Abstract
MicroRNA (miR)-140-5p is associated with the growth and metastasis of various tumor cell types, yet its role in multiple myeloma (MM) remains unclear. Therefore, the present study aimed to investigate the regulatory effect of miR-140-5p on MM. Reverse transcription-quantitative PCR analysis demonstrated that miR-140-5p was downregulated in MM cell lines, particularly in U266 and RPMI 8226 cells. A Cell Counting Kit-8, wound healing and Transwell assays, as well as flow cytometry indicated that a miR-140-5p mimic could suppress cell viability, migration and invasion. In addition, the mimic promoted apoptosis of U266 and RPMI 8226 cells. Western blot data demonstrated that transfection with miR-140-5p mimic significantly reduced the expression levels of Ki-67, cyclin D1, vimentin, Snail, matrix metalloproteinase (MMP)-2 and MMP-3. Moreover, as predicted by TargetScan7.2 and verified by luciferase activity assay, it was demonstrated that vascular endothelial growth factor A (VEGFA) was targeted by miR-140-5p. Further experiments indicated that VEGFA overexpression promoted cell viability, migration and invasion and suppressed apoptosis of MM cells, and that the miR-140-5p mimic partially reversed the effects of VEGFA overexpression. Therefore, miR-140-5p suppressed MM progression by targeting VEGFA. The present findings provide insight into potential therapeutic strategies for the treatment of MM.
Keywords: miR-140-5p, multiple myeloma, VEGFA, proliferation, migration, invasion
Introduction
Multiple myeloma (MM) is a type of hematological malignancy characterized by malignant clonal hyperplasia of plasma cells in the bone marrow, often resulting in susceptibility to infection, impaired renal function, anemia and hypercalcemia (1). According to research statistics, the incidence of MM is the second highest to non-Hodgkin's lymphoma (2), and the incidence of MM accounts for ~13% of all hematological tumors (3). With the development of molecular biology technology, progress has been made in broadening the understanding of MM pathogenesis and in the development of new drugs (4). In particular, gene-targeted therapy has long been the focus of the study and treatment of cancer (5). However, there are no curative therapies for MM. It is therefore urgent to develop new and more effective treatment methods (6,7). Therefore, further understanding of the molecular mechanism underlying MM progression may provide new insight into the development of novel therapeutic strategies.
MicroRNAs (miRNAs) are non-coding small RNAs ~22 nucleosides in length, which serve important roles in the regulation of gene expression (8). miRNAs participate in regulation of gene expression by directly binding to the 3′-untranslated region (UTR) of specific target genes, thereby affecting cell differentiation, development, apoptosis, proliferation and other biological activities (9–11). Recent evidence has indicated that miRNAs are dysregulated in several types of tumor and could modulate tumor progression (12,13). For instance, previous studies demonstrated that miR-450a inhibited the metastasis of ovarian cancer cells (14), and that miR-365-3p could promote metastasis, cancer stem cell-like properties and drug resistance in oral squamous cells (15). Moreover, miR-140-5p is considered a tumor suppressor for various types of tumor, including in retinoblastoma and ovarian cancer (16–18). Another previous study also indicated that miR-140-5p upregulation could inhibit autophagy and chemoresistance of myeloma cells (16). However, whether miR-140-5p regulates the growth and metastasis of MM remains unknown.
Vascular endothelial growth factor A (VEGFA) is a key regulator of tumor growth. As a result, VEGFA represents a potential therapeutic target in cancer treatment (19). Integrated genomic analysis indicated that VEGFA was commonly overexpressed in adenocarcinoma (20). Notably, VEGFA expression is correlated with the survival of patients with MM; for instance patients whose MM cells express low levels of VEGFA have the longest survival (21). VEGFA is the target of multiple miRNAs, such as miR-29c in lung adenocarcinoma (22), and miR-199a-5p in endometrial mesenchymal stem cells (23). A previous study demonstrated that miR-16 targeted a large number of downstream target genes, including VEGFA in MM (24). However, whether miR-140-5p can also target VEGFA in MM is unclear.
Therefore, the aim of the present study was to explore the function served by miR-140-5p in MM and to determine whether the potential tumor suppressive effects of miR-140-5p in MM are mediated through VEGFA.
Materials and methods
Cell culture
Human normal plasma cells were separated from the peripheral blood of three men and three women with a median age of 47 years using flow cytometry (25). All donors were healthy volunteers who had not previously received any drugs associated with immunological diseases. The blood (2 ml/person) was collected at the Jingzhou Central Hospital between 2016.03 and 2017.03. Informed consent was provided by all volunteers, and the present study was approved by the Ethics Committee of Jingzhou Central Hospital (approval no. JCH20150608EZ023). Briefly, peripheral blood mononuclear cells were separated by Ficoll®-Hypaque centrifugation from peripheral blood. Plasma cells were isolated from mononuclear blood cells using CD138 microbeads (Miltenyi Biotec, Inc.) according to the manufacturer's instructions (26). The isolated plasma cells were then cultured. The RPMI 8226, OPM2, U266 (TIB-196), H929, MM1.S and KMS11 MM cell lines were purchased from the American Type Culture Collection. RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.) containing 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.) and 1% dual antibiotics (penicillin 100 U/ml; streptomycin 0.1 mg/m1; Sigma-Aldrich; Merck KGaA) was used to culture all cells at 37°C with 5% CO2.
Transfection
The RPMI 8226 and U266 cell lines were used for transfection experiments, since these two cell lines displayed low miR-140-5p expression. Cells were seeded on a 6-well plate at a density of 1×106 cells/ml and cultured overnight at 37°C. The next day, cells were transfected with miR-140-5p mimic or a VEGFA overexpression plasmid. A total of 100 pmol of the miR-140-5p mimic (sense: 5′-ACCAUAGGGUAAAACCACUGUU-3′), mimic control (5′-UUGUACUACACAAAAGUACUG-3′), VEGFA overexpression plasmid (VEGFA-pcDNA3.1) and VEGFA negative control (NC) were synthesized by Sangon Biotech Co., Ltd.. The miR-140-5p mimic, VEGFA-pcDNA3.1 plasmid or their respective controls (100 pmol), were then dissolved in 50 µl Opti-MEM (HyClone; GE Healthcare Life Sciences). In addition, 1 µl Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) transfection reagent was then separately diluted in 50 µl DMEM, then incubated for 5 min at room temperature. After incubation, the nucleic acid was mixed with the diluted Lipofectamine® and the mixture was added to the cells. Cells were cultured at 37°C with 5% CO2 and the medium was replaced after 24 h. Then, 72 h after changing the medium, the cells were harvested for subsequent experiments.
Luciferase activity assay
The VEGFA 3′-UTR containing putative miR-140-5p binding sites was inserted into a pmirGLO dual luciferase vector (Promega Corporation) to generate wild-type (WT) pmirGLO-VEGFA. The mutant (MUT) 3′-UTR of VEGFA was inserted into the pmirGLO luciferase reporter vector at the XhoI/XbaI sites. RPMI 8226 and U266 cells were co-transfected with pmirGLO vector containing the WT or MUT VEGFA 3′-UTR and miR-140-5p mimic using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). After incubation for 48 h (27), relative luciferase activity in the cells was assessed using the Dual-Luciferase Reporter Assay kit (Promega Corporation), according to the manufacturer's protocol. The Renilla luciferase activity was used for reference.
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The concentration of RNA was measured using a NanoDrop™ spectrophotometer (Thermo Fisher Scientific, Inc.), then diluted to 500 ng/µl. For reverse transcription, 1 µg total RNA was converted into cDNA using the Superscript II First-Strand cDNA Synthesis kit (Invitrogen; Thermo Fisher Scientific, Inc.). The mRNA expression levels were determined with SYBR-Green PCR Master Mix (Thermo Fisher Scientific, Inc) using the 7500 Real-Time PCR system (Thermo Fisher Scientific, USA). The thermocycling conditions were as follows: i) Initial denaturation at 95°C for 1 min; ii) 40 cycles at 95°C for 30 sec, 58°C for 20 sec, and 70°C for 20 sec; and iii) a final extension at 72°C for 7 min. The samples were then maintained at 4°C. The expression levels in the RT-qPCR products were determined using the 2−ΔΔCq method (28). All primer sequences are listed in Table I.
Table I.
Primers used for reverse transcription-quantitative PCR.
| Target | Primer sequence | Species |
|---|---|---|
| miR-140-5p | Forward, 5′-GAGTGTCAGTGGTTTTACCCT-3′ | |
| Reverse, 5′-GCAGGGTCCGAGGTATTC-3′ | Human | |
| VEGFA | Forward, 5′-GGCCTCCGAAACCATGAACT-3′ | |
| Reverse, 5′-TCGTGATGATTCTGCCCTCC-3′ | Human | |
| U6 | Forward, 5′-TGACTTCCAAGTACCATCGCCA-3′ | |
| Reverse, 5′-TTGTAGAGGTAGGTGTGCAGCAT-3′ | Human | |
| GAPDH | Forward, 5′-GGTGAAGGTCGGAGTCAACG-3′ | |
| Reverse, 5′-CAAAGTTGTCATGGATGTACC-3′ | Human |
miR, microRNA; VEGFA, vascular endothelial growth factor A.
Cell viability detection
Cells were seeded into a 96-well plate (4×104), and 10 µl Cell Counting Kit-8 (CCK-8; cat. no. 96992, Sigma-Aldrich, Merck KGaA) solution was added into each well. Cells were then incubated for 4 h at 37°C. Absorbance was read at 450 nm using a Multiskan GO multiplate reader (Thermo Fisher Scientific, Inc.) Data were recorded as the average of triplicate wells for each condition.
Cell apoptosis
Cells were harvested 24 h after transduction and re-suspended at 1×106 cells/ml in 1X Annexin binding buffer (cat. no. 422201; BioLegend, Inc.). Next, 5 µl of fluorescein isothiocyanate (FITC)-conjugated Annexin V (cat. no. C1062S, Beyotime Institute of Biotechnology) and 1 µl of 100 µg/ml propidium iodide (cat. no. P1304MP; Thermo Fisher Scientific, Inc.) solution in 300 µl 1X Annexin binding buffer were added to the cell suspension for 15 min at room temperature. Stained samples were acquired using a FACSCalibur flow cytometer (BD Biosciences) and analyzed using FlowJo version 10.0 (FlowJo LLC).
Wound healing assay
Transfected cells were plated at a density of 5×105 cells/well on a 6-well plate. Cells were allowed to adhere to the surface of the well for 24 h, then, a uniform-width scratch was quickly made on the cell layer. The suspended cells were removed, and the remainder was cultured at low serum concentration (1%). Cell migration across the scratch was recorded at 0 and 48 h (light microscope; magnification, ×100), and the migration distance was measured using ImageJ software version 1.8.0 (National Institutes of Health). To eliminate the differences in initial width at 0 h among the different experimental groups, a relative migration rate was calculated as (0 h scratch width-48 h scratch width)/0 h scratch width × 100%. In order to ensure that the same location was observed at 0 and 48 h, a cross symbol was drawn at within the scratches and images obtained at both time-points were aligned with these signs.
Transwell Matrigel™ assay
Cells were harvested 24 h after transfection and re-suspended in FBS-free medium at a concentration of 1×106 cells/ml. The cell suspension was then added into the upper chamber of Transwell plates (Corning Inc.) pre-coated with Matrigel™ (Thermo Fisher Scientific, Inc.). Medium containing 10% FBS serum was added into the lower chamber. After a 24-h incubation at 37°C, the migratory cells were stained using 0.5% crystal violet at room temperature for 15 min, while those remaining in the upper chamber were removed using a cotton swab. Invading cells were counted under a light microscope, magnification ×200.
Western blotting
Total protein was extracted from cells using RIPA buffer (cat. no. 9806; Cell Signaling Technology, Inc.) then boiled for 5 min at 100°C for denaturation. Protein concentration was examined using bicinchoninic acid Assay kit (Thermo Fisher Scientific, Inc.). Proteins (20 µg/lane) were then separated using 12% SDS-PAGE, and then transferred to polyvinylidene fluoride membranes (EMD Millipore). The membranes were blocked with 5% milk at room temperature for 1 h. The blots were then probed with primary antibodies against Ki-67 (rabbit; 1:1,000; product code ab16667), cyclin D1 (rabbit; 1:1,000; product code ab16663), vimentin (rabbit; 1:1,000; product code ab193555), Snail (rabbit; 1:1,000; product code ab229701), matrix metalloproteinase (MMP)-2 (rabbit; 1:1,000; product code ab215986), MMP3 (rabbit; 1:1,000; product code ab52915) and GAPDH (rabbit; 1:2,000; product code ab181602) at 4°C overnight. All primary antibodies were obtained from Abcam. The membranes were then washed with PBS three times, then incubated with a horseradish peroxidase-conjugated Goat Anti-Rabbit IgG (H+L) secondary antibody (cat. no. SA00001-2; 1:2,000; ProteinTech Group, Inc.) at 37°C for 2 h. The protein bands were detected using the ECL western blotting kit (cat. no. 93-K820-500, Multi Sciences; http://www.liankebio.com/index.html) and analyzed using ImageJ version 4.7 (National Institutes of Health).
Statistical analysis
Statistical analysis was carried out using GraphPad Prism version 6.01 (GraphPad Software, Inc.). Data are presented as the mean ± SD of at least three independent experiments. Student's t-test was used for pairwise comparisons between the means of continuous variables. Multi-group comparisons were carried out using ANOVA, followed by Bonferroni correction. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-140-5p expression in MM cell lines and its effect on the viability and apoptosis of MM cells
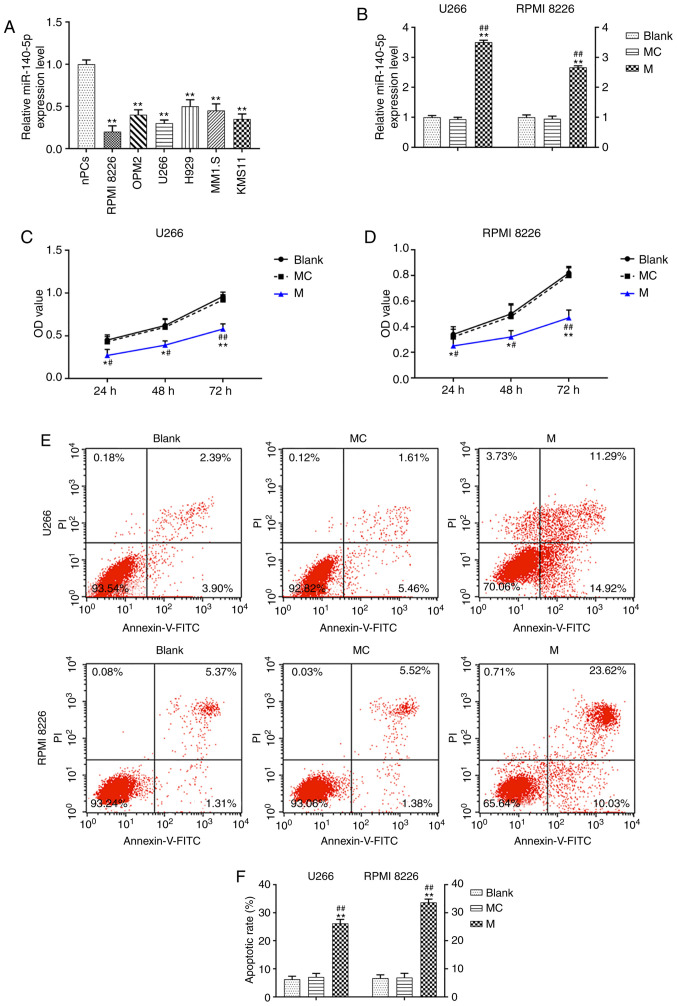
The expression of miR-140-5p in MM cells was significantly lower than in normal plasma cells (P<0.001; Fig. 1A). The U266 and RPMI 8226 cells were transfected with a miR-140-5p mimic, resulting in a significant increase in miR-140-5p expression, compared with the mimic control and blank control cells (P<0.001; Fig. 1B). Moreover, overexpression of miR-140-5p significantly decreased the viability of U266 and RPMI 8226 cells over time (P<0.001; Fig. 1C and D). Transfection with the miR-140-5p mimic also resulted in a significant increase in cell apoptosis (P<0.001; Fig. 1E and F).
Figure 1.
miR-140-5p overexpression affects the viability and apoptosis of MM cells. (A) RT-qPCR was used to detect the expression of miR-140-5p in MM cell lines and nPCs. n=3. **P<0.001, vs. nPCs. (B) Transfection efficiency of miR-140-5p in U266 and RPMI 8226 cell lines was detected by RT-qPCR. U6 was the internal reference. (C and D) Cell Counting Kit-8 was used to evaluate the effect of miR-140-5p transfection on cell viability. (E and F) Cell apoptosis was detected by flow cytometry. n=3. *P<0.05, **P<0.001, vs. Blank; #P<0.05, ##P<0.001, vs. MC. MM, multiple myeloma; miR, microRNA; MC, mimic control; M, mimic; nPCs, normal plasma cells; RT-qPCR, reverse transcription-quantitative PCR; OD, optical density; PI, propidium iodide; FITC, fluorescein isothiocyanate.
Effects of miR-140-5p overexpression on the migration and invasion of MM cells
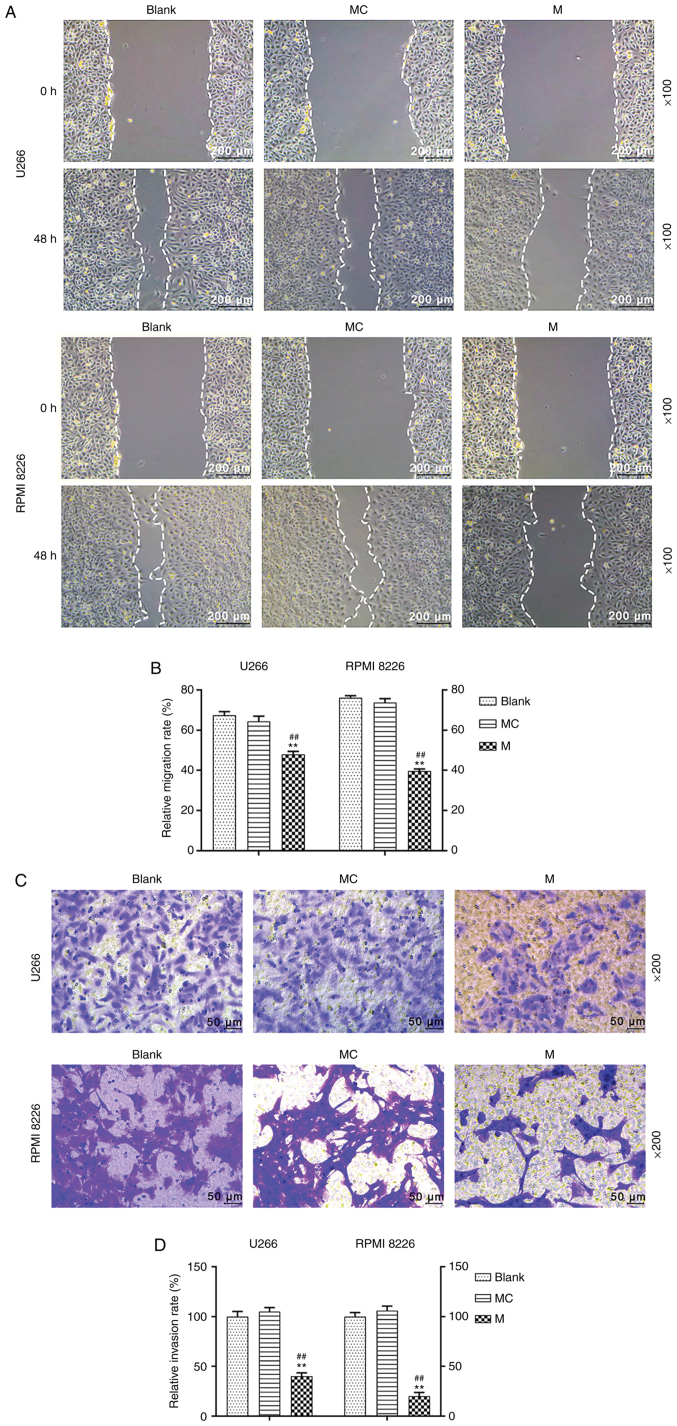
In order to evaluate cell migration in U266 and RPMI 8226 cells following transfection with miR-140-5p, a wound healing assay was carried out. Overexpression of miR-140-5p significantly inhibited cell migration, compared with the mimic control and blank cells (P<0.001; Fig. 2A and B). A Transwell Matrigel™ assay was also performed to assess cell invasion. Transfection with miR-140-5p resulted in significant inhibition of cell invasion, compared with the mimic control and blank cells (P<0.001; Fig. 2C and D).
Figure 2.
Effects of miR-140-5p overexpression on the migration and invasion of MM cells. (A and B) Cell migration was evaluate using a wound healing assay. Scale bar, 200 µm. Magnification, ×100. (C and D) Cell invasion was assessed using a Transwell Matrigel™ assay. Scale bar, 50 µm. Magnification, ×200. n=3. **P<0.001, vs. Blank; ##P<0.001, vs. MC. miR, microRNA; MM, multiple myeloma; MC, mimic control; M, mimic.
Effects of miR-140-5p overexpression on the expression levels of genes related to proliferation, migration and invasion
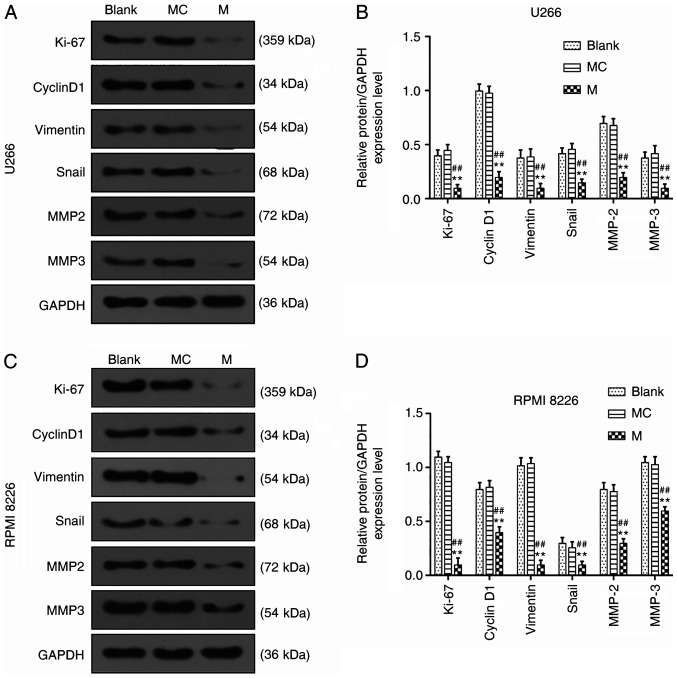
To determine the mechanism of miR-140-5p in MM cells, western blot assays were performed to determine the expression levels of proteins related to the proliferation, migration and invasion of U266 and RPMI 8226 cells. After miR-140-5p mimic had been transfected into the U266 and RPMI 8226 cells, the results revealed that overexpressed miR-140-5p inhibited the expression levels of Ki-67, cyclin D1, vimentin, Snail, MMP2 and MMP3 in U266 cells (P<0.001, Fig. 3A-D).
Figure 3.
Effects of miR-140-5p overexpression on the expression of genes related to proliferation, migration and invasion. (A and B) Western blotting was performed to detect the expression levels of Ki-67, cyclin D1, vimentin, Snail, MMP2 and MMP3 in U266 cells. (C and D) Western blotting was used to detect the expression levels of Ki-67, cyclin D1, vimentin, Snail, MMP2 and MMP3 in RPMI 8226 cells. n=3. **P<0.001, vs. Blank; ##P<0.001, vs. MC. miR, microRNA; MC, mimic control; M, mimic; MMP, matrix metalloproteinase.
VEGFA directly targets miR-140-5p in MM cell lines
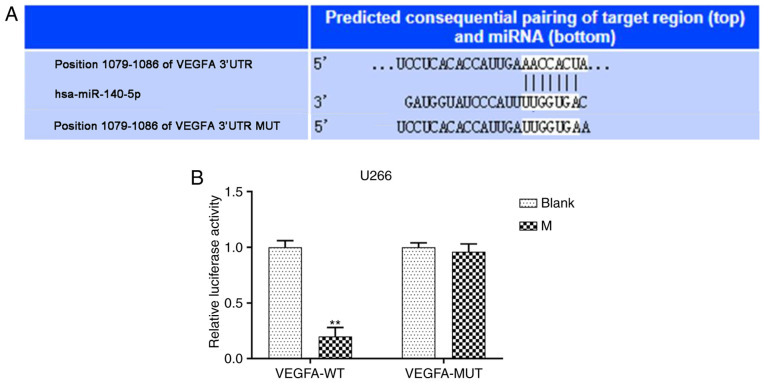
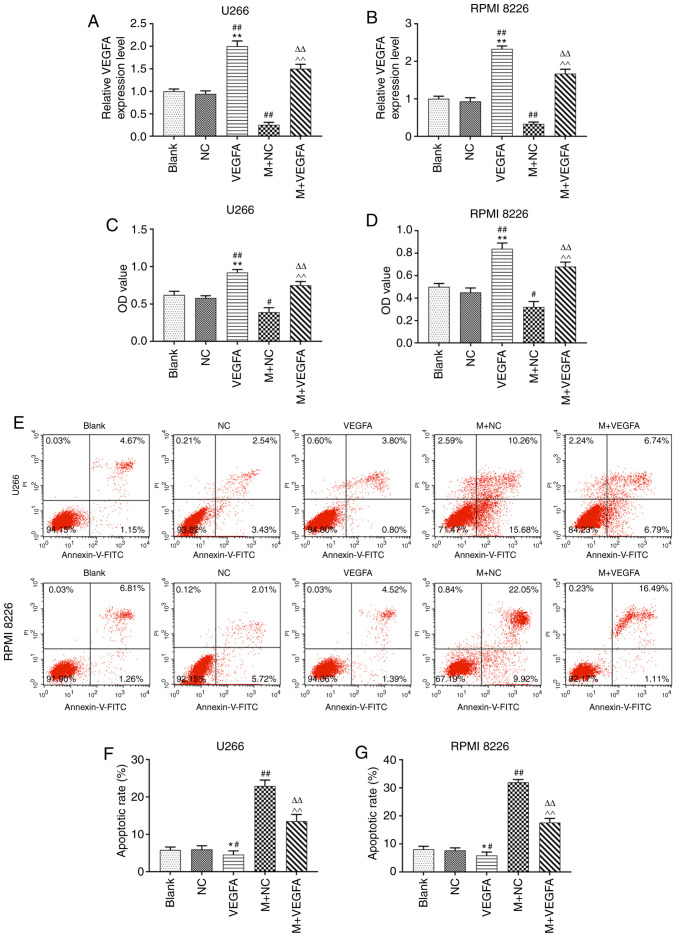
TargetScan software predicted that VEGFA was the potential target gene of miR-140-5p (Fig. 4A). Furthermore, pmirGLO dual luciferase reporter vectors containing VEGFA-WT or VEGFA-MUT 3-UTR sequences were constructed, and co-transfected with miR-140-5p mimic into MM cell lines. Relative luciferase activity following co-transfection with VEGFA-WT and mimic significantly decreased, compared with the blank, while VEGFA-MUT had no effect (P<0.001, Fig. 4B). Moreover, transfection VEGFA-pcDNA3.1 increased VEGFA expression following U266 and RPMI 8226 cell transfection. However, co-transfection of VEGFA-pcDNA3.1 with the miR-140-5p mimic significantly reduced VEGFA expression, compared with transfection with VEGFA-pcDNA3.1 alone (P<0.001; Fig. 5A and B).
Figure 4.
VEGFA directly targets miR-140-5p in MM cell lines. (A) Targetscan7.2 was used to predict the possible target genes for miR-140-5p. (B) Dual luciferase reporter assay was used to detect the fluorescence activity of VEGFA in overexpressed miR-140-5p. n=3. **P<0.001, vs. Blank. VEGFA, vascular endothelial growth factor A; miR/miRNA, microRNA; MM, multiple myeloma; MC, mimic control; M, mimic; UTR, untranslated region; WT, wild-type; MUT, mutant.
Figure 5.
Effects of miR-140-5p and VEGFA the on viability and apoptosis of MM cell lines. (A and B) Reverse transcription-quantitative PCR was used to detect the expression of VEGFA in U266 and RPMI 8226 cells. GAPDH was the internal reference. (C and D) Cell Counting Kit-8 was used to assess the effect of VEGFA and miR-140-5p on cell viability. (E-G) Cell apoptosis was evaluated using flow cytometry. n=3. *P<0.05, **P<0.001, vs. Blank; #P<0.05, ##P<0.001, vs. NC; ^^P<0.001, vs. VEGFA, ΔΔP<0.001, vs. mimic + NC. VEGFA, vascular endothelial growth factor A; miR, microRNA; MM, multiple myeloma; M, mimic; NC, negative control; OD, optical density; PI, propidium iodide; FITC, fluorescein isothiocyanate.
Effects of miR-140-5p on viability and apoptosis of MM cell lines by targeting VEGFA
VEGFA overexpression led to a significant increase in cell viability. However, co-transfection of VEGFA-pcDNA3.1 with the miR-140-5p mimic significantly reduced cell viability, compared with transfection with VEGFA-pcDNA3.1 alone (P<0.001; Fig. 5C and D). Moreover, apoptosis was significantly reduced following VEGFA overexpression, but increased following co-transfection with by VEGFA-pcDNA3.1 and the miR-140-5p mimic (P<0.001, Fig. 5E-G).
Effects of miR-140-5p and VEGFA on the migration and invasion of MM cell lines
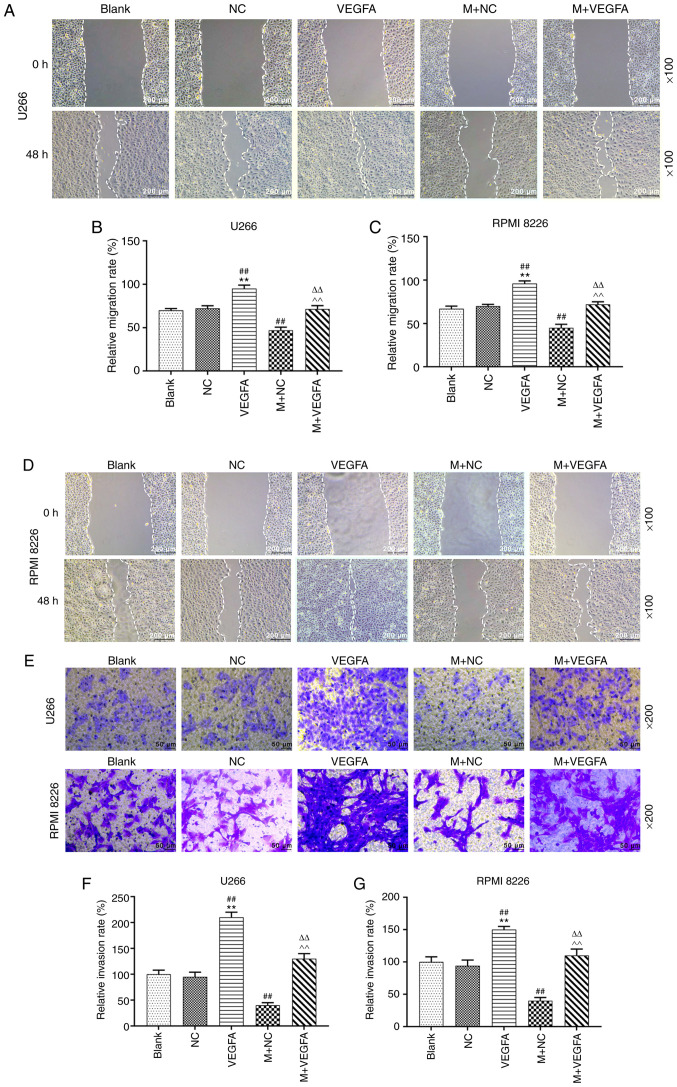
MM cell lines were transfected with VEGFA-pcDNA3.1, alone or with miR-140-5p mimic into the cells, and cell migration and invasion were assessed. VEGFA overexpression promoted cell migration, compared with blank cells. However, co-transfection with VEGFA-pcDNA3.1 and miR-140-5p mimic reduced the migration rate, compared with VEGFA overexpression alone (P<0.001; Fig. 6A-D). Moreover, overexpression of VEGFA was also promoted cell invasion, but this effect was reversed following co-transfection with the miR-140-5p mimic (P<0.001, Fig. 6E-G).
Figure 6.
Effects of miR-140-5p and VEGFA on migration and invasion of MM cell lines. (A and B) U266 cell migration was measured using a wound healing scratch assay. (C and D) RPMI 8226 cell migration was measured using a wound healing assay. Scale bar, 200 µm. Magnification, ×100. (E-G) Cell invasion was detected using a Transwell Matrigel™ assay. Scale bar, 50 µm. Magnification, ×200. n=3. **P<0.001, vs. Blank; ##P<0.001, vs. NC; ^^P<0.001, vs. VEGFA, ΔΔP<0.001, vs. mimic + NC. MiR, microRNA; VEGFA, vascular endothelial growth factor A; MM, multiple myeloma; M, mimic; NC, negative control.
Discussion
MM often results in susceptibility to infection, impaired renal function, anemia and hypercalcemia, which severely impacts on patient quality of life (29,30). To date, the pathogenesis of MM remains unclear, and specific treatment avenues for MM are still the focus of research (31). Previous studies have indicated that miRNAs are aberrantly expressed in different types of tumor, and that these molecules could act as tumor suppressor genes through genetic regulation of cell proliferation, migration, cell cycle and differentiation during tumorigenesis (12,32). For instance, previous studies demonstrated that miR-196-b-5p and miR-99-a-5p served important roles in the regulation of autophagy and apoptosis of MM cells (33), while miR-125-b and miR-34a affected their viability (34). Moreover, miR-720, miR-1246, miR-16 and miR-25 could serve as biological markers in MM prognostic evaluation (35,36). Therefore, miRNA function represents an important research area in the study of MM pathogenesis.
miR-140-5p is a newly discovered small-molecule coding RNA. Previous studies indicated that miR-140-5p played an important regulatory role in human gastric cancer (37), glioma (38) and non-small cell lung cancer (39). Another study demonstrated that miR-140-5p could regulate vitamin D levels by targeting MAPK, which provided new insight into the treatment of bone diseases (40). In addition, miR-140-5p could be involved in autophagy and drug resistance mechanisms in MM (41). However, the specific role of miR-140-5p in the occurrence and development of MM still remains unclear. Thus, the present study aimed to explore the role of miR-140-5p in MM.
miR-140-5p is significantly downregulated in gastric cancer (37) and breast cancer (42), compared with normal tissues. Similarly, in the present study, the expression of miR-140-5p in MM cell lines was significantly lower than in normal plasma cells. In a previous study, high miR-140-5p expression promoted cell viability of human osteosarcoma cells decreased by tumor drugs (43), and overexpressing miR-140-5p could promote apoptosis in chronic myeloid leukemia (44). Furthermore, the migration and invasion of lung cancer cells significantly decreased following miR-140-5p overexpression (39). In the present study, MM cell lines were transfected with a miR-140-5p in order to further investigate the biological effects of miR-140-5p overexpression on MM. miR-140-5p overexpression significantly reduced the viability and migration of MM cells, while increasing apoptosis. These results indicated that miR-140-5p could regulate MM progression, and that overexpression of miR-140-5p inhibited the development of MM.
miRNAs are known to bind to the 3′-UTR of specific target genes to jointly regulate tumorigenesis and tumor development (10). miR-140-5p binds several target genes and regulates the progression of numerous diseases. For example, miR-140-5p inhibited the growth of ovarian cancer by targeting platelet-derived growth factor receptor α (17). In addition, miR-140-5p could also promote proliferation and inhibit apoptosis of human pulmonary artery smooth muscle cells during hypoxia by targeting DNA methyltransferase 1 (45). Based on these previous findings, it was determined that miR-140-5p bound to a gene to jointly regulate the disease progression of MM. Targetscan7.2 software predicted that VEGFA was a possible target gene for miR-140-5p, and a dual luciferase assay also indicated that VEGFA interacted with miR-140-5p directly. Moreover, transfection with a VEGFA overexpression plasmid along with a miR-140-5p mimic confirmed that miR-140-5p overexpression could inhibit the expression of VEGFA in MM cells.
VEGFA can increase microvascular permeability, and promote the proliferation, vascular construction and the migration of endothelial cells from different tissues (46). The VEGFA signaling pathway is an important target for the inhibition of tumor angiogenesis (47). Studies demonstrated that miR-140-5p could inhibit cell proliferation, migration and invasion of lung cancer and colorectal cancer cells by targeting VEGFA (48,49). In the present study, overexpression of VEGFA could promote cell viability, invasion and migration, and inhibit cell apoptosis. Moreover, miR-140-5p overexpression partially reversed this effect and promoted cell viability, migration and invasion, while suppressing apoptosis. Consistent with previous studies, these results indicated that miR-140-5p could regulate MM progression through VEGFA (48).
In conclusion, the present study demonstrated that miR-140-5p suppressed MM progression by targeting VEGFA, and suggests that miR-140-5p could be a potential therapeutic target for the treatment of MM. However, a limitation of the present study was the lack of animal experiments and thus these results require further validation in vivo.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- MM
multiple myeloma
- RT-qPCR
reverse transcription-quantitative PCR
- miR
MicroRNA
- VEGFA
vascular endothelial growth factor A
- CCK-8
cell counting kit-8
- RPMI
Roswell Park Memorial Institute
- NC
negative control
- WT
wildtype
- MUT
mutant
- UTR
untranslated region
- FITC
fluorescein isothiocyanate
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
ML and HL made substantial contributions to the conception and design of the study. HL, JZ and ZY acquired, analyzed and interpreted the data. ML and HL drafted the article and critically revised it for important intellectual content. All authors agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of the work are appropriately investigated and resolved. All authors approved the final version of the manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Jingzhou Central Hospital (approval no. JCH20150608EZ023). All procedures involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. All volunteers provided informed consent.
Patient consent for participation
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Röllig C, Knop S, Bornhäuser M. Multiple myeloma. Lancet. 2015;385:2197–2208. doi: 10.1016/S0140-6736(14)60493-1. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 3.Ghobrial IM, Siegel DS, Vij R, Berdeja JG, Richardson PG, Neuwirth R, Patel CG, Zohren F, Wolf JL. TAK-228 (formerly MLN0128), an investigational oral dual TORC1/2 inhibitor: A phase I dose escalation study in patients with relapsed or refractory multiple myeloma, non-Hodgkin lymphoma, or Waldenstrom's macroglobulinemia. Am J Hematol. 2016;91:400–405. doi: 10.1002/ajh.24300. [DOI] [PubMed] [Google Scholar]
- 4.Agarwal A, Mahadevan D. Novel targeted therapies and combinations for the treatment of multiple myeloma. Cardiovasc Hematol Disord Drug Targets. 2013;13:2–15. doi: 10.2174/1871529X11313010002. [DOI] [PubMed] [Google Scholar]
- 5.Hao S, Du X, Song Y, Ren M, Yang Q, Wang A, Wang Q, Zhao H, Du Z, Zhang G. Targeted gene therapy of the HSV-TK/hIL-12 fusion gene controlled by the hSLPI gene promoter of human non-small cell lung cancer in vitro. Oncol Lett. 2018;15:6503–6512. doi: 10.3892/ol.2018.8148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morris LG, Chan TA. Therapeutic targeting of tumor suppressor genes. Cancer. 2015;121:1357–1368. doi: 10.1002/cncr.29140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov. 2013;12:847–865. doi: 10.1038/nrd4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mohr AM, Mott JL. Overview of microRNA biology. Semin Liver Dis. 2015;35:3–11. doi: 10.1055/s-0034-1397344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liang S, Zhang N, Deng Y, Chen L, Zhang Y, Zheng Z, Luo W, Lv Z, Li S, Xu T. miR-663 promotes NPC cell proliferation by directly targeting CDKN2A. Mol Med Rep. 2017;16:4863–4870. doi: 10.3892/mmr.2017.7129. [DOI] [PubMed] [Google Scholar]
- 10.Shi J. Regulatory networks between neurotrophins and miRNAs in brain diseases and cancers. Acta Pharmacol Sin. 2015;36:149–157. doi: 10.1038/aps.2014.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petri R, Jakobsson J. Identifying miRNA targets using AGO-RIPseq. Methods Mol Biol. 2018;1720:131–140. doi: 10.1007/978-1-4939-7540-2_9. [DOI] [PubMed] [Google Scholar]
- 12.Trionfini P, Benigni A. MicroRNAs as master regulators of glomerular function in health and disease. J Am Soc Nephrol. 2017;28:1686–1696. doi: 10.1681/ASN.2016101117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Orso F, Quirico L, Dettori D, Coppo R, Virga F, Ferreira LC, Paoletti C, Baruffaldi D, Penna E, Taverna D. Role of miRNAs in tumor and endothelial cell interactions during tumor progression. Semin Cancer Biol. 2020;60:214–224. doi: 10.1016/j.semcancer.2019.07.024. [DOI] [PubMed] [Google Scholar]
- 14.Muys BR, Sousa JF, Plaça JR, de Araújo LF, Sarshad AA, Anastasakis DG, Wang X, Li XL, de Molfetta GA, Ramão A, et al. miR-450a acts as a tumor suppressor in ovarian cancer by regulating energy metabolism. Cancer Res. 2019;79:3294–3305. doi: 10.1158/0008-5472.CAN-19-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang WC, Jang TH, Tung SL, Yen TC, Chan SH, Wang LH. A novel miR-365-3p/EHF/keratin 16 axis promotes oral squamous cell carcinoma metastasis, cancer stemness and drug resistance via enhancing β5-integrin/c-met signaling pathway. J Exp Clin Cancer Res. 2019;38:89. doi: 10.1186/s13046-019-1091-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miao X, Wang Z, Chen B, Chen Y, Wang X, Jiang L, Jiang S, Hao K, Zhang W. miR-140-5p suppresses retinoblastoma cell proliferation, migration, and invasion by targeting CEMIP and CADM3. Cell Mol Biol (Noisy-le-grand) 2018;64:42–47. doi: 10.14715/cmb/2018.64.6.8. [DOI] [PubMed] [Google Scholar]
- 17.Lan H, Chen W, He G, Yang S. miR-140-5p inhibits ovarian cancer growth partially by repression of PDGFRA. Biomed Pharmacother. 2015;75:117–122. doi: 10.1016/j.biopha.2015.07.035. [DOI] [PubMed] [Google Scholar]
- 18.Zhang D, Yang Y, Wu M, Zhao X, Sun Y, Xie H, Li H, Li Y, Wang K, Zhang J, et al. The moderating effect of social support on the relationship between physical health and suicidal thoughts among Chinese rural elderly: A nursing home sample. Int J Ment Health Nurs. 2018;27:1371–1382. doi: 10.1111/inm.12436. [DOI] [PubMed] [Google Scholar]
- 19.Ferrara N, Adamis AP. Ten years of anti-vascular endothelial growth factor therapy. Nat Rev Drug Discov. 2016;15:385–403. doi: 10.1038/nrd.2015.17. [DOI] [PubMed] [Google Scholar]
- 20.Cancer Genome Atlas Research Network; Analysis Working Group; Asan University; BC Cancer Agency; Brigham and Women's Hospital; Broad Institute; Brown University; Case Western Reserve University; Dana-Farber Cancer Institute; Duke University, et al, corp-author. Integrated genomic characterization of oesophageal carcinoma. Nature. 2017;541:169–175. doi: 10.1038/nature20805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Botta C, Di Martino MT, Ciliberto D, Cucè M, Correale P, Rossi M, Tagliaferri P, Tassone P. A gene expression inflammatory signature specifically predicts multiple myeloma evolution and patients survival. Blood Cancer J. 2016;6:e511. doi: 10.1038/bcj.2016.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu L, Bi N, Wu L, Ding X, Men Y, Zhou W, Li L, Zhang W, Shi S, Song Y, Wang L. MicroRNA-29c functions as a tumor suppressor by targeting VEGFA in lung adenocarcinoma. Mol Cancer. 2017;16:50. doi: 10.1186/s12943-017-0620-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsu CY, Hsieh TH, Tsai CF, Tsai HP, Chen HS, Chang Y, Chuang HY, Lee JN, Hsu YL, Tsai EM. miRNA-199a-5p regulates VEGFA in endometrial mesenchymal stem cells and contributes to the pathogenesis of endometriosis. J Pathol. 2014;232:330–343. doi: 10.1002/path.4295. [DOI] [PubMed] [Google Scholar]
- 24.Sun CY, She XM, Qin Y, Chu ZB, Chen L, Ai LS, Zhang L, Hu Y. miR-15a and miR-16 affect the angiogenesis of multiple myeloma by targeting VEGF. Carcinogenesis. 2013;34:426–435. doi: 10.1093/carcin/bgs333. [DOI] [PubMed] [Google Scholar]
- 25.Liu D, Lin P, Hu Y, Zhou Y, Tang G, Powers L, Medeiros LJ, Jorgensen JL, Wang SA. Immunophenotypic heterogeneity of normal plasma cells: Comparison with minimal residual plasma cell myeloma. J Clin Pathol. 2012;65:823–829. doi: 10.1136/jclinpath-2012-200881. [DOI] [PubMed] [Google Scholar]
- 26.Horst A, Hunzelmann N, Arce S, Herber M, Manz RA, Radbruch A, Nischt R, Schmitz J, Assenmacher M. Detection and characterization of plasma cells in peripheral blood: Correlation of IgE+ plasma cell frequency with IgE serum titre. Clin Exp Immunol. 2002;130:370–378. doi: 10.1046/j.1365-2249.2002.02025.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liang B, Yin JJ, Zhan XR. MiR-301a promotes cell proliferation by directly targeting TIMP2 in multiple myeloma. Int J Clin Exp Pathol. 2015;8:9168–9174. [PMC free article] [PubMed] [Google Scholar]
- 28.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 29.Fakhri B, Vij R. Clonal evolution in multiple myeloma. Clin Lymphoma Myeloma Leuk. 2016;16(Suppl):S130–S134. doi: 10.1016/j.clml.2016.02.025. [DOI] [PubMed] [Google Scholar]
- 30.Kiely F, Cran A, Finnerty D, O'Brien T. Self-reported quality of life and symptom burden in ambulatory patients with multiple myeloma on disease-modifying treatment. Am J Hosp Palliat Care. 2017;34:671–676. doi: 10.1177/1049909116646337. [DOI] [PubMed] [Google Scholar]
- 31.Kuroda J, Chinen Y. Multiple myeloma: Pathophysiology and progress in management. Rinsho Ketsueki. 2017;58:487–497. doi: 10.11406/rinketsu.58.487. [DOI] [PubMed] [Google Scholar]
- 32.Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol Med. 2014;20:460–469. doi: 10.1016/j.molmed.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 33.Shang J, Chen ZZ, Wang ZH, Wei TN, Wu WB, Chen WM. Association of miRNA-196b-5p and miRNA-99a-5p with autophagy and apoptosis in multiple myeloma cells. Zhonghua Xue Ye Xue Za Zhi. 2018;39:766–772. doi: 10.3760/cma.j.issn.0253-2727.2018.09.013. (In Chinese) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abdi J, Rastgoo N, Li L, Chen W, Chang H. Role of tumor suppressor p53 and micro-RNA interplay in multiple myeloma pathogenesis. J Hematol Oncol. 2017;10:169. doi: 10.1186/s13045-017-0538-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ren Y, Li X, Wang W, He W, Wang J, Wang Y. Expression of peripheral blood miRNA-720 and miRNA-1246 can be used as a predictor for outcome in multiple myeloma patients. Clin Lymphoma Myeloma Leuk. 2017;17:415–423. doi: 10.1016/j.clml.2017.05.010. [DOI] [PubMed] [Google Scholar]
- 36.Rocci A, Hofmeister CC, Pichiorri F. The potential of miRNAs as biomarkers for multiple myeloma. Expert Rev Mol Diagn. 2014;14:947–959. doi: 10.1586/14737159.2014.946906. [DOI] [PubMed] [Google Scholar]
- 37.Fang Z, Yin S, Sun R, Zhang S, Fu M, Wu Y, Zhang T, Khaliq J, Li Y. miR-140-5p suppresses the proliferation, migration and invasion of gastric cancer by regulating YES1. Mol Cancer. 2017;16:139. doi: 10.1186/s12943-017-0708-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang HL, Gao YM, Zhao JA. miR-140-5p inhibits human glioma cell growth and invasion by targeting JAG1. Mol Med Rep. 2017;16:3634–3640. doi: 10.3892/mmr.2017.6951. [DOI] [PubMed] [Google Scholar]
- 39.Flamini V, Jiang WG, Cui Y. Therapeutic role of MiR-140-5p for the treatment of non-small cell lung cancer. Anticancer Res. 2017;37:4319–4327. doi: 10.21873/anticanres.11825. [DOI] [PubMed] [Google Scholar]
- 40.Luo W, Liu L, Yang L, Dong Y, Liu T, Wei X, Liu D, Gu H, Kong J, Yuan Z, Zhao Q. The vitamin D receptor regulates miR-140-5p and targets the MAPK pathway in bone development. Metabolism. 2018;85:139–150. doi: 10.1016/j.metabol.2018.03.018. [DOI] [PubMed] [Google Scholar]
- 41.Lu D, Yang C, Zhang Z, Cong Y, Xiao M. Knockdown of Linc00515 inhibits multiple myeloma autophagy and chemoresistance by upregulating miR-140-5p and downregulating ATG14. Cell Physiol Biochem. 2018;48:2517–2527. doi: 10.1159/000492690. [DOI] [PubMed] [Google Scholar]
- 42.Lu Y, Qin T, Li J, Wang L, Zhang Q, Jiang Z, Mao J. MicroRNA-140-5p inhibits invasion and angiogenesis through targeting VEGF-A in breast cancer. Cancer Gene Ther. 2017;24:386–392. doi: 10.1038/cgt.2017.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wei R, Cao G, Deng Z, Su J, Cai L. miR-140-5p attenuates chemotherapeutic drug-induced cell death by regulating autophagy through inositol 1,4,5-trisphosphate kinase 2 (IP3k2) in human osteosarcoma cells. Biosci Rep. 2016;36:e00392. doi: 10.1042/BSR20160238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nie ZY, Liu XJ, Zhan Y, Liu MH, Zhang XY, Li ZY, Lu YQ, Luo JM, Yang L. miR-140-5p induces cell apoptosis and decreases Warburg effect in chronic myeloid leukemia by targeting SIX1. Biosci Rep. 2019;39:BSR20190150. doi: 10.1042/BSR20190150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y, Xu J. MiR-140-5p regulates hypoxia-mediated human pulmonary artery smooth muscle cell proliferation, apoptosis and differentiation by targeting Dnmt1 and promoting SOD2 expression. Biochem Biophys Res Commun. 2016;473:342–348. doi: 10.1016/j.bbrc.2016.03.116. [DOI] [PubMed] [Google Scholar]
- 46.Barquet LA. Role of VEGF in diseases of the retina. Arch Soc Esp Oftalmol. 2015;90(Suppl 1):S3–S5. doi: 10.1016/S0365-6691(15)30002-2. (In Spanish) [DOI] [PubMed] [Google Scholar]
- 47.Claesson-Welsh L, Welsh M. VEGFA and tumour angiogenesis. J Intern Med. 2013;273:114–127. doi: 10.1111/joim.12019. [DOI] [PubMed] [Google Scholar]
- 48.Yang P, Xiong J, Zuo L, Liu K, Zhang H. miR-140-5p regulates cell migration and invasion of non-small cell lung cancer cells through targeting VEGFA. Mol Med Rep. 2018;18:2866–2872. doi: 10.3892/mmr.2018.9291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang W, Zou C, Pan L, Xu Y, Qi W, Ma G, Hou Y, Jiang P. MicroRNA-140-5p inhibits the progression of colorectal cancer by targeting VEGFA. Cell Physiol Biochem. 2015;37:1123–1133. doi: 10.1159/000430237. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.