cDNA has been definitely confirmed by the successful results obtained in a clinical trial for the LAMB3-deficient form of junctional epidermolysis bullosa (48). RNA interference is a universal mechanism of post-transcriptional gene silencing induced by siRNAs, which are double-stranded non-coding RNAs of 21–22 nucleotides in length generated from longer double strand RNA by the action of Dicer (49). siRNA is able to prevent target gene expression by inducing the degradation of specific mRNA (50). In the present study, Runx2 siRNA was designed to knockdown Runx2 mRNA and transfected into HKFs. The results revealed that the biological functions (including proliferation, migration and ECM deposition) of HKFs were significantly inhibited by si-Runx2 transfection, suggesting that siRNA treatment could be used for the treatment of keloids.
Nonetheless, there are several limitations to the current study. Firstly, mTOR is a key component of the PI3K/AKT/mTOR signaling pathway and is considered as an attractive therapeutic target in the pathological process of keloids (46). However, the present study only investigated the PI3K and AKT phosphorylation levels and did not further confirm the potential mechanism of mTOR in regulating the progression of keloids. In addition, relatively few patients with keloids need to undergo surgical resection. Therefore, it is difficult to collect a large number of samples, which is a general limitation of keloid research. For this reason, the sample number (9 pairs) in the present study is small, and further large-scale analysis will be required in the future.
In conclusion, the results of the current study indicated that Runx2 expression levels may be upregulated in keloid tissues. Silencing Runx2 in HKFs inhibited the cell proliferation, migration, expression levels of ECM-related proteins through the suppression of the PI3K/AKT signaling pathway. Meanwhile, we found that silencing Runx2 inhibited the expression of α-SMA in HKFs and α-SMA was a marker of myofibroblasts, indicating that silencing Runx2 inhibited the differentiation into myofibroblasts. Therefore, Runx2 may be a potential therapeutic target for keloids and could be applied in the clinic as a novel therapeutic strategy.
Supplementary Material
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
WL, MW and YR designed and performed the experiments, and contributed to the writing of the manuscript. QZ and YW made substantial contributions to the conception and design of the present study, analysis and interpretation of data. XL and WH were involved in revising the manuscript critically for important intellectual content and the acquisition of data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the ethics committee of Tongji Hospital of Huazhong University of Science and Technology. Written, informed consent was obtained from all participants. The collection and use of samples were in accordance with the Declaration of Helsinki principles.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Ud-Din S, Bayat A. New insights on keloids, hypertrophic scars, and striae. Dermatol Clin. 2014;32:193–209. doi: 10.1016/j.det.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 2.de Oliveira GV, Gold MH. Hydrocolloid dressings can be used to treat hypertrophic scars: An outpatient dermatology service devoted to treat keloids and challenging scars. J Cosmet Dermatol. 2020 Oct 26; doi: 10.1111/jocd.13792. (Epub ahead of print). doi: 10.1111/jocd.13792 2020. [DOI] [PubMed] [Google Scholar]
- 3.Li Y, Liu H, Liang Y, Peng P, Ma X, Zhang X. DKK3 regulates cell proliferation, apoptosis and collagen synthesis in keloid fibroblasts via TGF-β1/Smad signaling pathway. Biomed Pharmacother. 2017;91:174–180. doi: 10.1016/j.biopha.2017.03.044. [DOI] [PubMed] [Google Scholar]
- 4.Lee JY, Yang CC, Chao SC, Wong TW. Histopathological differential diagnosis of keloid and hypertrophic scar. Am J Dermatopathol. 2004;26:379–384. doi: 10.1097/00000372-200410000-00006. [DOI] [PubMed] [Google Scholar]
- 5.Lee YS, Liang YC, Wu P, Kulber DA, Tanabe K, Chuong CM, Widelitz R, Tuan TL. STAT3 signalling pathway is implicated in keloid pathogenesis by preliminary transcriptome and open chromatin analyses. Exp Dermatol. 2019;28:480–484. doi: 10.1111/exd.13923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Q, Cai L, Wang M, Ke X, Zhao X, Huang Y. Identification of a novel mutation in the mechanoreceptor-encoding gene CXCR1 in patients with keloid. Arch Dermatol Res. 2018;310:561–566. doi: 10.1007/s00403-018-1847-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu J, Zhu H, Wang H, Li J, Han F, Liu Y, Zhang W, He T, Li N, Zheng Z, et al. Methylation of secreted frizzled-related protein 1 (SFRP1) promoter downregulates Wnt/β-catenin activity in keloids. J Mol Histol. 2018;49:185–193. doi: 10.1007/s10735-018-9758-3. [DOI] [PubMed] [Google Scholar]
- 8.Bijlard E, Kouwenberg CA, Timman R, Hovius SE, Busschbach JJ, Mureau MA. Burden of Keloid Disease: A Cross-sectional Health-related Quality of Life Assessment. Acta Derm Venereol. 2017;97:225–229. doi: 10.2340/00015555-2498. [DOI] [PubMed] [Google Scholar]
- 9.Chen L, Zhang YH, Wang S, Zhang Y, Huang T, Cai YD. Prediction and analysis of essential genes using the enrichments of gene ontology and KEGG pathways. PLoS One. 2017;12:e0184129. doi: 10.1371/journal.pone.0184129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu W, Huang X, Liang X, Zhou Y, Li H, Yu Q, Li Q. Identification of Key Modules and Hub Genes of Keloids with Weighted Gene Coexpression Network Analysis. Plast Reconstr Surg. 2017;139:376–390. doi: 10.1097/PRS.0000000000003014. [DOI] [PubMed] [Google Scholar]
- 11.Fagone P, Mazzon E, Cavalli E, Bramanti A, Petralia MC, Mangano K, Al-Abed Y, Bramati P, Nicoletti F. Contribution of the macrophage migration inhibitory factor superfamily of cytokines in the pathogenesis of preclinical and human multiple sclerosis: In silico and in vivo evidences. J Neuroimmunol. 2018;322:46–56. doi: 10.1016/j.jneuroim.2018.06.009. [DOI] [PubMed] [Google Scholar]
- 12.Presti M, Mazzon E, Basile MS, Petralia MC, Bramanti A, Colletti G, Bramanti P, Nicoletti F, Fagone P. Overexpression of macrophage migration inhibitory factor and functionally-related genes, D-DT, CD74, CD44, CXCR2 and CXCR4, in glioblastoma. Oncol Lett. 2018;16:2881–2886. doi: 10.3892/ol.2018.8990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mevel R, Draper JE, Lie ALM, Kouskoff V, Lacaud G. RUNX transcription factors: orchestrators of development. Development. 2019;146:dev148296. doi: 10.1242/dev.148296. [DOI] [PubMed] [Google Scholar]
- 14.Mümmler C, Burgy O, Hermann S, Mutze K, Günther A, Königshoff M. Cell-specific expression of runt-related transcription factor 2 contributes to pulmonary fibrosis. FASEB J. 2018;32:703–716. doi: 10.1096/fj.201700482R. [DOI] [PubMed] [Google Scholar]
- 15.Herreño AM, Ramírez AC, Chaparro VP, Fernandez MJ, Cañas A, Morantes CF, Moreno OM, Brugés RE, Mejía JA, Bustos FJ, et al. Role of RUNX2 transcription factor in epithelial mesenchymal transition in non-small cell lung cancer lung cancer: Epigenetic control of the RUNX2 P1 promoter. Tumour Biol. 2019;41 doi: 10.1177/1010428319851014. 1010428319851014. [DOI] [PubMed] [Google Scholar]
- 16.Fagone P, Mangano K, Pesce A, Portale TR, Puleo S, Nicoletti F. Emerging therapeutic targets for the treatment of hepatic fibrosis. Drug Discov Today. 2016;21:369–375. doi: 10.1016/j.drudis.2015.10.015. [DOI] [PubMed] [Google Scholar]
- 17.Fagone P, Mangano K, Mammana S, Pesce A, Pesce A, Caltabiano R, Giorlandino A, Portale TR, Cavalli E, Lombardo GA, et al. Identification of novel targets for the diagnosis and treatment of liver fibrosis. Int J Mol Med. 2015;36:747–752. doi: 10.3892/ijmm.2015.2264. [DOI] [PubMed] [Google Scholar]
- 18.Chen J, Lin Y, Sun Z. Deficiency in the anti-aging gene Klotho promotes aortic valve fibrosis through AMPKα-mediated activation of RUNX2. Aging Cell. 2016;15:853–860. doi: 10.1111/acel.12494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tchetina EV, Demidova NV, Markova GA, Taskina EA, Glukhova SI, Karateev DE. Increased baseline RUNX2, caspase 3 and p21 gene expressions in the peripheral blood of disease-modifying anti-rheumatic drug-naïve rheumatoid arthritis patients are associated with improved clinical response to methotrexate therapy. Int J Rheum Dis. 2017;20:1468–1480. doi: 10.1111/1756-185X.13131. [DOI] [PubMed] [Google Scholar]
- 20.Hahn JM, Glaser K, McFarland KL, Aronow BJ, Boyce ST, Supp DM. Keloid-derived keratinocytes exhibit an abnormal gene expression profile consistent with a distinct causal role in keloid pathology. Wound Repair Regen. 2013;21:530–544. doi: 10.1111/wrr.12060. [DOI] [PubMed] [Google Scholar]
- 21.Chen YT, Yao JN, Qin YT, Hu K, Wu F, Fang YY. Biological role and clinical value of miR-99a-5p in head and neck squamous cell carcinoma (HNSCC): A bioinformatics-based study. FEBS Open Bio. 2018;8:1280–1298. doi: 10.1002/2211-5463.12478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang SU, Kim YS, Kim YE, Park JK, Lee YS, Kang HY, Jang JW, Ryeo JB, Lee Y, Shin YS, et al. Opposite effects of non-thermal plasma on cell migration and collagen production in keloid and normal fibroblasts. PLoS One. 2017;12:e0187978. doi: 10.1371/journal.pone.0187978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garcia-Morales V, Friedrich J, Jorna LM, Campos-Toimil M, Hammes HP, Schmidt M, Krenning G. The microRNA-7-mediated reduction in EPAC-1 contributes to vascular endothelial permeability and eNOS uncoupling in murine experimental retinopathy. Acta Diabetol. 2017;54:581–591. doi: 10.1007/s00592-017-0985-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fang B, Liu Y, Zheng D, Shan S, Wang C, Gao Y, Wang J, Xie Y, Zhang Y, Li Q. The effects of mechanical stretch on the biological characteristics of human adipose-derived stem cells. J Cell Mol Med. 2019;23:4244–4255. doi: 10.1111/jcmm.14314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Zhang M, Zeng J, Zhao Z, Liu Z. Loss of MiR-424-3p, not miR-424-5p, confers chemoresistance through targeting YAP1 in non-small cell lung cancer. Mol Carcinog. 2017;56:821–832. doi: 10.1002/mc.22536. [DOI] [PubMed] [Google Scholar]
- 27.Rotgers E, Nurmio M, Pietilä E, Cisneros-Montalvo S, Toppari J. E2F1 controls germ cell apoptosis during the first wave of spermatogenesis. Andrology. 2015;3:1000–1014. doi: 10.1111/andr.12090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gene Ontology C, Gene Ontology Consortium Gene Ontology Consortium: Going forward. Nucleic Acids Res. 2015;43D:D1049–D1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45D1:D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cavalli E, Battaglia G, Basile MS, Bruno V, Petralia MC, Lombardo SD, Pennisi M, Kalfin R, Tancheva L, Fagone P, et al. Exploratory Analysis of iPSCS-Derived Neuronal Cells as Predictors of Diagnosis and Treatment of Alzheimer Disease. Brain Sci. 2020;10:166. doi: 10.3390/brainsci10030166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, Cheng C, Wang S, Xu M, Zhang D, Zeng W. Knockdown of FOXM1 inhibits activation of keloid fibroblasts and extracellular matrix production via inhibition of TGF-β1/Smad pathway. Life Sci. 2019;232:116637. doi: 10.1016/j.lfs.2019.116637. [DOI] [PubMed] [Google Scholar]
- 32.Berman B, Maderal A, Raphael B. Keloids and Hypertrophic Scars: Pathophysiology, Classification, and Treatment. Dermatol Surg. 2017;43(Suppl 1):S3–S18. doi: 10.1097/DSS.0000000000000819. [DOI] [PubMed] [Google Scholar]
- 33.Bock O, Schmid-Ott G, Malewski P, Mrowietz U. Quality of life of patients with keloid and hypertrophic scarring. Arch Dermatol Res. 2006;297:433–438. doi: 10.1007/s00403-006-0651-7. [DOI] [PubMed] [Google Scholar]
- 34.Liu Q, Wang X, Jia Y. Heat Shock Protein 90 Inhibitor Decreases Collagen Synthesis of Keloid Fibroblasts and Attenuates the Extracellular Matrix on the Keloid Spheroid Model. Plast Reconstr Surg. 2016;137:759e–760e. doi: 10.1097/PRS.0000000000002039. [DOI] [PubMed] [Google Scholar]
- 35.Philandrianos C, Kerfant N, Jaloux C, Jr, Martinet L, Bertrand B, Casanova D. Keloid scars (part I): Clinical presentation, epidemiology, histology and pathogenesis. Ann Chir Plast Esthet. 2016;61:128–135. doi: 10.1016/j.anplas.2015.09.005. (In French) [DOI] [PubMed] [Google Scholar]
- 36.Chen J, Liu K, Liu Y, Wang X, Zhang Z. Targeting mTORC1/2 with OSI-027 inhibits proliferation and migration of keloid keratinocytes. Exp Dermatol. 2019;28:270–275. doi: 10.1111/exd.13882. [DOI] [PubMed] [Google Scholar]
- 37.Ogawa R. Keloid and Hypertrophic Scars Are the Result of Chronic Inflammation in the Reticular Dermis. Int J Mol Sci. 2017;18:606. doi: 10.3390/ijms18030606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Komori T. Regulation of Proliferation, Differentiation and Functions of Osteoblasts by Runx2. Int J Mol Sci. 2019;20:1694. doi: 10.3390/ijms20071694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li G, Li YY, Sun JE, Lin WH, Zhou RX. ILK-PI3K/AKT pathway participates in cutaneous wound contraction by regulating fibroblast migration and differentiation to myofibroblast. Lab Invest. 2016;96:741–751. doi: 10.1038/labinvest.2016.48. [DOI] [PubMed] [Google Scholar]
- 40.Evangelisti C, Evangelisti C, Chiarini F, Lonetti A, Buontempo F, Bressanin D, Cappellini A, Orsini E, McCubrey JA, Martelli AM. Therapeutic potential of targeting mTOR in T-cell acute lymphoblastic leukemia (review) Int J Oncol. 2014;45:909–918. doi: 10.3892/ijo.2014.2525. [DOI] [PubMed] [Google Scholar]
- 41.Sharma VR, Gupta GK, Sharma AK, Batra N, Sharma DK, Joshi A, Sharma AK. PI3K/Akt/mTOR Intracellular Pathway and Breast Cancer: Factors, Mechanism and Regulation. Curr Pharm Des. 2017;23:1633–1638. doi: 10.2174/1381612823666161116125218. [DOI] [PubMed] [Google Scholar]
- 42.Steelman LS, Martelli AM, Cocco L, Libra M, Nicoletti F, Abrams SL, McCubrey JA. The therapeutic potential of mTOR inhibitors in breast cancer. Br J Clin Pharmacol. 2016;82:1189–1212. doi: 10.1111/bcp.12958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ge F, Wang F, Yan X, Li Z, Wang X. Association of BAFF with PI3K/Akt/mTOR signaling in lupus nephritis. Mol Med Rep. 2017;16:5793–5798. doi: 10.3892/mmr.2017.7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fagone P, Ciurleo R, Lombardo SD, Iacobello C, Palermo CI, Shoenfeld Y, Bendtzen K, Bramanti P, Nicoletti F. Transcriptional landscape of SARS-CoV-2 infection dismantles pathogenic pathways activated by the virus, proposes unique sex-specific differences and predicts tailored therapeutic strategies. Autoimmun Rev. 2020;19:102571. doi: 10.1016/j.autrev.2020.102571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nicoletti F, Fagone P, Meroni P, McCubrey J, Bendtzen K. mTOR as a multifunctional therapeutic target in HIV infection. Drug Discov Today. 2011;16:715–721. doi: 10.1016/j.drudis.2011.05.008. [DOI] [PubMed] [Google Scholar]
- 46.Tu T, Huang J, Lin M, Gao Z, Wu X, Zhang W, Zhou G, Wang W, Liu W. CUDC 907 reverses pathological phenotype of keloid fibroblasts in vitro and in vivo via dual inhibition of PI3K/Akt/mTOR signaling and HDAC2. Int J Mol Med. 2019;44:1789–1800. doi: 10.3892/ijmm.2019.4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aragona M, Blanpain C. Gene therapy: Transgenic stem cells replace skin. Nature. 2017;551:306–307. doi: 10.1038/nature24753. [DOI] [PubMed] [Google Scholar]
- 48.Mavilio F, Pellegrini G, Ferrari S, Di Nunzio F, Di Iorio E, Recchia A, Maruggi G, Ferrari G, Provasi E, Bonini C, et al. Correction of junctional epidermolysis bullosa by transplantation of genetically modified epidermal stem cells. Nat Med. 2006;12:1397–1402. doi: 10.1038/nm1504. [DOI] [PubMed] [Google Scholar]
- 49.Wang W, Qu M, Xu L, Wu X, Gao Z, Gu T, Zhang W, Ding X, Liu W, Chen YL. Sorafenib exerts an anti-keloid activity by antagonizing TGF-β/Smad and MAPK/ERK signaling pathways. J Mol Med (Berl) 2016;94:1181–1194. doi: 10.3892/ijmm.2016.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tian Y, Jin L, Zhang W, Ya Z, Cheng Y, Zhao H. AMF siRNA treatment of keloid through inhibition signaling pathway of RhoA/ROCK1. Genes Dis. 2018;6:185–192. doi: 10.1016/j.gendis.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.