Abstract
Background
The objective of this study was to evaluate the virulence of P. aeruginosa ventilator-associated pneumonia (VAP) strains (cases) in terms of biofilm production and other phenotypic and genotypic virulence factors compared to P. aeruginosa strains isolated from other infections (controls).
Methods
Biofilm production was tested to assess biomass production and metabolic activity using crystal violet binding assay and XTT assay, respectively. Pigment production (pyocyanin and pyoverdine) was evaluated using cetrimide agar. Virulence genes were detected by conventional multiplex PCR and virulence was tested in an in vivo model in Galleria mellonella larvae.
Results
We did not find statistically significant differences between VAP and no-VAP strains (p > 0.05) regarding biofilm production. VAP strains had no production of pyocyanin after 24 h of incubation (p = 0.023). The distribution of virulence genes between both groups were similar (p > 0.05). VAP strains were less virulent than non-VAP strains in an in vivo model of G. mellonella (p < 0.001).
Conclusion
The virulence of VAP-Pseudomonas aeruginosa does not depend on biofilm formation, production of pyoverdine or the presence of some virulence genes compared to P. aeruginosa isolated from non-invasive locations. However, VAP strains showed attenuated virulence compared to non-VAP strains in an in vivo model of G. mellonella.
Supplementary Information
Supplementary information accompanies this paper at 10.1186/s12879-020-05534-1.
Keywords: Pseudomonas aeruginosa, Ventilator-associated pneumonia, Virulence genes, Biofilm, Galleria mellonella
Background
Pseudomonas aeruginosa is one of the most common causes of ventilator-associated pneumonia (VAP), with high mortality rates (approximately 13%), prolonged hospital stays and increasing hospital costs [1]. VAP is a nosocomial lung infection that appears 48 h after intubation characterised by new lung infiltrates, signs of systematic infection, changes in the appearance of sputum, leukocytosis and decline in oxygenation [2, 3]. The incidence of VAP ranges between 2 and 16 episodes per ventilator days being around 44% of the episodes caused by P. aeruginosa [4].
VAP is caused mainly by multidrug-resistant and extremely drug-resistant strains of P. aeruginosa, leading to an increase in treatment failure [5]. Moreover, the activity of antibiotics is restricted by P. aeruginosa biofilms that form rapidly on the surface of the endotracheal tube [6]. These microbial communities constitute one of the most important virulence factors of P. aeruginosa. However, many more virulence factors are involved in VAP development, such as type III secretory proteins (exoT, exoS, exoY, and exoU), which control the expression of exotoxins, quorum-sensing (QS) system proteins (lasR/lasI and rhlR/rhlI) allowing the communication between cells, elastases (lasA and lasB) that disrupt tight junctions between host epithelial cells, alginate (alg genes), and pigments such as pyoverdine, which up-regulates the transcription of some kinds elastases, and pyocyanin, involved in oxidative stress promoting the alteration of mitochondrial electron transport of the host [1, 7, 8].
Diagnosis and treatment of VAP are still a challenge in clinical settings. Understanding the molecular basis of pathogenicity may lead to new treatment targets and new biomarkers for improving management of VAP. Hence, the aim of this study was to compare the virulence of P. aeruginosa strains causing VAP in terms of various factors with that of P. aeruginosa strains isolated from other sources of infection.
Methods
This multicentre study was conducted in the Clinical Microbiology Laboratory of Hospital General Universitario Gregorio Marañón, Madrid, Spain. A total of 90 P. aeruginosa strains were collected and they were distributed as follows: 40 different (1 per patient) cases from lower respiratory tract samples of patients with VAP obtained from 5 institutions (4 from Istituti Fisioterapici Ospitalieri, Rome, Italy; 3 from Hospital Ramón y Cajal, Madrid, Spain; 1 from Hospital la Paz, Madrid, Spain; 11 from Hospital Clinic, Barcelona, Spain, and 21 from Hospital Universitario Gregorio Marañón, Spain) and 50 controls from various sites (central venous catheter, urinary tract, blood, faeces, cerebrospinal fluid, joint fluid, cornea, abscesses, skin ulcers, and nephrostomy tubes) obtained from Istituti Fisioterapici Ospitalieri (n = 15) and Hospital General Universitario Gregorio Marañón (n = 35). Control samples were collected randomly both retrospectively and prospectively. VAP-samples were collected from patients with clinical and microbiological diagnosis of VAP when the last one was confirmed.
All the assays were performed in the Clinical Microbiology Laboratory of Hospital General Universitario Gregorio Marañón, Madrid, Spain. The other centres were responsible of collecting samples and clinical and microbiology data of the samples.
Biofilm production
Biofilms were assessed as described by our group, with some modifications [9]. Briefly, a loopful of fresh medium with P. aeruginosa was inoculated in 20 ml of BHI broth (Brain Heart Infusion) and incubated with shaking at 37 °C for 24 h in an orbital shaker. After 3 cycles of centrifugation-resuspension with PBS (phosphate buffer saline), inoculums were adjusted to 0.5 McFarland (108 cfu/ml) in BHI broth. One hundred microliters were then added to each well of a 96-well plate, and plates were incubated at 37 °C for 24 h. Wells were washed 3 times with PBS, and, once the plates were completely dry, the crystal violet (CV) and the tetrazolium salt XTT [2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide] assays were applied.
For biomass quantification, after PBS washes, 200 μl of 99% methanol was used to fix biofilms for 10 min at room temperature. Methanol was discarded, and 125 μl of CV was added for 15 min. The plates were washed with distilled water, and 125 μl of 30% acetic acid was added for 15 min. Absorbance was measured at 550 nm in a spectrophotometer (Biochrom EZ Read 400).
The metabolic activity of the biofilms was evaluated using XTT. Menadione and XTT were mixed immediately before each experiment at a proportion of 1 μl per 10 ml of XTT, and 100 μl was added to each of the wells after PBS wash. Plates were incubated at 37 °C for 2 h in darkness. Absorbance was measured at 492 nm using a spectrophotometer (Biochrom EZ Read 400).
All strains were tested in triplicate using P. aeruginosa PA01 (a biofilm-producing strain) as the positive control and sterile BHI broth as the negative control.
Strains were classified as low, moderate, or high biofilm producers using the optical density (OD) as follow: low biofilm producer, OD ≤ 2 × ODc; moderate biofilm producer, 2 × ODc < OD < 4 × ODc; and high biofilm producer, OD ≥ 4 × ODc, where ODc represents 3 standard deviations above the mean of the negative control and OD indicates the mean optical density of the strain [10].
Pyocyanin and pyoverdine production
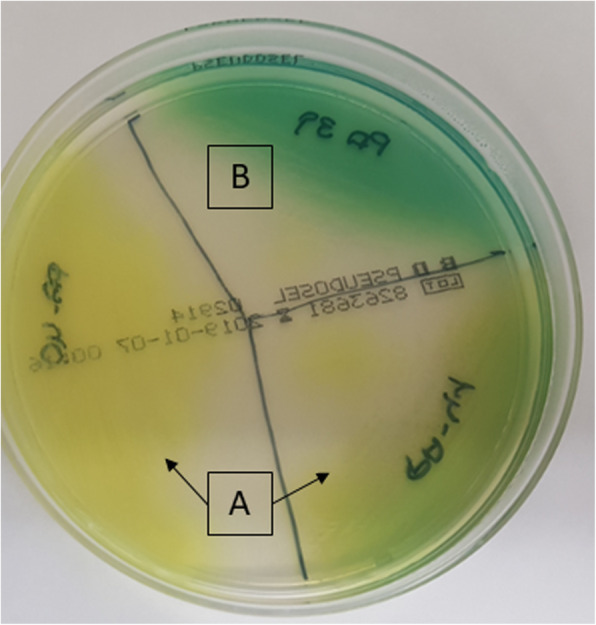
Pigment production was evaluated in cetrimide agar (Pseudosel Agar, BD) by qualitative observation. Inoculated plates were incubated at 37 °C for 24 h. Colonies that appeared fluorescent green, yellow, or brown in colour were considered to be pyoverdine producers. Dark green or blue colonies were considered to be pyocyanin producers (Fig. 1).
Fig 1.
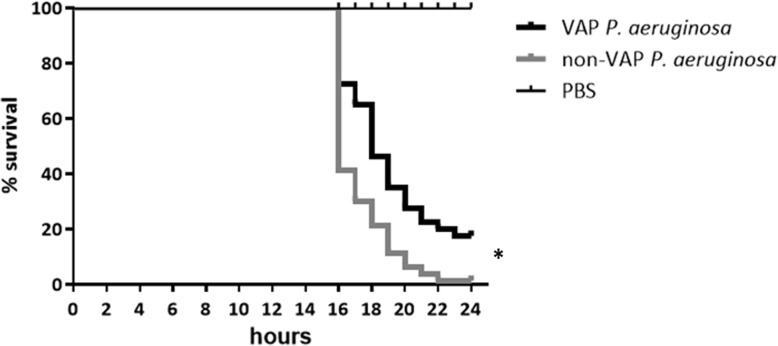
Survival curves for Galleria mellonella larvae after infection with Pseudomonas aeruginosa strains isolated from patients with VAP (VAP P. aeruginosa, n = 8) and from patients with other infections (non-VAP P. aeruginosa, n = 8). PBS was used as a positive control for larvae survival. *Statistically differences were found between VAP and non-VAP P. aeruginosa (p < 0.001) using the log-rank test
Virulence genes
DNA extraction
Strain DNA was extracted using the QIAamp® DNA Mini Kit (250) (QIAGEN) following the manufacturer’s instructions. DNA was eluted in 100 μl of elution buffer and stored at 4 °C.
Polymerase chain reaction
We tested the presence or absence of 14 virulence genes—exoT, exoY, exoS, exoU, algD, algU, plcH, plcN, apr, rhII, rhIR, lasI, lasR, and lasB—by conventional multiplex and uniplex PCR, as described elsewhere [11–13]. Primer sequences [11, 13], amplicon size, and PCR conditions are shown in Table 1. exoU primers were designed in this study using the online Primer-Blast program of the National Center for Biotechnology Information. The PCR master mix was composed of nuclease-free water, Phusion U Green Multiplex PCR Master Mix (1X) (Thermo Scientific™), and primers at a final concentration of 0.3 μM. PCR was performed in a thermocycler (GeneAmp PCR system 9700, Applied Biosystems). Strain PA01 was used as a positive control. PA14, which was kindly provided by Dr. Oliver, was used as a positive control for detection of exoU.
Table 1.
Distribution of the presence of virulence genes from Pseudomonas aeruginosa strains isolated from lower respiratory tract samples of patients with VAP (cases) and patients with non-VAP infection (controls)
| algD N (%) | algU N (%) | plcH N (%) | plcN N (%) | exoS N (%) | exoY N (%) | exoT N (%) | exoU N (%) | apr N (%) | lasB N (%) | lasI N (%) | lasR N (%) | rhII N (%) | rhRI N (%) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Controls | 50 (100.0) | 41 (82.0) | 50 (100.0) | 50 (100.0) | 37 (74.0) | 45 (90.0) | 50 (100.0) | 15 (30.0) | 50 (100.0) | 50 (100.0) | 48 (96.0) | 48 (96.0) | 38 (76.0) | 37 (74.0) |
| Cases | 40 (100.0) | 33 (82.5) | 40 (100.0) | 40 (100.0) | 31 (77.5) | 39 (97.5) | 40 (100.0) | 13 (32.5) | 39 (97.5) | 40 (100.0) | 40 (100.0) | 39 (97.5) | 36 (90.0) | 36 (90.0) |
| Total | 90 (100.0) | 74 (82.2) | 90 (100.0) | 90 (100.0) | 68 (75.6) | 84 (93.3) | 90 (100.0) | 28 (31.1) | 89 (98.9) | 90 (100.0) | 88 (97.8) | 87 (96.7) | 74 (82.0) | 73 (81.0) |
N Number of strains that showed the gene; % Percentage of strains that possess the gene
No statistically differences in the presence of virulence genes were found between control and case strains (p > 0.05, chi-square)
Electrophoresis
Fragments were detected by agarose electrophoresis in 2% agarose gels at 170 V for 30 min. exoS, plcN, lasB, and lasR amplicons were detected using 4% agarose gels at 170 V for 90 min. Ready-to-use GeneRuler 1-kb and 100-bp DNA ladders (Thermo ScientificTM) were applied to size DNA products.
Sequencing
Sequencing was performed in PA01 strain to confirm that amplicons corresponded to the studied genes. DNA was purified using the QIAquick® PCR Purification Kit (250) (QIAGEN) following the manufacturer’s instructions. Forward strains were sequenced in the Genomics Department of Hospital General Universitario Gregorio Marañón. Sequences were aligned using the BLAST database to identify the genes.
In vivo virulence assay
Of the total of 40 cases and 50 controls, we were only able to test 8 randomly chosen strains of each (cases: PA18, 19 21, 29, 35, 38, 52, and 57; controls: PA03 from skin ulcer, PA27, and PA64 from catheters, PA39 from bone biopsy, PA40 from a rectal exudate, PA50 from a nephrostomy, PA56 from cornea, and PA61 from urine) in the G. mellonella model owing to difficulties in the supply of larvae.
The Galleria mellonella (G. mellonella) infection model was applied as described by Marcos-Zambrano et al., with some modifications [14]. Briefly, an inoculum of 103 cfu/ml of P. aeruginosa was injected into the hemocoel via the last left proleg of the instar larvae of G. mellonella (Bichosa, Salceda de Caselas, Spain) using a Hamilton syringe. Infected larvae were incubated at 37 °C for 24 h. Each experimental group was composed of 10 randomly chosen larvae (330 ± 20 mg) cleaned with 70% ethanol prior to each experiment. Larvae injected and not injected with PBS were used as controls to monitor injection injury. Larvae death was recorded every hour from 16 h after infection to 24 h. Completely melanised larvae and/or completely immobilized larvae were considered dead.
Statistical analysis
Quantitative variables were expressed as mean (SD), and qualitative variables were reported as percentages. Categorical variables (distribution of high, moderate and low biofilm producer between case and control groups and the presence or absence of virulence genes between groups) were compared using the chi-square test, and continuous variables (absorbance values for CV and XTT assays between control and case groups) were compared using the t test. Statistical significance was set at p < 0.05. All tests were performed using the statistical program SPSS 21.0 (IBM).
Killing curves were analysed using the Kaplan-Meier method with GraphPad Prism 5.02 software (GraphPad, La Jolla, CA, USA). Differences in survival were compared using the log-rank (Mantel-Cox) test. Statistical significance was set at p < 0.05.
Ethics statement
The strains collected during this study were obtained by regular diagnostic procedures of the involved institutions. This study was approved by the Ethics Committee of the Hospital General Universitario Gregorio Marañón (code: MICRO.HGUGM.2015–075). No informed consent was required.
Results
Biofilm production
Of the 90 strains studied, 76 (84.4%), 13 (14.5%), and 1 (1.1%) strains were high, moderate, and low biomass producers, respectively. As for the metabolic activity of biofilms, the distribution of high, moderate, and low producers was, 17 (18.9%), 34 (37.8%), and 39 (43.3%), respectively. Distribution of strains by biofilm production classified by cases and controls is reported in Table 2. No statistically significant differences in biofilm production were found with respect to biomass production or metabolic activity between cases and controls (p > 0.05).
Table 2.
Primer sequences, fragment size, and PCR conditions for detection of virulence genes in Pseudomonas aeruginosa
| Gene | Primer sequence (5′➔3′) | Fragment size (bp) | Annealing T (°C) |
|---|---|---|---|
| algD | F: AAGGCGGAAATGCCATCTCC | 290 | 60 |
| R: AGGGAAGTTCCGGGCGTTTG | |||
| exoY | F: TATCGACGGTCATCGTCAGGT | 1035 | |
| R: TTGATGCACTCGACCAGCAAG | |||
| plcH | F: GCACGTGGTCATCCTGATGC | 608 | |
| R: TCCGTAGGCGTCGACGTAC | |||
| rhIR | F: TGCATTTTATCGATCAGGGC | 133 | 57 |
| R: CACTTCCTTTTCCAGGACG | |||
| lasI | F: CGTGCTCAAGTGTTCAAGG | 295 | |
| R: TACAGTCGGAAAAGCCCAG | |||
| apr | F: TGTCCAGCAATTCTCTTGC | 1017 | |
| R: CGTTTTCCACGGTGACC | |||
| exoT | F: CAATCATCTCAGCAGAACCC | 1159 | |
| R: TGTCGTAGAGGATCTCCTG | |||
| rhII | F: TTCATCCTCCTTTAGTCTTCCC | 155 | |
| R: TTCCAGCGATTCAGAGAGC | |||
| exoS | F: CGTCGTGTTCAAGCAGATGGTGCTG | 444 | 65 |
| R: CCGAACCGCTTCACCAGGC | |||
| plcN | F: TCCGTTATCGCAACCAGCCCTACG | 481 | |
| R: TCGCTGTCGAGCAGGTCGAAC | |||
| lasB | F: GGAATGAACGAAGCGTTCTCCGAC | 284 | |
| R: TGGCGTCGACGAACACCTCG | |||
| lasR | F: AAGTGGAAAATTGGAGTGGAG | 130 | |
| R: GTAGTTGCCGACGACGATGAAG | |||
| algU | F: CGCGAACCGCACCATCGCTC | 1500 | 66 |
| R: GCCGCACGTCACGAGC | |||
| exoU | F: TACCAGGTACGGCCATGTTC | 575 | 59 |
| R: ACGCTCTGAAGCCTGAAGAC |
Bp Base pairs; F Forward; R Reverse; T Temperature
PCR was performed following the manufacturer’s instructions
Pyocyanin and pyoverdine production
Pigment production is another virulence factor of P. aeruginosa. In our collection, 74.4% of the strains secreted pyoverdine, 7.8% pyocyanin, 10% both pigments, and 7.8% neither. Pyoverdine was produced more frequently than pyocyanin in VAP strains (92.5% vs 7.5%, respectively) (p < 0.001). Production of pyocyanin was more often found in non-VAP isolates (26.0%) than in VAP isolates (7.5%) (p = 0.023). All the strains that produced both pigments or neither pigment belonged to the control group.
Virulence genes
All the strains presented at least 10 virulence genes. algD, lasB, plcN, plcH, and exoT were present in all the strains. Apr, lasI, lasR, and exoY were found in 99, 98, 97, and 93% of the strains, respectively. A lower incidence was obtained for rhII (82%), rhRI (81%), algU (82%), and exoS (76%). exoU was present in only 28 (31.1%) strains. One strain presented lasI but not lasR. The same phenomenon was observed in another strain, in which rhII was present, although its regulatory gene (rhIR) was absent. The distribution of virulence genes did not differ significantly between cases and controls (p > 0.05) (Table 3). However, a tendency towards absence of rhlI and rhlR was observed in control strains (p = 0.084 and p = 0.054, respectively).
Table 3.
Distribution and classification of the whole collection of Pseudomonas aeruginosa strains according to their capacity to form biofilms in terms of biomass production and metabolic activity
| CV | XTT | |||||
|---|---|---|---|---|---|---|
| Low producers | Moderate producers | High producers | Low producers | Moderate producers | High producers | |
| Controls, N (%) | 0 (0.0) | 8 (8.9) | 42 (46.7) | 18 (20.0) | 20 (22.2) | 12 (13.3) |
| Cases, N (%) | 1 (1.1) | 5 (5.6) | 34 (37.7) | 21 (23.0) | 14 (15.6) | 5 (5.6) |
| Total, N (%) | 1 (1.1) | 13 (14.5) | 76 (84.4) | 39 (43.3) | 34 (37.8) | 17 (18.9) |
N Number of strains; % Percentage of strains; CV Crystal violet; XTT Tetrazolium salt
In vivo virulence assay
Although only 8 strains from each group were compared in terms of virulence and no correlation between genetic background and the phenotype in vivo was observed (data not shown), we found significant differences between larvae mortality (p < 0.001). The median (IQR) time of survival for cases was 18 h (3.0) and for control strains 16 h (2.0) (Fig. 2). This means that P. aeruginosa strains causing VAP were less virulent than those isolated from other origins.
Fig 2.
Classification of pyoverdine and pyocyanin producer by qualitative observational method. a, pyoverdine producer strain; b, pyocyanin producer strain
Discussion
Our results show that the underlying pathogenicity mechanism of P. aeruginosa VAP is not restricted to biofilm production and specific virulence genes. However, VAP isolates did not produce pyocyanin after 24 h of incubation and showed lower virulence in the G. mellonella model than non-VAP strains.
Many studies have focused on the management of VAP owing to its impact in clinical settings [15–17]. However, few studies investigate the pathogenesis of P. aeruginosa VAP [7, 18, 19]. Hence, this study is the first to characterise a large sample of P. aeruginosa strains isolated from patients with VAP in comparison with strains from other sources from various hospitals in Europe, thus contributing to our understanding of the role of virulence mechanisms of P. aeruginosa in the pathogenesis of VAP.
P. aeruginosa is characterised by its capacity to form robust biofilms that play a substantial role in many human infections such as cystic fibrosis (CF), chronic wound infection, chronic otitis media, and implanted medical device infection [20]. One of the main risk factors for VAP is the presence of an endotracheal tube into which P. aeruginosa forms biofilms and enters the lower respiratory tract to cause infection [6]. We showed that in terms of biomass, high-biofilm-producing strains are implicated not only in VAP, but also in many other kinds of infections, although with no statistically significant differences, as reported by Rybtke et al. [20]. Moreover, the finding that most of the strains showed moderate or low metabolic activity when grown in biofilms may be considered a characteristic of P. aeruginosa biofilms. This characteristic confers intrinsic antibiotic resistance to biofilms, especially for antibiotics that target cellular processes associated with metabolic activity and active growth, thus rendering P. aeruginosa infections difficult to treat [20, 21].
Production of pigments such as pyoverdine and pyocyanin also play a key role in the virulence of P. aeruginosa [7]. Strains isolated from VAP patients showed a correlation with the absence of production of pyocyanin, and all the strains secreted pyoverdine after 24 h of incubation. Pyoverdine is a siderophore that chelates the iron needed for survival of P. aeruginosa [7]. The role of pyoverdine could explain why it is secreted by all VAP strains, as it is necessary for biofilm formation and hence for colonization of the endotracheal tube. However, pyocyanin is a redox compound involved in damage to lung epithelium and has been widely studied in cystic fibrosis [22, 23]. Although pyocyanin was not produced in VAP strains in our study, we hypothesized that pigment production could be a 2-step process, as pyocyanin has been reported to play an essential role in the pathogenesis of pneumonia [23]. Pyoverdine is necessary for colonization and biofilm formation and could be secreted initially, before synthetization of pyocyanin, leading to lung damage and pneumonia.
As for virulence factors, biofilm formation is regulated by many genes, such as algR (algU and algD), psl, and pel, which play a role in mucoidity and robustness of the extracellular matrix [21]. After detecting algU and algD, both of which are involved in alginate production, we observed that algD was present in all the strains, whereas the presence of algU was lower. algU is an extracytoplasmic sigma factor that is normally sequestered in the cytoplasmic membrane. Under specific conditions, it is released and activates overproduction of alginate [24]. This virulence factor plays a key role in the pathogenesis of cystic fibrosis by contributing to the mucoid phenotype of strains [24].
In addition to biofilm formation, the principal virulence factors of P. aeruginosa are elastase, phospholipase C, protease A, exotoxins and cytotoxins, flagella and pili, pigment production, and QS regulatory system proteins, which regulate both virulence factor transcription and biofilm formation [25]. Although it is well established that lung damage is caused by these proteins during pneumonia, their importance as virulence factors in VAP is unknown [8, 26]. We observed that there were no statistically significant differences in the presence of virulence genes between case and control strains. Our results corroborate the conclusion of Wolfgang et al., who maintained that the P. aeruginosa genome is highly conserved, at least for the 14 genes of this study, independently of the source of isolation [27].
Finally, we investigated the virulence of P. aeruginosa in an in vivo model based on G. mellonella larvae, which have been used widely to analyse the virulence of many microorganisms owing to their specific advantages [28]. We observed that VAP strains were less virulent than strains isolated from other infections, as previously reported by Wang et al. in an outbreak of VAP in a hospital in China, even though all strains presented similar patterns of virulence genes [29], indicating that virulence may be controlled by transcriptional regulation. Our results suggest that depending on the site of infection, particularly in the lung in VAP, modification of some virulence genes of P. aeruginosa combined with mutation of QS genes, as reported by Köhler et al., may play a key role in adaptation by bacteria [30]. Furthermore, although lung immune response is still under research, it plays a key role in the infection of P. aeruginosa in VAP patients [31]. Hence, alternative approaches to the diagnosis and treatment of VAP may rely on the development of an exhaustive epigenetic profile of VAP strains and a deep study of the lung immune system in mechanical ventilator.
Although ours is the first study to provide important information on the distribution of virulence factors in a wide range of VAP isolates of P. aeruginosa collected from hospitals in Spain and Italy, it is subject to limitations such as the functionality of the genes was not investigated by sequencing. However, the most important was the low number of strains tested in the in vivo model. However, although we only tested 8 strains in each group, we were able to find statistically significant differences. Increasing the sample size would only have increased the statistical power.
Conclusion
We demonstrated that the virulence and pathogenesis of P. aeruginosa in VAP infection does not depend on the presence of virulence genes or biofilm formation. However, these isolates were less virulent in vivo. Future studies are needed to elucidate how virulence is regulated during VAP and hence, improve the diagnosis and treatment of the disease.
Supplementary Information
Additional file 1. Supplementary material
Acknowledgments
We thank Thomas O’Boyle for writing assistance. We also thank Miss Cristina Iglesias-Arriba, Dr. Mercedes Marín, and Dr. Adoración Martín-Lopez for their help performing the PCR and Dr. Pablo Martín-Rabadán for providing the samples. Finally, we thank Dr. Jesús Guinea, Dr. Pilar Escribano, Judith Díaz-García and colleagues for providing G. mellonella larvae and for their help in the optimization of the in vivo model for P. aeruginosa.
Abbreviation
- VAP
Ventilator-associated pneumonia
Authors’ contributions
All authors have read and approved the manuscript. BA has made major contributions to the conception and design of the study, the acquisition, analysis and interpretation of the data and writing of the manuscript. LFB, EGDD, MM, EC, IM, MP and PM have made major contributions the acquisition of the data and writing of the manuscript. MG has made major contributions to the conception and design of the study and writing of the manuscript.
Funding
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
M. Guembe is supported by the Miguel Servet Program (ISCIIIMICINN, MS13/00268) from the Health Research Fund (FIS) of the Carlos III Health Institute (ISCIII), Madrid, Spain. B. Alonso was supported by the Consejería de Educación, Juventud y Deporte de la Comunidad de Madrid and Fondo Social Europeo (PEJ15/BIO/AI-0406). The study was partially financed by grants of the ISCIII (PI18/00045) and the European Regional Development Fund (FEDER) “A way of making Europe”.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
The strains collected during this study were obtained by regular diagnostic procedures of the involved institutions. This study was approved by the Ethic Committee of the Hospital General Universitario Gregorio Marañón (code: MICRO.HGUGM.2015–075). The Ethics Committee waived the need for consent due to neither the diagnosis nor the treatment were altered. Moreover the data of the patient was not exposed.
The study was in accordance with the provisions of the Biomedical Research Act 14/2007. The specifications of the OM SAS/3470 of 2009 were respected, as well as the provisions of the Law on Biomedical Research of 2007 and the Organic Law 15/1999, on the Protection of Personal Data.
Consent for publication
Not Applicable.
Competing interests
The authors declare that they have no conflicts of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Beatriz Alonso, Email: beaafm@hotmail.com.
María Guembe, Email: mariaguembe@hotmail.com.
References
- 1.Aykac K, Ozsurekci Y, Tanir BS. Future directions and molecular basis of ventilator associated pneumonia. Can Respir J. 2017;2017:2614602. doi: 10.1155/2017/2614602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kalanuria AA, Ziai W, Mirski M. Ventilator-associated pneumonia in the ICU. Crit Care. 2014;18(2):208. doi: 10.1186/cc13775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kalil AC, Metersky ML, Klompas M, Muscedere J, Sweeney DA, Palmer LB, et al. Management of Adults with Hospital-acquired and Ventilator-associated Pneumonia: 2016 clinical practice guidelines by the Infectious Diseases Society of America and the American Thoracic Society. Clin Infect Dis. 2016;63(5):e61–e111. doi: 10.1093/cid/ciw353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sarda C, Fazal F, Rello J. Management of ventilator-associated pneumonia (VAP) caused by resistant gram-negative bacteria: which is the best strategy to treat? Expert Rev Respir Med. 2019;13(8):787–798. doi: 10.1080/17476348.2019.1632195. [DOI] [PubMed] [Google Scholar]
- 5.Maurice NM, Bedi B, Sadikot RT. Pseudomonas aeruginosa biofilms: host response and clinical implications in lung infections. Am J Respir Cell Mol Biol. 2018;58(4):428–439. doi: 10.1165/rcmb.2017-0321TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fernandez-Barat L, Torres A. Biofilms in ventilator-associated pneumonia. Future Microbiol. 2016;11:1599–610. [DOI] [PubMed]
- 7.Berra L, Schmidt U, Wiener-Kronish J. Relationship between virulence factors and outcome of ventilator-associated pneumonia related to Pseudomonas aeruginosa. Curr Respir Med Rev. 2010;6(1):19–25. doi: 10.2174/157339810790820458. [DOI] [Google Scholar]
- 8.Gellatly SL, Hancock RE. Pseudomonas aeruginosa: new insights into pathogenesis and host defenses. Pathog Disease. 2013;67(3):159–173. doi: 10.1111/2049-632X.12033. [DOI] [PubMed] [Google Scholar]
- 9.Alonso B, Lucio J, Perez-Granda MJ, Cruces R, Sanchez-Carrillo C, Bouza E, et al. Does biomass production correlate with metabolic activity in Staphylococcus aureus? J Microbiol Methods. 2016;131:110–12. [DOI] [PubMed]
- 10.Stepanovic S, Vukovic D, Hola V, Di Bonaventura G, Djukic S, Cirkovic I, et al. Quantification of biofilm in microtiter plates: overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS. 2007;115(8):891–899. doi: 10.1111/j.1600-0463.2007.apm_630.x. [DOI] [PubMed] [Google Scholar]
- 11.Fazeli N, Momtaz H. Virulence gene profiles of multidrug-resistant Pseudomonas aeruginosa isolated from Iranian hospital infections. Iran Red Crescent Med J. 2014;16(10):e15722. doi: 10.5812/ircmj.15722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gholami A, Majidpour A, Talebi-Taher M, Boustanshenas M, Adabi M. PCR-based assay for the rapid and precise distinction of Pseudomonas aeruginosa from other Pseudomonas species recovered from burns patients. J Prev Med Hyg. 2016;57(2):E81–E85. [PMC free article] [PubMed] [Google Scholar]
- 13.Sabharwal N, Dhall S, Chhibber S, Harjai K. Molecular detection of virulence genes as markers in Pseudomonas aeruginosa isolated from urinary tract infections. Int J Mol Epidemiol Genet. 2014;5(3):125–134. [PMC free article] [PubMed] [Google Scholar]
- 14.Marcos-Zambrano LJ, Puig-Asensio M, Perez-Garcia F, Escribano P, Sanchez-Carrillo C, Zaragoza O, et al. Candida guilliermondii complex is characterized by high antifungal resistance but low mortality in 22 cases of candidemia. Antimicrob Agents Chemother. 2017;61(7):e00099–17. [DOI] [PMC free article] [PubMed]
- 15.Weiss E, Essaied W, Adrie C, Zahar JR, Timsit JF. Treatment of severe hospital-acquired and ventilator-associated pneumonia: a systematic review of inclusion and judgment criteria used in randomized controlled trials. Crit Care. 2017;21(1):162. doi: 10.1186/s13054-017-1755-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Landelle C, Nocquet Boyer V, Abbas M, Genevois E, Abidi N, Naimo S, et al. Impact of a multifaceted prevention program on ventilator-associated pneumonia including selective oropharyngeal decontamination. Intensive Care Med. 2018;44(11):1777–1786. doi: 10.1007/s00134-018-5227-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tokmaji G, Vermeulen H, Muller MC, Kwakman PH, Schultz MJ, Zaat SA. Silver-coated endotracheal tubes for prevention of ventilator-associated pneumonia in critically ill patients. Cochrane Database Syst Rev. 2015(8):Cd009201. [DOI] [PMC free article] [PubMed]
- 18.Sadikot RT, Blackwell TS, Christman JW, Prince AS. Pathogen-host interactions in Pseudomonas aeruginosa pneumonia. Am J Respir Crit Care Med. 2005;171(11):1209–1223. doi: 10.1164/rccm.200408-1044SO. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kohler T, Guanella R, Carlet J, van Delden C. Quorum sensing-dependent virulence during Pseudomonas aeruginosa colonisation and pneumonia in mechanically ventilated patients. Thorax. 2010;65(8):703–710. doi: 10.1136/thx.2009.133082. [DOI] [PubMed] [Google Scholar]
- 20.Rybtke M, Hultqvist LD, Givskov M, Tolker-Nielsen T. Pseudomonas aeruginosa biofilm infections: community structure, antimicrobial tolerance and immune response. J Mol Biol. 2015;427(23):3628–3645. doi: 10.1016/j.jmb.2015.08.016. [DOI] [PubMed] [Google Scholar]
- 21.Lee K, Yoon SS. Pseudomonas aeruginosa biofilm, a programmed bacterial life for fitness. J Microbiol Biotechnol. 2017;27(6):1053–1064. doi: 10.4014/jmb.1611.11056. [DOI] [PubMed] [Google Scholar]
- 22.Rada B, Leto TL. Pyocyanin effects on respiratory epithelium: relevance in Pseudomonas aeruginosa airway infections. Trends Microbiol. 2013;21(2):73–81. doi: 10.1016/j.tim.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lau GW, Hassett DJ, Ran H, Kong F. The role of pyocyanin in Pseudomonas aeruginosa infection. Trends Mol Med. 2004;10(12):599–606. doi: 10.1016/j.molmed.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 24.Stacey SD, Pritchett CL. Pseudomonas aeruginosa AlgU contributes to posttranscriptional activity by increasing rsmA expression in a mucA22 strain. J Bacteriol. 2016;198(13):1812–1826. doi: 10.1128/JB.00133-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Azam MW, Khan AU. Updates on the pathogenicity status of Pseudomonas aeruginosa. Drug Discov Today. 2019;24(1):350–59. [DOI] [PubMed]
- 26.Sawa T. The molecular mechanism of acute lung injury caused by Pseudomonas aeruginosa: from bacterial pathogenesis to host response. J Intensive Care. 2014;2(1):10. doi: 10.1186/2052-0492-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wolfgang MC, Kulasekara BR, Liang X, Boyd D, Wu K, Yang Q, et al. Conservation of genome content and virulence determinants among clinical and environmental isolates of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2003;100(14):8484–8489. doi: 10.1073/pnas.0832438100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsai CJ, Loh JM, Proft T. Galleria mellonella infection models for the study of bacterial diseases and for antimicrobial drug testing. Virulence. 2016;7(3):214–229. doi: 10.1080/21505594.2015.1135289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang K, Chen YQ, Salido MM, Kohli GS, Kong JL, Liang HJ, et al. The rapid in vivo evolution of Pseudomonas aeruginosa in ventilator-associated pneumonia patients leads to attenuated virulence. Open Biol. 2017;7(9):170029. [DOI] [PMC free article] [PubMed]
- 30.Kohler T, Buckling A, van Delden C. Cooperation and virulence of clinical Pseudomonas aeruginosa populations. Proc Natl Acad Sci U S A. 2009;106(15):6339–6344. doi: 10.1073/pnas.0811741106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Winter FHR, s Jongers B Bielen K, Mancuso D, Timbermont L, Lammens C, et al. Mechanical ventilation impairs IL-17 cytokine family expression in ventilator-associated pneumonia. 2019;20(20):5072. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Supplementary material
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.