Abstract
Biochemical reactions in eukaryotic cells occur in subcellular, membrane-bound compartments called organelles. Each kind of organelle is characterized by a unique lumenal chemical composition whose stringent regulation is vital to proper organelle function. Disruption of the lumenal ionic content of organelles is inextricably linked to disease. Despite their vital roles in cellular homeostasis, there are large gaps in our knowledge of organellar chemical composition largely from a lack of suitable probes. In this Outlook, we describe how, using organelle-targeted ratiometric probes, one can quantitatively image the lumenal chemical composition and biochemical activity inside organelles. We discuss how excellent fluorescent detection chemistries applied largely to the cytosol may be expanded to study organelles by chemical imaging at subcellular resolution in live cells. DNA-based reporters are a new and versatile platform to enable such approaches because the resultant probes have precise ratiometry and accurate subcellular targeting and are able to map multiple chemicals simultaneously. Quantitatively mapping lumenal ions and biochemical activity can drive the discovery of new biology and biomedical applications.
Short abstract
The chemical environment within most organelles is still unknown. Quantitative chemical imaging in live cells with DNA-based fluorescent reporters is revealing new chemistry and biology of organelles.
Introduction
We are poised at a very interesting time-point in cell biology, where we are discovering how the rich biology of single cells can drive emergent phenomena based on the coordinated responses of many connected cells. This renewed interest in cell biology has been empowered by successful molecular and chemical technologies. For example, genetic engineering has given us fluorescent proteins of any desired wavelength.1−3 With gene editing,4 one can now make previously prohibitively difficult cellular expression systems tractable.5−7 Super-resolution microscopy, expansion microscopy, and cryoelectron microscopy are now revealing subcellular architecture and organelle connectivity with unprecedented resolution.8−12 With computational algorithms and deep learning applied to big data sets, we can extract any desired parameter from thousands of images.13 When overlaid with proteomics, these approaches provide a molecular picture of organelle form and function that was simply impossible to visualize even two decades ago.14−16 Indeed, disruptions in organelle movement, morphology, fusion, fission, and trafficking are tied to protein composition of the organelle.17−19 These methods are converging to reveal the different ways in which organelle physiology regulates cell physiology.
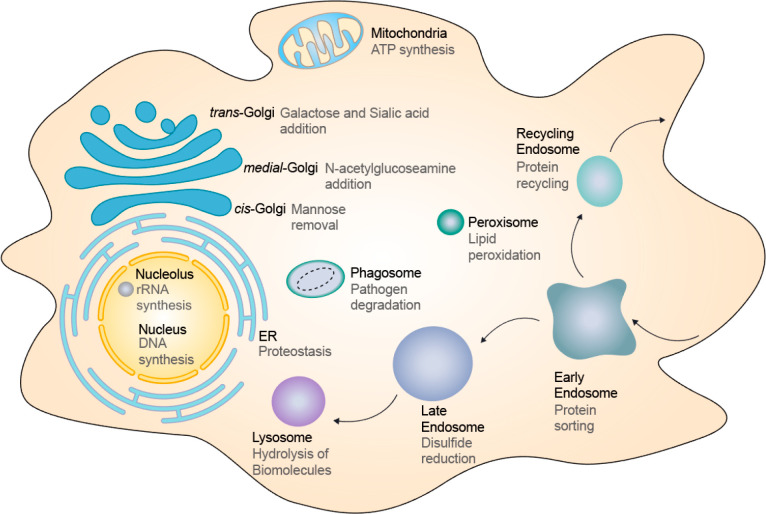
One of the aspects of the cell that we still do not know very much about is the lumenal chemical environment of organelles. This knowledge could potentially add a new layer to inform how proteins might function in organelles or how whole organelles might function. In terms of fundamental chemistry, organelles can be considered as different chemical reactors. Within these chemical reactors, the lumenal environment or “solution conditions” within promote specific chemistries unique to the organelle while simultaneously disfavoring other biochemistries that occur in related organelles.20 Indeed, in the endoplasmic reticulum, peptide bonds are being formed, and in the Golgi, proteins are being glycosylated; in the peroxisome, long fatty acid chains are broken down into shorter ones, and in the lysosome, proteins are degraded into amino acids.21 Therefore, we posit that the set of all ion channels and transporters on a given organelle gives rise to an emergent property—sculpting a lumenal ionic environment uniquely suited to the internal biochemistry of the organelle (Figure 1).
Figure 1.
Organelles and their lumenal chemistries. Schematic of various organelles within the eukaryotic cell (black font). Organelles are specialized subunits, and each organelle executes specific biochemistries (gray font) enabled by its unique lumenal chemical environment to maintain cellular homeostasis.
Excellent chemical and genetically encodable probes for pH reveal that when organelles malfunction, the lumenal pH is often disrupted.22,23 However, we have no knowledge of what happens to other ions that make up the lumenal microenvironment. This is important particularly in organelles because the level of every major ion such as H+, Ca2+, Cl–, Na+, or K+ is dependent on the level of at least one other ion. These ions are considered the major ions as they are the most abundant; their transport is crucial for setting up membrane potential, and they are also well-accepted second messengers that initiate signaling pathways in development and disease. For many diseases, especially neurodegenerative diseases, organ dysfunction can be traced right down to a dysfunctional organelle in the cells comprising that tissue.24,25 Lumenal ionic imbalances can be caused by dysfunctional ion channels or transporters that fail to bring in specific ions with the right efficiencies,26,27 or defective membrane proteins that change the permeability of the organelle membrane to different ions.28 The altered lumenal environment would broadly impact biochemical reactions within the lumen and impede cell function. Thus, if we could develop ways to measure the levels of organellar ions in living cells, it would help us identify molecules that could reset the balance of key ions in dysregulated organelles, rescue the affected organellar biochemistries, and ameliorate disease.29
In this Outlook, we describe the development of an emerging class of reporters based on DNA specifically suited to quantitatively image chemicals within organelles that was previously inaccessible to precise measurement. Since this DNA-based, chemical imaging technology is modular, most new chemical sensors can be integrated to image the relevant analyte in a range of organelles currently accessible to the technology. At the same time, accessing a new organelle opens up the possibility to study a range of new chemistries for which DNA reporters are already available. Using DNA reporters, one can image ions, reactive second messengers, and even enzymatic activity in organelles.30−32 We will discuss how one can construct a quantitative “chemical heatmap” of subcellular organelles starting from the development of a small-molecule dye that can fluorescently report a given chemical. As to how the DNA reporters can be directed to other organelle lumens, the readers are directed to a forthcoming sister review in Nature Reviews Materials. For this Outlook, the reader is advised to simply note that DNA reporters can be targeted to a specific organelle.
Principle of Ratiometry and Early Reporter Technologies for Chemical Imaging
Ratiometry Enables Quantitation
The 1:1 ratio of sensing
fluorophore and a reference fluorophore is crucial for accurate quantitation
if the sensing dye is not intrinsically ratiometric. At fixed temperature
and standard experimental conditions, the absolute intensity of the
sensing fluorophore (Is) is affected by
both analyte concentration, [a], and sensor concentration, [s] (Figure 2a). Thus, Is ∝ [s][a]k where k is a value that is determined by the nature
of the sensing mechanism, where k > 0 for a turn-on
sensor and k < 0 for a turn-off sensor. It is
impractical to control [s] across different live cells due to their
intrinsic heterogeneity in endocytic uptake and relative internal
trafficking efficiencies. However, adding another fluorophore, that
is insensitive to environmental chemicals in a 1:1 ratio to the sensing
fluorophore, can normalize for the contribution of [s] to the signal
intensity, Is. The intensity of such a
normalizing fluorophore, In, is proportional
only to the amount of sensor, i.e., In ∝ [s]. Thus, the ratio Is/In yields a normalized intensity 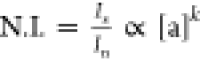 that
is proportional only to the amount
of analyte and can be used to derive [a] quantitatively. This principle
can be demonstrated with and without ratiometry regardless of analyte
concentration (Figure 2a).
that
is proportional only to the amount
of analyte and can be used to derive [a] quantitatively. This principle
can be demonstrated with and without ratiometry regardless of analyte
concentration (Figure 2a).
Figure 2.
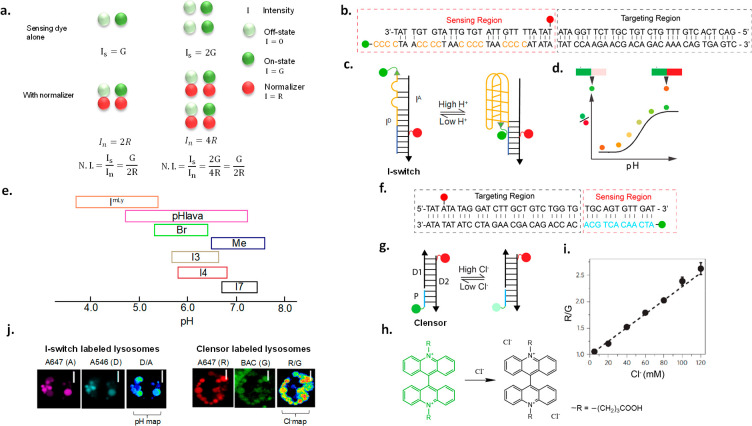
Ratiometric quantification and its application to DNA reporters. (a) Ratiometric imaging corrects for sensing fluorophore intensity differences which arise due to cell-to-cell heterogeneity. (b, c) Sequence and structure of I-switch. Alexa 546 is the donor dye on ID, and Alexa647 is the acceptor dye on IA. (d) D/A ratio of I-switch increases with the increase in pH. (e) Available and characterized I-switch variants and their pH-sensing regimes. (f, g) Sequence and structure of Cl– sensing DNA probe, Clensor. Schematic of Clensor showing Alexa647 as a normalizing fluorophore (red) and BAC as a sensing fluorophore (green) attached to a PNA strand (blue). (h) Clensor reports on Cl– by collisional quenching of fluorophore BAC. (i) Clensor ratiometric response (R/G) increases with Cl– concentration. (j) Construction of heatmaps of I-switch and Clensor in lysosomes of C. elegans obtained by dividing D to A images and R to G images, respectively.
Small-Molecule-Based Chemical Reporters
Several excellent examples exist of sensitive and specific sensors that are based on small molecules that report on diverse ions, reactive species second messengers, and other cellular analytes.33−35 Small-molecule sensors are bright and have better cell permeability than biological macromolecules, high selectivity to the analyte that they report, and high responsivity or dynamic range. By integrating excellent detection chemistries to well-known fluorophore cores, one can achieve high molar brightness that surpasses most fluorescent proteins.34,36 The high molar brightness that turns on due to either the relief of photoinduced electron transfer (PeT) or uncaging leads to dynamic ranges that are simply not accessible to fluorescent proteins.37,38 The introduction of cleavable ester groups can enhance the cell permeability of most small-molecule sensors.39 However, most small-molecule sensors are single wavelength, which limits the ability to quantitate analyte levels in cells. Further, they diffuse rapidly and homogeneously throughout the cell which obscures valuable spatial information.
Genetically Encodable Fluorescent Proteins
In terms of spatial addressability, nothing supersedes genetically encodable fluorescent proteins (FPs).5,40 In general, plasmids encoding FPs must be transfected or genomically integrated and then undergo transcription and translation to yield the fluorescent reporter. FPs can be targeted to subcellular locations within the cell or tissue of choice in vivo(41−43) and therefore provide spatial and temporal information on cell or tissue morphology or protein expression. However, FPs are limited in terms of the chemical information they can provide. Organelle-targeted photoactivatable FPs provide information on their biogenesis and maturation,44,45 but measures of absolute protein activity within organelles have not yet been achieved.46 Most FPs are pH sensitive in the acidic regime and therefore have been used as effective reporters of acidic pH by utilizing a second fluorescent protein that is less sensitive to pH as a ratiometric signal.47 Genetically encoded Ca2+ indicators and genetically encoded voltage indicators have also been realized, but these cannot yet report on absolute values of calcium or membrane potential.48−50 Recently, a few FP reporters have been realized for K+, reactive oxygen species, and nitric oxide (NO) by fusing two different FPs, and this enables ratiometric imaging.42,51−53 However, the pH sensitivity of these reporters limits their deployment in organelles, whose lumens are frequently acidic.
Hybrid Reporters
The disconnect between small molecules and FPs was somewhat solved by a distinct class of reporters that are hybrids between fluorophores that are sensitive to an analyte, e.g., pH or Cl–, and macromolecules such as dextrans or proteins, where the latter act as endocytic tracers.19,54 Fluorescein-labeled dextran is, in fact, the most widely used reagent for imaging pH in endocytic organelles.55 Tagging fluorescein to transferrin, a ligand for the transferrin receptor, has been used to label and measure the pH of the recycling endosome.55 Conjugating pH or chloride-sensitive fluorophores to endocytic tracer proteins such as choleratoxin B (CTxB) has been used to estimate pH and chloride in organelles such as the Golgi apparatus in the secretory pathway.56 In such probes, the small-molecule reporter provides the chemical imaging capability whereas the protein or macromolecule provides stable and robust spatial confinement within the cell, so that the lumen of the organelle can be chemically imaged.
Generally, reporter dyes are attached to the protein of interest using N-hydroxysuccinimide or thiol-maleimide chemistries by conjugation in vitro. This labels a fraction of all the exposed lysines or cysteines respectively on the protein. This leads to sensors with batch-to-batch variability in terms of reporter stoichiometry which further complicates quantitative imaging due to the presence of only a single fluorophore. The alternative is the introduction of a second external reference dye, frequently by comixing with a protein labeled with a second ion-insensitive fluorophore to give a two-component reporter system.19,56 This further increases the variability, since both components can distribute nonuniformly across organelles within the same cell. In fact, images of organelles that contain only one of the components are excluded from subsequent analyses.19,57 However, these quasiratiometric approaches are important as they provided pioneering estimates of organelle ionic composition.19,54
DNA-Based Ratiometric Reporters
DNA-based reporters integrate the specificity, sensitivity, and photophysical properties of small-molecule fluorescent probes with the stable subcellular localization afforded by biologics.58,59 DNA synthesis is facile, pure, and economical. Further, the 1:1 ratio of Watson–Crick–Franklin base-pairing in a DNA duplex makes it an ideal scaffold to develop ratiometric reporters. This makes DNA-based ratiometric reporters molecularly precise, have negligible batch to batch variation, and specifically modifiable at any given nucleotide position.60,61 Ratiometric DNA reporters can be easily made by annealing complementary oligonucleotides each conjugated to a sensing and a reference fluorophore, respectively. A good reference fluorophore should be insensitive to ions and reactive species in the cellular environment. It is always present in known stoichiometry, typically 1:1, with the sensing fluorophore in the DNA reporter, and therefore also in every organelle containing the reporter.62 It can hence be considered an internal reference for normalization. Ratiometric imaging corrects for sensor intensity differences arising due to nonuniform probe uptake, trafficking differences among cells, and other parameters that cause inhomogeneous probe distribution due to cell-to-cell heterogeneity and enables quantitation.63
The modularity of the DNA scaffold enables one to integrate different modules with independent and dependent functions, with stoichiometric precision on a single scaffold.64 There are a library of DNA and RNA sequences encompassing an array of sensing capabilities that can be used as independently functioning modules.30,58,65−69 Thus, precharacterized modules may be integrated or substituted to give a new device.70,71 This opens up a panoply of combinations for sensing, normalizing, and targeting, facilitating the quantitation of any analyte for which detection chemistry already exists, in a range of biological contexts. Thus, ratiometric DNA-based reporters are targetable, modular, and programmable and present a versatile platform for selective, subcellular quantitative imaging of ions and small molecules in cells and in vivo.
Subcellular Targeting of DNA Nanodevices
DNA nanodevices localize inside a given organelle by displaying specific targeting motifs. Thus, in addition to a reporter module, DNA devices frequently incorporate a targeting module.58,72 The targeting module is a domain displayed on the DNA reporter scaffold that encodes trafficking information. Thus, when added to the cell culture milieu, the DNA reporter binds a specific cell-surface protein through its targeting module and gets transported to a specific endocytic organelle. Even in Nature, pathogens such as bacteria, fungi, viruses, and their secreted toxins have designated destinations within specific cells.58,59,73,74 They contact the cell membrane, dock onto specific cell-surface trafficking proteins, and coopt the associated trafficking route of the latter to reach a target organelle.75−77 DNA reporters can display these same molecular signals to traffic into specific organelles.78
DNA is a natural endocytic ligand for scavenger receptors, also known as anion ligand binding receptors (ALBRs). The polyanionic phosphate backbone binds ALBRs and enters the endolysosomal pathway by receptor-mediated endocytosis. Therefore, DNA sensors can be specifically targeted to label every organelle stage on the endolysosomal pathway such as early endosomes, late endosomes, or lysosomes in a time-dependent manner.58 A broader framework and more general molecular logic underlying various targeting strategies are described in a forthcoming review in Nature Reviews Materials in 2021.
Quantitative Imaging with Reversible Sensors
Selection of a Compatible Sensing Fluorophore
Many fluorophores do not change their chemical sensing properties upon conjugation to the DNA backbone.65,71 However, most fluorophores get quenched to different degrees thus changing their sensitivity to the analyte of interest. A few photophysical properties like quenching by PeT or collisional quenching are affected by proximity to nucleobases or the negatively charged phosphate backbone. These issues can be circumvented by separating the fluorophore and the DNA assembly by an inert linker (e.g., PNA and PEG).31,65,79 Integrating fluorophores to DNA nanodevices could expand the use of reporters to a variety of biological contexts. For example, two-photon active dyes could enable deep tissue imaging.80 FLIM-compatible fluorophores would enable FLIM imaging in systems with high autofluorescence,81 and positron emission tomography (PET) active small molecules could be used for in vivo imaging in larger vertebrates.82
Design and Working Principle of DNA-Based Single Ion Reporters
In this section, two different strategies are described to demonstrate how one can use DNA reporters for quantitative chemical imaging. One is a pH reporter where pH changes the conformation of DNA leading to a fluorescence change, and the other is a Cl– sensor based on collisional quenching of a chloride-sensitive dye.65,72 Whether DNA is actively involved in the sensing mechanism or acts as a passive scaffold that positions various dye molecules, ratiometric quantification allows the unambiguous determination of environmental pH or chloride levels.
DNA-Based pH Reporter
The I-switch is a ∼60 bp DNA duplex comprising two oligonucleotides ID and IA that, when hybridized, can report environmental pH. The I-switch comprises two domains: a sensing domain and a targeting domain (Figure 2b).72
Sensing Domain
ID has a cytosine-rich 5′ region (orange font, Figure 2b) which folds into a four-stranded structural motif called an i-motif at acidic pH.83 This pH-driven conformational change is transduced into a change in fluorescence properties by incorporating a FRET pair on ID and IA.84,85 ID bears a donor fluorophore (Alexa 488, D) at its 5′ end. An acceptor (Alexa647N, A) is optimally positioned on IA such that when the I-switch “closes” at ∼pH 5, D and A are proximal and show high FRET (Figure 2c). At pH 7, it is in an “open” conformation with no FRET. The sensor domain is a mismatched duplex that, at pH 7.0, positions D and A out of FRET distance.
The pH responsive regime of an I-switch is determined from an in vitro calibration curve corresponding to the ratio of D/A intensities as a function of pH. The linear region where D/A changes with pH corresponds to the pH regime in which a pH reporter may be deployed (Figure 2d). The pH responsive regime in I-switches may be tuned by (i) changing the number of cytosines, (ii) using chemically modified cytosines, or (iii) changing the number and position of mismatches in the sensor domain (Figure 2e). The tunability of DNA has led to a range of I-switch variants that cover different pH regimes.86,87 Further, since the I-switch works by FRET, it is not limited to specific dyes, and one can use FRET pairs of different wavelengths that are compatible with diverse fluorescent protein backgrounds. One can envisage other pH sensors based on DNA that use pH-sensing dyes such as pHrodo or Oregon Green 488 as seen in pHlava or ImLy (Figure 2e).24,31
Targeting domain
The lower half of the I-switch is a 27-mer duplex (Figure 2a). This domain prevents the dissociation of ID and IA when the sensor domain forms the “closed” state and also harbors organelle targeting moieties (Figure 2b).
DNA-Based Chloride Reporter
Clensor is a pH-independent chloride sensor. It comprises three sequences: a 12-mer peptide nucleic acid (PNA, P), a 26-mer DNA (D1), and a 38-mer DNA (D2). Clensor too comprises sensing and targeting domains (Figure 2f).65
Sensing Domain
This is a 12 mer PNA–DNA duplex of P and D2, where P bears a Cl–-sensitive dye called 10,10′-bis[3-carboxypropyl]-9,9′-biacridinium dinitrate (BAC), and a D2 bears a Cl–-insensitive, normalizing dye Alexa 647 (Figure 2g). BAC is collisionally quenched by Cl– ions (Figure 2h).56,88 PNA conjugation moderately preserves the Cl– sensing abilities of BAC which are very badly affected when conjugated to DNA.65,89 The high-melting PNA–DNA duplex positions the normalizing dye, Alexa647N, out of FRET range from BAC, and the duplex DNA domain incorporates the targeting module. An in vitro calibration plot is generated from the ratio of fluorescence emission intensities of BAC (G) to Alexa647N (R) as a function of Cl– concentration (Figure 2i).
Constructing Chemical Heatmaps with Ratiometric DNA Reporters
This is a two-step process. First, the performance characteristics of DNA-based probes in cells or in vivo are determined through rigorous calibration. Next, these calibration profiles are used to quantitate lumenal ions in organelles in situ.
In situ calibration profiles for DNA reporters in organelles yield the expected values of R/G or D/A for Clensor or an I-switch variant when organellar Cl– or pH is fixed to known values, in a given in vivo context and in a given instrumental setup. Typically, endosomes of the cells of interest are first labeled by any of the targeting mechanisms. The labeled endosomes are then “clamped” by artificially setting the lumenal ionic composition to known pH or Cl– values.30,62,65 Typically this is achieved by soaking labeled cells or nematodes for 70 min in clamping buffer of designated pH (or designated Cl–) containing a mixture of ionophores that act to equilibrate the lumenal pH (or Cl–) value to that in the clamping buffer.62,65 Note that nematodes or cells that are pH or Cl– clamped are not in their physiological state. However, the purpose of the in vivo clamping experiment is to evaluate probe performance and check probe integrity, at fixed pH or Cl– values, prior to chemically imaging the living system in a physiological scenario.
Fluorescence images of clamped cells are acquired in the donor (D) and acceptor (A) channels for pH imaging with the I-switch or the BAC (G) and the normalizing dye (R) channels for Cl– imaging with Clensor (Figure 2j). In order to construct a pH map of I-switch-labeled endosomes, the ratio of the D and A intensities at every pixel is computed and converted into a spatial D/A image, with the values of D/A pseudocolored according to their magnitude to generate a pseudocolor pH map, also called a pH heatmap. A typical example of a pH image is shown in Figure 2j. A similar operation on R and G images with Clensor yields an R/G or Cl– heatmap. The R/G or D/A values of ∼50 endosomes are computed for each value of pH or Cl– and plotted as a function of pH or Cl– to yield the in-cell or in vivo calibration profile for the DNA reporter. The fold change (FC) in the signal is defined by the ratio of the maximum to the minimum values of D/A for pH or R/G for Cl–. If the integrity of the DNA reporter is maintained within the living system, then the in vivo or in-cell FC would match the in vitro value.
In situ chemical maps of organelles in the desired context are constructed by first labeling with the I-switch or Clensor and then imaging in the D and A channel for pH or R and G channel for Cl–. The obtained D/A or R/G values are compared with the in-cell calibration profile which provides the pH or Cl– value in the labeled organelle.
Combination DNA Reporters and Two-Ion Measurement (2-IM)
Accurately measuring key physiological cations such as Ca2+, Na+, K+, etc. within organelles is not feasible unless one can simultaneously measure pH for the following reasons. First, the entry of a given ion into an organelle is tied to the pH within that organelle. For example, Ca2+ entry into the lysosome or Na+ entry into the recycling endosome is coupled to its lumenal pH.90−92 Thus, if the entry or exit of a given ion is affected, the lumenal pH is concomitantly affected. Second, many ion-sensitive fluorophores sense the relevant ion via protonatable groups that coordinate the ion via electron pairs that quench a fluorophore core via photoinduced electron transfer (PeT).35,38,93,94 The binding of the specific ion engages the lone pairs and thus turns on the fluorescence by relieving the PeT. For example, Ca2+-sensitive fluorophores have carboxylic acid groups that coordinate Ca2+.1,3,34,37 Changes in environmental pH in the acidic regime change the extent of PeT efficiency, rendering the fluorescence readout ambiguous.34 Finally, the presence of protonatable groups in the sensing moiety also changes the binding affinity (Kd) toward the specific ion. For example, protonation of carboxylic acid groups in the Ca2+ sensing fluorophore increases the Kd as the acidity of the milieu increases. This makes quantitation by the dye inaccurate because, apart from fluorescence intensity, one needs to account and correct for the contribution of pH in order to accurately estimate Ca2+ concentration. Therefore, the ideal approach is to simultaneously measure two coupled ions in the same organelle, with single organelle resolution.
Two-Ion Measurement (2-IM).
With combination DNA reporters, one can now measure two different ions in the same organelle simultaneously, such that the information on concentration of each ion is addressable at the single organelle level.65,72 This new imaging modality is denoted two-ion measurement (2-IM), and although described first for pH and Cl–, it can be applied to any two ions. Typically, a 2-IM probe harbors at least two reporter domains; one for each ion reports on each ion quantitatively, and the readout from one reporter domain does not interfere with the other. For example, the 2-IM probe ChloropHore is a combination sensor for pH and Cl–, where sensing mechanisms of either ion are independent of each other (Figure 3a).70
Figure 3.
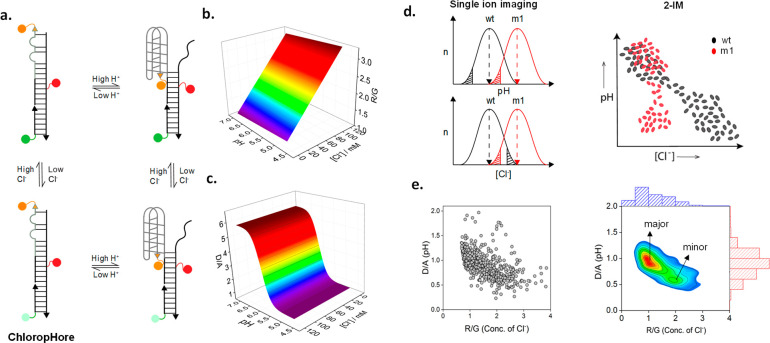
Two-ion measurement (2-IM) using combination ion sensors. (a) ChloropHore measures pH and Cl– simultaneously to chemically resolve organelle states. (b) R/G reports Cl– independently of pH. (c) D/A reports pH independently of Cl–. (d) Single ion imaging of organelle pH or Cl– mask organelle populations due to averaging effects, but 2-IM resolves them. (e) Raw 2-IM data showing D/A (pH) and R/G (Cl–) from ∼600 lysosomes in ∼30 fibroblasts, converted into a density plot (right) showing major and minor lysosome populations.
The DNA scaffold is modular and enables one to display an I-switch reporter module for pH sensing and the Clensor reporter module for Cl– sensing into a single DNA assembly as in ChloropHore. Importantly, the in vitro pH and Cl– sensing characteristics in ChloropHore are identical to those of the corresponding I-switch and Clensor, respectively.65,72 Thus, neither module affects the performance of the other when integrated into a single assembly. To characterize a 2-IM reporter in vitro, the concentration of both constituent ions must be fixed. The pH sensitivity of the I-switch domain must be tested at a range of Cl– concentrations covering the physiological regime, and the Cl– sensitivity of the Clensor domain must be tested over the complete physiological pH regime. Thus, 2-IM probes will yield calibration surfaces rather than calibration curves (Figure 3b,c). Similarly, in-cell calibrations must use a full range of clamping buffers to assess probe characteristics across the entire physiological regime of pH and Cl–.70
One of the applications of 2-IM probes is to chemically resolve organelles into subpopulations. Organelle subpopulations are difficult, if not impossible, to resolve in live cells by mapping only one ion because of averaging effects (Figure 3d). As in bulk biophysics, averaging effects tend to obscure the contribution of different functional subpopulations.95 However, single molecule biophysics resolves functional conformational populations and thereby yields insights into structural mechanisms of biomolecules that are simply not accessible with bulk biophysics.96 Similarly, 2-IM enables organelle subpopulations to be chemically resolved in situ because the information on ion-levels is addressable at the single organelle level (Figure 3d).
As an illustrative example, ChloropHore-labeled lysosomes could be resolved by 2-IM. Lysosomal activity is facilitated by its unique ionic milieu maintained, in turn, by a specific lysosome membrane protein composition. Many cell types have specialized lysosomes that perform distinct functions, in addition to normal lysosomes. For example, skin cells have melanosomes,97 neutrophils have azurophil granules,98 and cytotoxic T-cells have secretory lysosomes.99 Specialized lysosomes are distinguished by specific membrane protein markers, revealing a slightly different protein composition and biochemical activity from normal lysosomes.100,101 Therefore, lysosomes can be chemically resolved into subpopulations based on their ion content. By imaging lysosomes in four channels corresponding to D, A, R, and G, one builds D/A maps corresponding to pH and R/G maps for the same lysosomes corresponding to Cl–. By plotting the value of lysosomal pH versus the corresponding value of Cl– per lysosome for about ∼600 lysosomes, one can generate a scatter plot (Figure 3e). Converting the scatter plot into a density plot yields a 2-IM profile for lysosomes (Figure 3e).70 Two distinct peaks in the density plot correspond to two different lysosome populations.
One can better understand why 2-IM is so effective at resolving organelle populations by analogy with 1D and 2D gel electrophoresis (2D-GE).102 In 2D-GE, proteins that cannot be separated in 1D by their isoelectric point are resolved in the second dimension by separation based on their mass. Similarly, organelle populations that are difficult to resolve using a single ion can be resolved in another dimension based on a second ion.
pH-Corrected Ion Maps
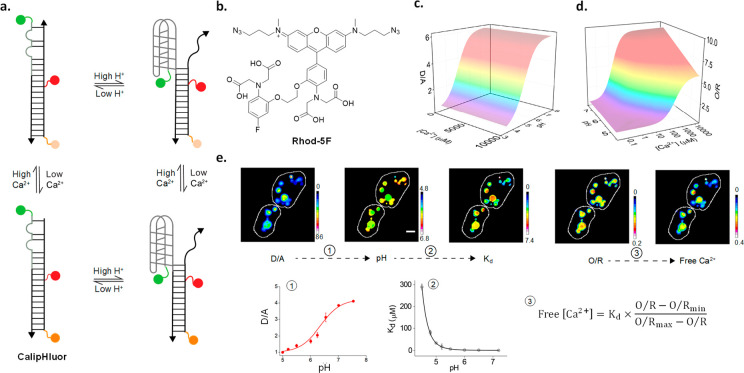
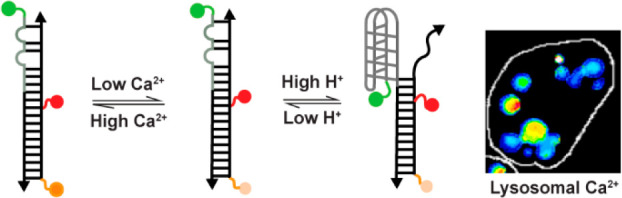
Many ion-sensitive dyes cannot be used in organelles, as they are pH-sensitive. Using a combination DNA reporter for Ca2+ and pH, denoted CalipHluor, we illustrate how pH-corrected maps of organellar ions can be obtained using such dyes (Figure 4a).71 This methodology can be applied to those organelles that can be labeled by DNA reporters. Lumenal Ca2+ regulates the function of several organelles. One can modify an I-switch of the appropriate pH sensitivity to the organelle of interest with a Ca2+-sensitive dye (Figure 4c,d). One should also have a prior rough estimate of the range of Ca2+ in the compartment and choose a Ca2+-sensitive dye of appropriate dissociation constant (Kd). For example, to map Ca2+ in lysosomes of nematodes, CalipHluor incorporates a novel Ca2+-sensitive fluorophore Rhod-5F which has a Kd of 330 μM at pH 5.0, which corresponds to lysosomal pH in Caenorhabditis elegans (Figure 4b).62
Figure 4.
Ion measurement using a pH-correctable paradigm. (a) Design and working principle of the pH-correctable Ca2+ reporter, CalipHluor. (b) Structure of the Ca2+ sensing fluorophore, Rhod-5F. 3D calibration surfaces of (c) D/A and (d) O/R as a function of pH and Ca2+. (e) Image analysis workflow for heatmaps of free Ca2+ in lysosomes. The pH map is obtained from the D/A versus pH curve as in 1. Then, the Kd map is generated from a function describing pH versus Kd as is 2. The Ca2+ map is obtained from the O/R map and Kd map using eq 3.
To obtain a map of pH-corrected lysosomal Ca2+, we first obtain images of CalipHluor-labeled lysosomes in the four channels, D, A, O, and R, where O and R correspond to the emission of Rhod-5F and the reference dye under direct excitation. From the D and A images, one can generate a D/A map. The D/A map can in turn be transformed to a “pH map” where the D/A value at every pixel is substituted with its corresponding pH value (Figure 4e).71 From the in vitro and in-cell calibration, surfaces at all physiologically relevant combinations of Ca2+ and pH provide information on how the Kd of Rhod-5F varies as a function of pH. Similarly, the pH map can be transformed to a Kd map (Figure 4e). Thereafter, once the values of the minimum and maximum values of O/R are experimentally determined, one can generate a pH-corrected Ca2+ map by taking the pixel-wise product of the Kd value and the O/R value from the Kd and the O/R maps, respectively, using the expression (O/R – O/Rmin)/(O/Rmax – O/R) (Figure 4e).71 Such an approach can be used to obtain ion heatmaps as well as absolute values of ion content and is generalizable across ions.
Quantitative Imaging with Irreversible Reporters
Quantitating Enzymatic Cleavage
Perturbations in organellar ionic concentrations affect the lumenal activity of enzymes residing in the relevant organelle.24,103 Thus, the ability to quantitate enzymatic activity exclusively in a given organelle would provide insight into how its activity impacts organelle function. It could be used as a diagnostic for diseases arising due to enzyme dysfunction and also as a tool to develop therapeutics for these diseases. It is extremely challenging to develop reversible reporters of enzymatic activity. This is because enzyme activity involves more than just reversible binding of a chemical entity; it usually converts the bound substrate into a different entity by a chemical reaction. Thus, most reporters of enzymatic activity are either irreversible or quasireversible.104
In this section, we outline a paradigm to quantitate enzymatic activity at subcellular resolution using irreversible reporters. Nearly 70% of all the enzymes in biosynthetic pathways are distributed into distinct subcellular pools.105 These distinct pools play different roles depending on their location. Thus, the ability to map their activity with subcellular resolution will help deconvolute their contribution to various biological processes.106 A well-developed protein-based reporter platform for enzymatic activity includes kinase activity probes that have been used to study the dynamics of diverse kinases such as PKA, PKC, ERK, and Src kinase in live cells.107,108 So far, FP-based sensors simultaneously report the overall activity within the cell. While such reporters can effectively track the activity of the major populations of enzyme, contributions from minor populations are overwhelmed in these scenarios and not amenable to investigation.
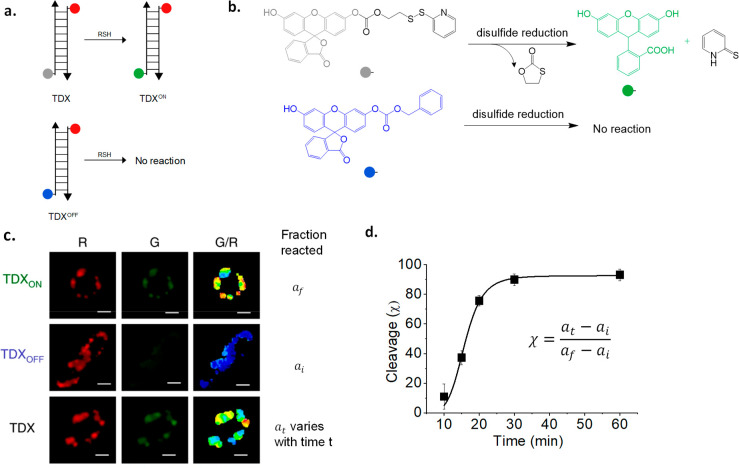
DNA reporters targeted to organelles can selectively address the activity of intraendosomal disulfide reductases. The major disulfide reductases protein disulfide isomerase-3 (PDI-3) and thioredoxin-1 (TRX-1) were shown to have minor populations present in endosomes using a DNA-based reporter for disulfide reduction, denoted TDX (Figure 5a).32 Disulfide reduction is sensed by a caged fluorescein derivative, where fluorescence is quenched by a carbonate linked to a 2-thiopyridyl group through a disulfide linkage (Figure 5b).109 Reduction of the disulfide bond chemically uncages the fluorescein moiety. The DNA reporter TDX uses Rhodamine Red X as a normalizing fluorophore.32,110
Figure 5.
Visualization of enzyme activity maps exclusively in organelles enabled by DNA reporters. (a) Design of the tripartite reporter system TDX, TDXON, and TDXOFF that reports on endosomal disulfide reduction. (b) Working principle of the disulfide-sensitive fluorophore. (c) Representative images of the ratiometric thioreductase activity maps in endosomes of C. elegans coelomocytes as G/R heatmaps. (d) Intraendosomal disulfide reduction as a function of time in late endosomes.
The TDX reporter is a tripartite system, that uses two additional
DNA reporters: one which reports the maximum value of G/R corresponding
to 100% disulfide reduction of the sensor, called TDXON, and a second that reports the minimum value of G/R corresponding
to basal levels of nonspecific uncaging of the sensor, called TDXOFF (Figure 5a,b). Worms or cells are treated separately with each of the three
reporters, TDXON, TDXOFF, and TDX; once they
label the organelle of interest, they are imaged in both channels,
and G/R values are computed for labeled endosomes (Figure 5c). The percentage response
of the reporter TDX in any context may be calculated using the formula 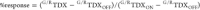 (Figure 5d).32
(Figure 5d).32
This method revealed that endosomal disulphide reduction was brought about mainly by two thioreductases, TRX-1 and PDI-3. Although TRX-1 and PDI-3 are mainly present in cytosol and ER, respectively, the tripartite reporter system is able to exclusively interrogate a minor population of these proteins localized in late endosomes.
Quantitating Reactive Species
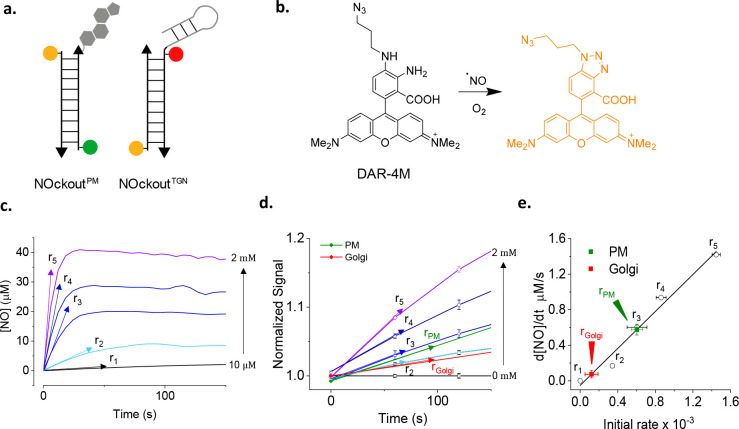
Note that genetically encoded reporters of enzymatic activity generally reveal steady state activity because they are expressed constitutively rather than on inducible promoters. Thus, unless the enzyme of interest can be activated by a biochemical cue, reporting cannot be initiated at a specific time. The advantage of an externally added reporter that undergoes enzymatic cleavage is that one can use a kinetic paradigm to quantitate enzyme activity. DNA reporters have the advantage of being extraneously introduced. Thus, the dynamic evolution of the reporter signal may be used to quantitate enzyme activity at basal levels or upon stimulation. A case in point is the utilization of DNA-based reporter NOckout that senses NO•, to quantitate eNOS or NOS3 activity (see later) at two different subcellular locations (Figure 6a).79
Figure 6.
Quantitatively mapping the reactive species, NO, at subcellular resolution. (a) Design of DNA reporters for NO, NOckout sensors. NOckoutPM reports NO at the plasma membrane while NOckoutTGN does so at the trans-Golgi network. (b) Working principle of the NO sensing dye, DAR-4M. (c) Rate of absolute NO production by known quantities of NO donor DEA-NONOate in buffer. (d) Rate of response of NOckoutPM in cells treated with identical amounts of DEA-NONOate as in plot c. (e) In-cell calibration plot of rates of NO production using plots c and d, and the measured rates of NO production at the TGN (red) and plasma membrane (green).
An outstanding challenge is to measure the in situ activities of enzymes that produce reactive species such as reactive oxygen species (ROS) or reactive nitrogen species (RNS). This is because they cannot be stored and released on demand and must be produced in the right amount, at the location, and on cue.111 They diffuse rapidly from the point of production, react with surrounding biomolecules, and can also travel in solution to have effects at locations far away from the site of production. Thus, reactive species have highly variable half-lives depending on the physiological context.112 The effective concentration of a reactive species changes with time, making it even more challenging to quantitate the activity of reactive species producing enzymes in situ. Hence, the activities of these enzymes are best quantified by the rate of production of reactive species. Some reactive species such as NO• are charge neutral and can cross the membranes easily while others such as H2O2 need specific membrane transporters.93 NO is a reactive nitrogen species produced by three NO synthase isoforms, iNOS or NOS2, eNOS or NOS3, and nNOS or NOS1.113 We describe a framework here on how DNA reporters enable the measurement of ROS or RNS production rates to measure the activities of nitric oxide synthase 3 (NOS3), an NO producing enzyme.74,79
The DNA reporter NOckout uses diaminorhodamine-4 M (DAR) as its NO sensing module (Figure 6b).36 Developed by Nagano et al, DAR is bright, photostable, highly specific to RNS, and pH insensitive.36 DAR fluorescence is quenched by the lone pair of electrons on its aromatic amine groups. Upon reaction with NO, the amine groups form a triazole that relieves PeT quenching (Figure 6b).36 Therefore, the normalizing fluorophores must be insensitive to reactive species, especially NO, e.g., Atto647N or Alexa488. NOckout was used to quantitate the activity of the enzyme NOS3 that is present at two distinct subcellular locations—the plasma membrane and the trans-Golgi network.114 Two NOckout variants, NOckoutPM and NOckoutTGN, that each localize at the plasma membrane and the Golgi, respectively, were used to track NO production at each of these locations and thereby quantitate NOS3 activity selectively at either location (Figure 6a).79
The activity of NOS3 at a given subcellular location was measured from the rate of NO production at that location (Figure 6c–e). Using the plasma membrane as an illustrative example, the plasma membrane of cells was labeled with NOckoutPM and fixed. To these cells, a known amount of NO was generated chemically by adding specific amounts of an NO donor such as DEA-NONOate, and the cells were continuously imaged in the DAR and normalizing dye channel (R). The rate of increase of the DAR signal yields a sigmoidal curve from which the rate of NO production (r) was computed (Figure 6c). A plot of the rate of NO production versus the concentration of NO yields a calibration curve (Figure 6e).79 Thereafter, a similar procedure on live cells with or without NOS3 activation, followed by measurement of r, yielded the activity of NOS3 basally or upon stimulation (Figure 6d,e).79 This methodology has been applied to measure the activity of NOS2 at the phagosome and can be extended to quantitate other reactive species such as HOCl, also produced in the phagosome.31,74
Conclusion
There are several examples of highly selective small-molecule modulators of ion channels and transporters that are resident on the plasma membrane, many of which have now become successful drugs.115 However, examples of small-molecule modulators of organellar channels and transporters are exceedingly rare,116,117 despite organelles having ∼10 times as many ion channels or transporters than the plasma membrane.27 Many are directly linked to diseases ranging from cancer to neurodegeneration. Thus, a chemical imaging platform for ions in organelles could make it possible to systematically target a desired organellar ion channel or transporter and potentially treat the relevant disease by restoring the disrupted ionic milieu in the dysfunctional organelle.
In a singular recent example in 2019, the therapeutic potential of restoring ion homeostasis in the lysosome was shown to stunning effect.118 An infant with a novel hyperactivating CLCN7 mutation—which hyperacidifies lysosomes—showed severe neurological impairments. The infant seemed to simultaneously manifest three different lysosomal diseases. Treatment with chloroquine, which alkalinizes lysosomes, reversed the impairments in a particularly dramatic fashion.118 This was a singular case of a lysosomal disease caused by hyperacidification, for which there is, fortunately, a therapeutic solution. For the vast majority of neurodegenerative diseases where imbalances in specific ions, e.g., low chloride, high pH, or suboptimal Ca2+ in lysosomes, are well-documented, a catalog of small molecules that could rebalance that specific ion is not available.
The ability to accurately estimate differences in ionic composition in organelles arising from protein activity modulated either by small molecules or by genetic intervention allows us to address fundamental biology of ion channels and transporters in organelles. For example, the localization and function of chloride transporters DmClC-c and DmClC-b in Drosophila melanogaster was unknown. By measuring chloride levels in specific endocytic organelles using Clensor, in wild-type and mutant cells, one could pinpoint DmClC-c action in early endosomes and in recycling endosomes, while DmClC-b acted in lysosomes.65 In a second example, much is known about proteins that release Ca2+ from lysosomes since one can assay their activity using reporters of cytosolic Ca2+. However, for a long time, there was no knowledge related to mechanisms of Ca2+ import into lysosomes, since there was no tool to measure Ca2+ in the acidic lysosome lumen. The development of CalipHluor made it practical to assay Ca2+ in acidic organelles, and thus, the first example of a protein that facilitated lysosomal Ca2+ import in the animal kingdom was identified. This protein, ATP13A2, is a major risk gene for Parkinson’s disease.71
Analyzing lysosomes by 2-IM revealed that lysosomes could be chemically resolved into subpopulations in live cells derived directly from patients suffering from Niemann–Pick diseases.70 Not only was it possible to chemically subtype the three different types of Niemann–Pick diseases, but also, 2-IM profiles in the presence of therapeutics for Niemann–Pick A, B, or C revealed responsivity to therapeutics at a cellular level. This was because the 2-IM profiles of each subtype shifted toward normal only when the cells were treated with the appropriate therapeutic and remained unchanged when they were treated with the wrong therapeutic. 2IM enables one to screen for drug molecules in an unbiased way; it could also be used to assess patient suitability for a specific therapeutic treatment or to monitor disease progression and patient responsivity at a cellular level.
Disruptions to the ionic milieu of an organelle are expected to alter the internal biochemistry of the organelle.119 Thus, the ability to assay enzyme activity selectively in organelles is a necessary complement that can also be studied using DNA reporters. The ability to study enzyme activity at subcellular resolution also opens up the avenues for discovering location-specific endogenous regulators as well as small-molecule modulators of the protein in a given organelle. NOckout can also act as a reporter for NOS3, that has activity both at the plasma membrane and at the Golgi.79 By reporting on NO generated specifically at the plasma membrane or at the Golgi, one can similarly identify protein or small-molecule modulators that have location specific activity. NOS3 is a well-known clinical target for cardiovascular disease and cancer. The ability to regulate its activity with subcellular resolution will offer new therapeutic avenues.
Measuring reactive nitrogen species such as NO in the phagosome now enables one to quantitate the strength of an immune response elicited by an immunogen. For example, since NOckout per se is nonimmunogenic, phagosomal NO in an immune cell acts as a direct readout of NOS2 activity.31 Thus, the introduction of a specific immunogenic motif or pathogen associated molecular pattern (PAMP) can reveal its immunogenicity and can also be used to identify the cognate pathogen recognition receptor (PRR) responsible for immune activation.120 For example, using NOckout, it was shown that pathogen-derived ssRNA could act as PAMPs in zebrafish, and TLR7 was identified as its cognate PRR. PRRs are known to act synergistically. For example, TLR9 and TLR21 or TLR2 and TLR3 can synergistically recognize CpG and lipoproteins/dsRNA, respectively, as PAMPs.121 By displaying the appropriate PAMP combinations with a precise stoichiometry on NOckout and measuring the amount of phagosomal NO, one can identify synergistic or antagonistic PRRs. Many TLRs in humans and various model organisms do not as yet have ligands identified. Further, new PAMPs are being continuously identified.122,123 NOckout could be used to identify the cognate PRRs of novel PAMPs.
Measuring reactive oxygen species and reactive nitrogen species in the phagosome at single cell resolution potentially enables one to assay the activity of single immune cells phenotypically. In conjunction with other single cell analysis methods such as scRNA-seq,124 or immunofluorescence,125 one can begin to construct quantitative models of the immune response.
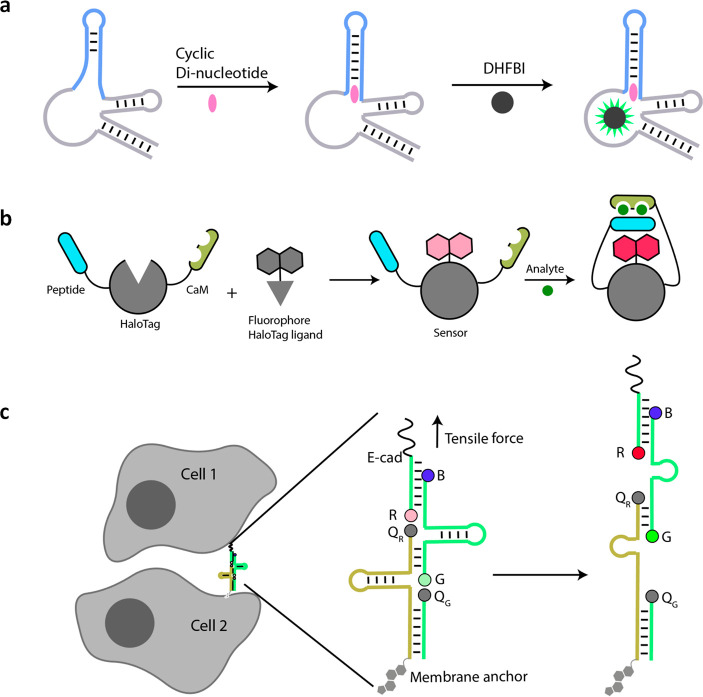
Outstanding challenges for DNA reporters in quantitative imaging are developing ways to sense and report on physical cues within the cell. These are challenging tasks even for protein-based reporters. However, given the remarkable engineerability of nucleic acids, it is likely that breakthroughs may emerge on these fronts. For example, DNA-based reporters have been used to sense force at the plasma membrane and membrane potential within organelles.126,127 The DNA-based tensile force sensor denoted DNAMeter (Figure 8c) senses tensile force by unfolding a DNA hairpin structure, causing a fluorescence change. Such devices offer great promise to measure extracellular and intracellular forces noninvasively if they can be targeted to desired locations.128
Figure 8.
New classes of hybrid probes. (a) Schematic for ligand responsive RNA-based fluorescent biosensors. Upon binding ligand (cyclic dinucleotide in this case), structural changes in the aptamer are transduced toward Spinach which forms a fluorescent complex with DHFBI. (b) Schematic showing domain organization and working principle chemigenetic indicators. (c) Schematic showing tensile force sensing using DNAMeter on live cells. Tensile forces separate quenchers (QG and QR from fluorophores G and R, respectively) increasing fluorescence. Cholesterol serves as a membrane anchor, and E-cadherin (E-cad) targets DNAMeter at the intercellular junctions (left).
The integration of advanced imaging modalities in live imaging significantly expanded the scope of genetically encoded reporters such as GFP. For example, anisotropy imaging of GFP-fused GPI-anchored proteins has revealed their nanoscale organization that controls cell signaling.129 Fluorescence lifetime imaging has been used to pinpoint protein interactions in live cells. We envisage that DNA reporters can be designed expressly for FLIM, anisotropy, Raman imaging, STED, STORM, or PALM by incorporating fluorophores especially suited to these imaging modalities. Single-molecule localization microscopy (SMLM) in the form of DNA-PAINT or its quantitative version, qPAINT, is already being deployed to count RNA and proteins, albeit in fixed cells and tissues.130 FLIM has already been used on DNA reporters, albeit to check device integrity, and not to elicit chemical information.63 Integration to imaging modalities such as lattice light sheet microscopy and two-photon excitation microscopy could enable longer imaging times and deeper tissue accessibility, respectively.131,132 Thus, DNA reporters are ripe for integration into more specialized imaging modalities to uncover new regimes of physicochemical information within the cell.
While there are excellent DNA reporters for temperature, they have yet to be deployed within living systems.133 Molecular reporters for current, viscosity, and osmolarity await creation and development. Novel reporters of physical parameters in subcellular spaces would give us new knowledge of the cell and likely reveal new biology.
The susceptibility of DNA devices to nucleases present in blood and their subsequent excretion via the renal system limit the time-window in which DNA nanodevices can act as reporters in vivo in vertebrate models. Chemically modified DNA or RNA that incorporates non-natural nucleotide analogues can potentially increase the in vivo half-life of DNA reporters. These include phosphodiester backbone modifications (e.g., L-DNA, LNA, or phosphorothioates), sugar modifications (e.g., 2′-fluoro and 2′-O-methyl), end-modified oligonucleotides (e.g., PEGylation or 3′ inverted thymidine), or modified nucleobases (e.g., SOMAmers).134
Quantitative imaging of biomacromolecules like RNA can provide spatial information on gene expression. Most current methods to achieve this are based on fluorescence in situ hybridization (FISH).135 Other techniques such as DNAzymes or gold nanoparticle mediated amplification have also been used to image RNA molecules.136 Short fluorescent oligonucleotides can hybridize reversibly and rapidly to endogenous target RNAs or to oligonucleotide-bearing antibodies bound to endogenous protein targets. Using points accumulation for imaging nanoscale topography (PAINT), one can quantitatively image endogenous RNAs or proteins at super-resolution, albeit in fixed samples.137
Although the quantitative chemical imaging we describe in this Outlook is focused on organelles, the imaging paradigm described in here is generalizable and adaptable to all parts of the cell. New classes of hybrid probes that could potentially enable quantitative chemical imaging are chemigenetic indicators and protein spherical nucleic acids (ProSNAs). ProSNAs use a protein core conjugated to several fluorescent oligonucleotides. They can potentially be modified to report on both chemical content and biochemical activity in the cytoplasm using small-molecule dyes and the protein scaffold, respectively.138,139 Chemigenetic indicators consist of intracellularly expressed HaloTagged proteins conjugated to small-molecule fluorophores that provide subcellular targeting (Figure 8b). Structural changes in the protein scaffold upon encountering an analyte reposition the fluorophore in a different microenvironment leading to a fluorescence change. Chemigenetic probes for Ca2+ and voltage have been realized but do not yet provide absolute measures.140 Chemigenetic indicators need to circumvent pH sensitivity of fluorescent protein modules to enable quantitative imaging in organelles with acidic lumens.
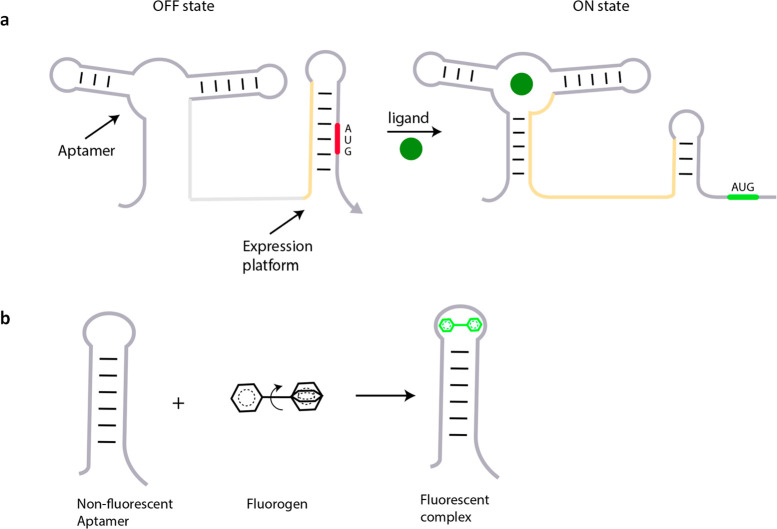
Despite the attractive properties of chemigenetic indicators, there is tremendous opportunity for genetically encoded RNA-based sensors that are making rapid improvements using powerful in vitro evolution methods.141,142 The emergence of Spinach, a chemigenetic RNA reporter for ligand localization, can be integrated to a palette of aptamers that sense cellular metabolites (Figure 8a).143 Newer riboswitches responsive to diverse small molecules using RNA sensing domains, e.g., cyclic di-GMP, thiamine, or flavin mononucleotide, are being continually discovered.144 A few, e.g., cyclic di-GMP and S-adenosylmethionine (SAM), have been integrated to Spinach to semiquantitatively map these molecules in the cytoplasm (Figure 8a).145,146 There are many more exciting riboswitches responsive to ions, amino acids, vitamins, and antibiotics,147 which could potentially be used to map their targets using the framework described here (Figure 7a,b). In fact, ratiometric DNA devices that use shape-shifting aptamer domains interfaced to Spinach variants have been used to construct cAMP maps, albeit in giant unilamellar vesicles.148 RNA can be anchored to specific locations via interactions with proteins, and the problem of probe diffusibility we expect will be surmounted. The introduction of ratiometry into Spinach technology promises to be the next leap for quantitative chemical imaging in the cytosol.149 We envisage that quantitative chemical imaging applied to ratiometric small-molecule-based probes and biomacromolecule-based scaffolds that provide stable spatial localization within the cell will enable the study of diverse biological processes at an entirely new level of molecular detail.
Figure 7.
Small-molecule recognition domains in riboswitches and their application in fluorescence imaging. (a) Schematic showing control of gene expression using aptamer domains in a riboswitch, where ligand binding to the aptamer motif unmasks a cryptic AUG codon (ON state). (b) An unbound weakly fluorescent molecule fluoresces intensely upon binding its aptamer, as in the RNA mimic of green fluorescence protein, Spinach.
Acknowledgments
This work was supported by the University of Chicago Women’s Board; the Mergel Funsky award; grant NS114428 from NINDS; the ONO Pharma Breakthrough Science Award and University of Chicago start-up funds to Y.K. We thank Kyle Lin for a critical reading of the manuscript.
Author Contributions
Y.K., J.Z., and M.S.J. developed the review outline. Y.K. wrote the manuscript. J.Z. and M.S.J. incorporated figures and references. All authors commented and gave input on the manuscript.
The authors declare the following competing financial interest(s): Y.K. is co-founder and Chief Science Officer of Esya Inc that uses DNA reporters.
References
- Zhang J.; Campbell R. E.; Ting A. Y.; Tsien R. Y. Creating New Fluorescent Probes for Cell Biology. Nat. Rev. Mol. Cell Biol. 2002, 3 (12), 906–918. 10.1038/nrm976. [DOI] [PubMed] [Google Scholar]
- Rodriguez E. A.; Campbell R. E.; Lin J. Y.; Lin M. Z.; Miyawaki A.; Palmer A. E.; Shu X.; Zhang J.; Tsien R. Y. The Growing and Glowing Toolbox of Fluorescent and Photoactive Proteins. Trends Biochem. Sci. 2017, 42 (2), 111–129. 10.1016/j.tibs.2016.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsien R. Y. Fluorescent Probes of Cell Signaling. Annu. Rev. Neurosci. 1989, 12 (1), 227–253. 10.1146/annurev.ne.12.030189.001303. [DOI] [PubMed] [Google Scholar]
- Hsu P. D.; Lander E. S.; Zhang F. Development and Applications of CRISPR-Cas9 for Genome Engineering. Cell 2014, 157 (6), 1262–1278. 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valm A. M.; Cohen S.; Legant W. R.; Melunis J.; Hershberg U.; Wait E.; Cohen A. R.; Davidson M. W.; Betzig E.; Lippincott-Schwartz J. Applying Systems-Level Spectral Imaging and Analysis to Reveal the Organelle Interactome. Nature 2017, 546 (7656), 162–167. 10.1038/nature22369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S.; Sekine S.; Pessino V.; Li H.; Leonetti M. D.; Huang B. Improved Split Fluorescent Proteins for Endogenous Protein Labeling. Nat. Commun. 2017, 8 (1), 370. 10.1038/s41467-017-00494-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y.; Li D.; Zhang S.; Yang Y.; Liu J.-J.; Wang X.; Liu C.; Milkie D. E.; Moore R. P.; Tulu U. S.; Kiehart D. P.; Hu J.; Lippincott-Schwartz J.; Betzig E.; Li D. Visualizing Intracellular Organelle and Cytoskeletal Interactions at Nanoscale Resolution on Millisecond Timescales. Cell 2018, 175 (5), 1430–1442. 10.1016/j.cell.2018.09.057. [DOI] [PubMed] [Google Scholar]
- Chen F.; Tillberg P. W.; Boyden E. S. Expansion Microscopy. Science 2015, 347 (6221), 543–548. 10.1126/science.1260088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betzig E.; Patterson G. H.; Sougrat R.; Lindwasser O. W.; Olenych S.; Bonifacino J. S.; Davidson M. W.; Lippincott-Schwartz J.; Hess H. F. Imaging Intracellular Fluorescent Proteins at Nanometer Resolution. Science 2006, 313 (5793), 1642–1645. 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- Hein B.; Willig K. I.; Hell S. W. Stimulated Emission Depletion (STED) Nanoscopy of a Fluorescent Protein-Labeled Organelle inside a Living Cell. Proc. Natl. Acad. Sci. U. S. A. 2008, 105 (38), 14271–14276. 10.1073/pnas.0807705105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A.; Rivera-Molina F.; Xi Z.; Toomre D.; Schepartz A. Endosome Motility Defects Revealed at Super-Resolution in Live Cells Using HIDE Probes. Nat. Chem. Biol. 2020, 16 (4), 408–414. 10.1038/s41589-020-0479-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D.; Fan X.; Zhang H.; Huang Z.; Tang Q.; Li W.; Bai J.; Lei X.; Chen X.. Click-ExM Enables Expansion Microscopy for All Biomolecules. bioRxiv, 2020, 2020.03.19.998039. https://www.biorxiv.org/content/10.1101/2020.03.19.998039v1. [DOI] [PubMed]
- Wu Y.; Rivenson Y.; Wang H.; Luo Y.; Ben-David E.; Bentolila L. A.; Pritz C.; Ozcan A. Three-Dimensional Virtual Refocusing of Fluorescence Microscopy Images Using Deep Learning. Nat. Methods 2019, 16 (12), 1323–1331. 10.1038/s41592-019-0622-5. [DOI] [PubMed] [Google Scholar]
- Thul P. J.; Åkesson L.; Wiking M.; Mahdessian D.; Geladaki A.; Blal H. A.; Alm T.; Asplund A.; Björk L.; Breckels L. M.; Bäckström A.; Danielsson F.; Fagerberg L.; Fall J.; Gatto L.; Gnann C.; Hober S.; Hjelmare M.; Johansson F.; Lee S.; Lindskog C.; Mulder J.; Mulvey C. M.; Nilsson P.; Oksvold P.; Rockberg J.; Schutten R.; Schwenk J. M.; Sivertsson Å.; Sjöstedt E.; Skogs M.; Stadler C.; Sullivan D. P.; Tegel H.; Winsnes C.; Zhang C.; Zwahlen M.; Mardinoglu A.; Pontén F.; Feilitzen K. v.; Lilley K. S.; Uhlén M.; Lundberg E. A Subcellular Map of the Human Proteome. Science 2017, 356 (6340), eaal3321. 10.1126/science.aal3321. [DOI] [PubMed] [Google Scholar]
- Gut G.; Herrmann M. D.; Pelkmans L. Multiplexed Protein Maps Link Subcellular Organization to Cellular States. Science 2018, 361 (6401), eaar7042. 10.1126/science.aar7042. [DOI] [PubMed] [Google Scholar]
- Moffitt J. R.; Hao J.; Wang G.; Chen K. H.; Babcock H. P.; Zhuang X. High-Throughput Single-Cell Gene-Expression Profiling with Multiplexed Error-Robust Fluorescence in Situ Hybridization. Proc. Natl. Acad. Sci. U. S. A. 2016, 113 (39), 11046–11051. 10.1073/pnas.1612826113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S. K.; Indig F. E.; Olivieri N.; Levine N. D.; Latterich M. Organelle Membrane Fusion: A Novel Function for the Syntaxin Homolog Ufe1p in ER Membrane Fusion. Cell 1998, 92 (5), 611–620. 10.1016/S0092-8674(00)81129-0. [DOI] [PubMed] [Google Scholar]
- Loss O.; Stephenson F. A. Developmental Changes in Trak-Mediated Mitochondrial Transport in Neurons. Mol. Cell. Neurosci. 2017, 80, 134–147. 10.1016/j.mcn.2017.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D. E.; Ostrowski P.; Jaumouillé V.; Grinstein S. The Position of Lysosomes within the Cell Determines Their Luminal PH. J. Cell Biol. 2016, 212 (6), 677–692. 10.1083/jcb.201507112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader M.; Godinho L. F.; Costello J. L.; Islinger M. The Different Facets of Organelle Interplay—an Overview of Organelle Interactions. Front. Cell Dev. Biol. 2015, 3, 56. 10.3389/fcell.2015.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satori C. P.; Henderson M. M.; Krautkramer E. A.; Kostal V.; Distefano M. M.; Arriaga E. A. Bioanalysis of Eukaryotic Organelles. Chem. Rev. 2013, 113 (4), 2733–2811. 10.1021/cr300354g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demaurex N.; Furuya W.; D’Souza S.; Bonifacino J. S.; Grinstein S. Mechanism of Acidification of the Trans-Golgi Network (TGN). In Situ Measurements of PH Using Retrieval of TGN38 and Furin from the Cell Surface. J. Biol. Chem. 1998, 273 (4), 2044–2051. 10.1074/jbc.273.4.2044. [DOI] [PubMed] [Google Scholar]
- Liu X.; Su Y.; Tian H.; Yang L.; Zhang H.; Song X.; Foley J. W. Ratiometric Fluorescent Probe for Lysosomal PH Measurement and Imaging in Living Cells Using Single-Wavelength Excitation. Anal. Chem. 2017, 89 (13), 7038–7045. 10.1021/acs.analchem.7b00754. [DOI] [PubMed] [Google Scholar]
- Chakraborty K.; Leung K.; Krishnan Y. High Lumenal Chloride in the Lysosome Is Critical for Lysosome Function. eLife 2017, 6, e28862 10.7554/eLife.28862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usenovic M.; Tresse E.; Mazzulli J. R.; Taylor J. P.; Krainc D. Deficiency of ATP13A2 Leads to Lysosomal Dysfunction, α-Synuclein Accumulation, and Neurotoxicity. J. Neurosci. 2012, 32 (12), 4240–4246. 10.1523/JNEUROSCI.5575-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadsby D. C.; Vergani P.; Csanády L. The ABC Protein Turned Chloride Channel Whose Failure Causes Cystic Fibrosis. Nature 2006, 440 (7083), 477–483. 10.1038/nature04712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H.; Martinoia E.; Szabo I. Organellar Channels and Transporters. Cell Calcium 2015, 58 (1), 1–10. 10.1016/j.ceca.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Károlyi L.; Koch M. C.; Grzeschik K.-H.; Seyberth H. W. The Molecular Genetic Approach to ”Bartter’s Syndrome. J. Mol. Med. 1998, 76 (5), 317–325. 10.1007/s001090050223. [DOI] [PubMed] [Google Scholar]
- Feng X.; Xiong J.; Lu Y.; Xia X.; Zhu M. X. Differential Mechanisms of Action of the Mucolipin Synthetic Agonist, ML-SA1, on Insect TRPML and Mammalian TRPML1. Cell Calcium 2014, 56 (6), 446–456. 10.1016/j.ceca.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modi S.; M G S.; Goswami D.; Gupta G. D.; Mayor S.; Krishnan Y. A DNA Nanomachine That Maps Spatial and Temporal PH Changes inside Living Cells. Nat. Nanotechnol. 2009, 4 (5), 325–330. 10.1038/nnano.2009.83. [DOI] [PubMed] [Google Scholar]
- Veetil A. T.; Zou J.; Henderson K. W.; Jani M. S.; Shaik S. M.; Sisodia S. S.; Hale M. E.; Krishnan Y. DNA-Based Fluorescent Probes of NOS2 Activity in Live Brains. Proc. Natl. Acad. Sci. U. S. A. 2020, 117 (26), 14694–14702. 10.1073/pnas.2003034117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan K.; Veetil A. T.; Chakraborty K.; Krishnan Y. DNA Nanodevices Map Enzymatic Activity in Organelles. Nat. Nanotechnol. 2019, 14 (3), 252–259. 10.1038/s41565-019-0365-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima H.; Nakatsubo N.; Kikuchi K.; Kawahara S.; Kirino Y.; Nagoshi H.; Hirata Y.; Nagano T. Detection and Imaging of Nitric Oxide with Novel Fluorescent Indicators: Diaminofluoresceins. Anal. Chem. 1998, 70 (13), 2446–2453. 10.1021/ac9801723. [DOI] [PubMed] [Google Scholar]
- Tsien R. Y. New Calcium Indicators and Buffers with High Selectivity against Magnesium and Protons: Design, Synthesis, and Properties of Prototype Structures. Biochemistry 1980, 19 (11), 2396–2404. 10.1021/bi00552a018. [DOI] [PubMed] [Google Scholar]
- Chang M. C. Y.; Pralle A.; Isacoff E. Y.; Chang C. J. A Selective, Cell-Permeable Optical Probe for Hydrogen Peroxide in Living Cells. J. Am. Chem. Soc. 2004, 126 (47), 15392–15393. 10.1021/ja0441716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima H.; Hirotani M.; Nakatsubo N.; Kikuchi K.; Urano Y.; Higuchi T.; Hirata Y.; Nagano T. Bioimaging of Nitric Oxide with Fluorescent Indicators Based on the Rhodamine Chromophore. Anal. Chem. 2001, 73 (9), 1967–1973. 10.1021/ac001136i. [DOI] [PubMed] [Google Scholar]
- Grynkiewicz G.; Poenie M.; Tsien R. Y. A New Generation of Ca2+ Indicators with Greatly Improved Fluorescence Properties. J. Biol. Chem. 1985, 260 (6), 3440–3450. [PubMed] [Google Scholar]
- Au-Yeung H. Y.; Chan J.; Chantarojsiri T.; Chang C. J. Molecular Imaging of Labile Iron(II) Pools in Living Cells with a Turn-On Fluorescent Probe. J. Am. Chem. Soc. 2013, 135 (40), 15165–15173. 10.1021/ja4072964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavis L. D.; Chao T.-Y.; Raines R. T. Synthesis and Utility of Fluorogenic Acetoxymethyl Ethers. Chem. Sci. 2011, 2 (3), 521–530. 10.1039/C0SC00466A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T.-L.; Upadhyayula S.; Milkie D. E.; Singh V.; Wang K.; Swinburne I. A.; Mosaliganti K. R.; Collins Z. M.; Hiscock T. W.; Shea J.; Kohrman A. Q.; Medwig T. N.; Dambournet D.; Forster R.; Cunniff B.; Ruan Y.; Yashiro H.; Scholpp S.; Meyerowitz E. M.; Hockemeyer D.; Drubin D. G.; Martin B. L.; Matus D. Q.; Koyama M.; Megason S. G.; Kirchhausen T.; Betzig E. Observing the Cell in Its Native State: Imaging Subcellular Dynamics in Multicellular Organisms. Science 2018, 360 (6386), eaaq1392. 10.1126/science.aaq1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaccolo M.; De Giorgi F.; Cho C. Y.; Feng L.; Knapp T.; Negulescu P. A.; Taylor S. S.; Tsien R. Y.; Pozzan T. A Genetically Encoded, Fluorescent Indicator for Cyclic AMP in Living Cells. Nat. Cell Biol. 2000, 2 (1), 25–29. 10.1038/71345. [DOI] [PubMed] [Google Scholar]
- Eroglu E.; Gottschalk B.; Charoensin S.; Blass S.; Bischof H.; Rost R.; Madreiter-Sokolowski C. T.; Pelzmann B.; Bernhart E.; Sattler W.; Hallström S.; Malinski T.; Waldeck-Weiermair M.; Graier W. F.; Malli R. Development of Novel FP-Based Probes for Live-Cell Imaging of Nitric Oxide Dynamics. Nat. Commun. 2016, 7, 10623. 10.1038/ncomms10623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L.; Hires S. A.; Mao T.; Huber D.; Chiappe M. E.; Chalasani S. H.; Petreanu L.; Akerboom J.; McKinney S. A.; Schreiter E. R.; Bargmann C. I.; Jayaraman V.; Svoboda K.; Looger L. L. Imaging Neural Activity in Worms, Flies and Mice with Improved GCaMP Calcium Indicators. Nat. Methods 2009, 6 (12), 875–881. 10.1038/nmeth.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbowski M.; Cleland M. M.; Roelofs B. Photoactivatable Green Fluorescent Protein-Based Visualization and Quantification of Mitochondrial Fusion and Mitochondrial Network Complexity in Living Cells. Methods Enzymol. 2014, 547, 57–73. 10.1016/B978-0-12-801415-8.00004-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runions J.; Brach T.; Kühner S.; Hawes C. Photoactivation of GFP Reveals Protein Dynamics within the Endoplasmic Reticulum Membrane. J. Exp. Bot. 2006, 57 (1), 43–50. 10.1093/jxb/eri289. [DOI] [PubMed] [Google Scholar]
- Kohnhorst C. L.; Schmitt D. L.; Sundaram A.; An S. Subcellular Functions of Proteins under Fluorescence Single-Cell Microscopy. Biochim. Biophys. Acta, Proteins Proteomics 2016, 1864 (1), 77–84. 10.1016/j.bbapap.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maulucci G.; Chiarpotto M.; Papi M.; Samengo D.; Pani G.; De Spirito M. Quantitative Analysis of Autophagic Flux by Confocal PH-Imaging of Autophagic Intermediates. Autophagy 2015, 11 (10), 1905–1916. 10.1080/15548627.2015.1084455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyawaki A.; Llopis J.; Heim R.; McCaffery J. M.; Adams J. A.; Ikura M.; Tsien R. Y. Fluorescent Indicators for Ca 2+ Based on Green Fluorescent Proteins and Calmodulin. Nature 1997, 388 (6645), 882–887. 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- Gong Y.; Wagner M. J.; Li J. Z.; Schnitzer M. J. Imaging Neural Spiking in Brain Tissue Using FRET-Opsin Protein Voltage Sensors. Nat. Commun. 2014, 5, 3674. 10.1038/ncomms4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T.-W.; Wardill T. J.; Sun Y.; Pulver S. R.; Renninger S. L.; Baohan A.; Schreiter E. R.; Kerr R. A.; Orger M. B.; Jayaraman V.; Looger L. L.; Svoboda K.; Kim D. S. Ultrasensitive Fluorescent Proteins for Imaging Neuronal Activity. Nature 2013, 499 (7458), 295–300. 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y.; Wu S.-Y.; Rancic V.; Aggarwal A.; Qian Y.; Miyashita S.-I.; Ballanyi K.; Campbell R. E.; Dong M. Genetically Encoded Fluorescent Indicators for Imaging Intracellular Potassium Ion Concentration. Commun. Biol. 2019, 2 (1), 1–10. 10.1038/s42003-018-0269-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belousov V. V.; Fradkov A. F.; Lukyanov K. A.; Staroverov D. B.; Shakhbazov K. S.; Terskikh A. V.; Lukyanov S. Genetically Encoded Fluorescent Indicator for Intracellular Hydrogen Peroxide. Nat. Methods 2006, 3 (4), 281–286. 10.1038/nmeth866. [DOI] [PubMed] [Google Scholar]
- Bischof H.; Rehberg M.; Stryeck S.; Artinger K.; Eroglu E.; Waldeck-Weiermair M.; Gottschalk B.; Rost R.; Deak A. T.; Niedrist T.; Vujic N.; Lindermuth H.; Prassl R.; Pelzmann B.; Groschner K.; Kratky D.; Eller K.; Rosenkranz A. R.; Madl T.; Plesnila N.; Graier W. F.; Malli R. Novel Genetically Encoded Fluorescent Probes Enable Real-Time Detection of Potassium in Vitro and in Vivo. Nat. Commun. 2017, 8 (1), 1422. 10.1038/s41467-017-01615-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seksek O.; Biwersi J.; Verkman A. S. Translational Diffusion of Macromolecule-Sized Solutes in Cytoplasm and Nucleus. J. Cell Biol. 1997, 138 (1), 131–142. 10.1083/jcb.138.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorkin A. D.; Teslenko L. V.; Nikolsky N. N. The Endocytosis of Epidermal Growth Factor in A431 Cells: A PH of Microenvironment and the Dynamics of Receptor Complex Dissociation. Exp. Cell Res. 1988, 175 (1), 192–205. 10.1016/0014-4827(88)90266-2. [DOI] [PubMed] [Google Scholar]
- Sonawane N. D.; Verkman A. S. Determinants of [Cl-] in Recycling and Late Endosomes and Golgi Complex Measured Using Fluorescent Ligands. J. Cell Biol. 2003, 160 (7), 1129–1138. 10.1083/jcb.200211098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canton J.; Grinstein S.. Measuring Lysosomal PH by Fluorescence Microscopy. In Methods in Cell Biology; Platt F., Platt N., Eds.; Lysosomes and Lysosomal Diseases; Academic Press, 2015; Vol. 126, Chapter 5, pp 85–99. [DOI] [PubMed] [Google Scholar]
- Chakraborty K.; Veetil A. T.; Jaffrey S. R.; Krishnan Y. Nucleic Acid-Based Nanodevices in Biological Imaging. Annu. Rev. Biochem. 2016, 85 (1), 349–373. 10.1146/annurev-biochem-060815-014244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jani M. S.; Veetil A. T.; Krishnan Y. Precision Immunomodulation with Synthetic Nucleic Acid Technologies. Nat. Rev. Mater. 2019, 4 (6), 451–458. 10.1038/s41578-019-0105-4. [DOI] [Google Scholar]
- Cho E. J.; Lee J.-W.; Ellington A. D. Applications of Aptamers as Sensors. Annu. Rev. Anal. Chem. 2009, 2 (1), 241–264. 10.1146/annurev.anchem.1.031207.112851. [DOI] [PubMed] [Google Scholar]
- Rosi N. L.; Mirkin C. A. Nanostructures in Biodiagnostics. Chem. Rev. 2005, 105 (4), 1547–1562. 10.1021/cr030067f. [DOI] [PubMed] [Google Scholar]
- Surana S.; Bhat J. M.; Koushika S. P.; Krishnan Y. An Autonomous DNA Nanomachine Maps Spatiotemporal PH Changes in a Multicellular Living Organism. Nat. Commun. 2011, 2, 340. 10.1038/ncomms1340. [DOI] [PubMed] [Google Scholar]
- Bhatia D.; Surana S.; Chakraborty S.; Koushika S. P.; Krishnan Y. A Synthetic Icosahedral DNA-Based Host-Cargo Complex for Functional in Vivo Imaging. Nat. Commun. 2011, 2, 339. 10.1038/ncomms1337. [DOI] [PubMed] [Google Scholar]
- Veetil A. T.; Chakraborty K.; Xiao K.; Minter M. R.; Sisodia S. S.; Krishnan Y. Cell-Targetable DNA Nanocapsules for Spatiotemporal Release of Caged Bioactive Small Molecules. Nat. Nanotechnol. 2017, 12 (12), 1183. 10.1038/nnano.2017.159. [DOI] [PubMed] [Google Scholar]
- Saha S.; Prakash V.; Halder S.; Chakraborty K.; Krishnan Y. A PH-Independent DNA Nanodevice for Quantifying Chloride Transport in Organelles of Living Cells. Nat. Nanotechnol. 2015, 10 (7), 645–651. 10.1038/nnano.2015.130. [DOI] [PubMed] [Google Scholar]
- Krishnan Y.; Simmel F. C. Nucleic Acid Based Molecular Devices. Angew. Chem., Int. Ed. 2011, 50 (14), 3124–3156. 10.1002/anie.200907223. [DOI] [PubMed] [Google Scholar]
- Dunn M. R.; Jimenez R. M.; Chaput J. C. Analysis of Aptamer Discovery and Technology. Nat. Rev. Chem. 2017, 1 (10), 1–16. 10.1038/s41570-017-0076. [DOI] [Google Scholar]
- Potyrailo R. A.; Conrad R. C.; Ellington A. D.; Hieftje G. M. Adapting Selected Nucleic Acid Ligands (Aptamers) to Biosensors. Anal. Chem. 1998, 70 (16), 3419–3425. 10.1021/ac9802325. [DOI] [PubMed] [Google Scholar]
- Stojanovic M. N.; Kolpashchikov D. M. Modular Aptameric Sensors. J. Am. Chem. Soc. 2004, 126 (30), 9266–9270. 10.1021/ja032013t. [DOI] [PubMed] [Google Scholar]
- Leung K.; Chakraborty K.; Saminathan A.; Krishnan Y. A DNA Nanomachine Chemically Resolves Lysosomes in Live Cells. Nat. Nanotechnol. 2019, 14 (2), 176–183. 10.1038/s41565-018-0318-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanaswamy N.; Chakraborty K.; Saminathan A.; Zeichner E.; Leung K.; Devany J.; Krishnan Y. A PH-Correctable, DNA-Based Fluorescent Reporter for Organellar Calcium. Nat. Methods 2019, 16, 1. 10.1038/s41592-018-0232-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modi S.; Nizak C.; Surana S.; Halder S.; Krishnan Y. Two DNA Nanomachines Map PH Changes along Intersecting Endocytic Pathways inside the Same Cell. Nat. Nanotechnol. 2013, 8 (6), 459–467. 10.1038/nnano.2013.92. [DOI] [PubMed] [Google Scholar]
- Fielden L. F.; Kang Y.; Newton H. J.; Stojanovski D. Targeting Mitochondria: How Intravacuolar Bacterial Pathogens Manipulate Mitochondria. Cell Tissue Res. 2017, 367 (1), 141–154. 10.1007/s00441-016-2475-x. [DOI] [PubMed] [Google Scholar]
- Thekkan S.; Jani M. S.; Cui C.; Dan K.; Zhou G.; Becker L.; Krishnan Y. A DNA-Based Fluorescent Reporter Maps HOCl Production in the Maturing Phagosome. Nat. Chem. Biol. 2019, 15 (12), 1165–1172. 10.1038/s41589-018-0176-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannes L.; Römer W. Shiga Toxins--from Cell Biology to Biomedical Applications. Nat. Rev. Microbiol. 2010, 8 (2), 105–116. 10.1038/nrmicro2279. [DOI] [PubMed] [Google Scholar]
- Chinnapen D. J.-F.; Chinnapen H.; Saslowsky D.; Lencer W. I. Rafting with Cholera Toxin: Endocytosis and Trafficking from Plasma Membrane to ER. FEMS Microbiol. Lett. 2007, 266 (2), 129–137. 10.1111/j.1574-6968.2006.00545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh M.; Helenius A. Virus Entry: Open Sesame. Cell 2006, 124 (4), 729–740. 10.1016/j.cell.2006.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia D.; Arumugam S.; Nasilowski M.; Joshi H.; Wunder C.; Chambon V.; Prakash V.; Grazon C.; Nadal B.; Maiti P. K.; Johannes L.; Dubertret B.; Krishnan Y. Quantum Dot-Loaded Monofunctionalized DNA Icosahedra for Single-Particle Tracking of Endocytic Pathways. Nat. Nanotechnol. 2016, 11 (12), 1112–1119. 10.1038/nnano.2016.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jani M. S.; Zou J.; Veetil A. T.; Krishnan Y. A DNA-Based Fluorescent Probe Maps NOS3 Activity with Subcellular Spatial Resolution. Nat. Chem. Biol. 2020, 16 (6), 660–666. 10.1038/s41589-020-0491-3. [DOI] [PubMed] [Google Scholar]
- Miller M. J.; Wei S. H.; Parker I.; Cahalan M. D. Two-Photon Imaging of Lymphocyte Motility and Antigen Response in Intact Lymph Node. Science 2002, 296 (5574), 1869–1873. 10.1126/science.1070051. [DOI] [PubMed] [Google Scholar]
- Sarder P.; Maji D.; Achilefu S. Molecular Probes for Fluorescence Lifetime Imaging. Bioconjugate Chem. 2015, 26 (6), 963–974. 10.1021/acs.bioconjchem.5b00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanaswami V.; Dahl K.; Bernard-Gauthier V.; Josephson L.; Cumming P.; Vasdev N. Emerging PET Radiotracers and Targets for Imaging of Neuroinflammation in Neurodegenerative Diseases: Outlook Beyond TSPO. Mol. Imaging 2018, 17, 153601211879231. 10.1177/1536012118792317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring K.; Leroy J.-L.; Guéron M. A Tetrameric DNA Structure with Protonated Cytosine-Cytosine Base Pairs. Nature 1993, 363 (6429), 561–565. 10.1038/363561a0. [DOI] [PubMed] [Google Scholar]
- Alberti P.; Mergny J.-L. DNA Duplex-Quadruplex Exchange as the Basis for a Nanomolecular Machine. Proc. Natl. Acad. Sci. U. S. A. 2003, 100 (4), 1569–1573. 10.1073/pnas.0335459100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D.; Balasubramanian S. A Proton-Fuelled DNA Nanomachine. Angew. Chem., Int. Ed. 2003, 42 (46), 5734–5736. 10.1002/anie.200352402. [DOI] [PubMed] [Google Scholar]
- Nesterova I. V.; Elsiddieg S. O.; Nesterov E. E. A Dual Input DNA-Based Molecular Switch. Mol. BioSyst. 2014, 10 (11), 2810–2814. 10.1039/C4MB00363B. [DOI] [PubMed] [Google Scholar]
- Lannes L.; Halder S.; Krishnan Y.; Schwalbe H. Tuning the PH Response of I-Motif DNA Oligonucleotides. ChemBioChem 2015, 16 (11), 1647–1656. 10.1002/cbic.201500182. [DOI] [PubMed] [Google Scholar]
- Lukacs G. L.; Chang X. B.; Kartner N.; Rotstein O. D.; Riordan J. R.; Grinstein S. The Cystic Fibrosis Transmembrane Regulator Is Present and Functional in Endosomes. Role as a Determinant of Endosomal PH. J. Biol. Chem. 1992, 267 (21), 14568–14572. [PubMed] [Google Scholar]
- Prakash V.; Saha S.; Chakraborty K.; Krishnan Y. Rational Design of a Quantitative, PH-Insensitive, Nucleic Acid Based Fluorescent Chloride Reporter. Chem. Sci. 2016, 7 (3), 1946–1953. 10.1039/C5SC04002G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohgaki R.; Matsushita M.; Kanazawa H.; Ogihara S.; Hoekstra D.; van IJzendoorn S. C. D. The Na+/H+ Exchanger NHE6 in the Endosomal Recycling System Is Involved in the Development of Apical Bile Canalicular Surface Domains in HepG2 Cells. Mol. Biol. Cell 2010, 21 (7), 1293–1304. 10.1091/mbc.e09-09-0767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchionda M.; Pittman J. K.; Mayor R.; Patel S. Ca2+/H+ Exchange by Acidic Organelles Regulates Cell Migration in Vivo. J. Cell Biol. 2016, 212 (7), 803–813. 10.1083/jcb.201510019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondapalli K. C.; Llongueras J. P.; Capilla-González V.; Prasad H.; Hack A.; Smith C.; Guerrero-Cázares H.; Quiñones-Hinojosa A.; Rao R. A Leak Pathway for Luminal Protons in Endosomes Drives Oncogenic Signalling in Glioblastoma. Nat. Commun. 2015, 6 (1), 6289. 10.1038/ncomms7289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller E. W.; Dickinson B. C.; Chang C. J. Aquaporin-3 Mediates Hydrogen Peroxide Uptake to Regulate Downstream Intracellular Signaling. Proc. Natl. Acad. Sci. U. S. A. 2010, 107 (36), 15681–15686. 10.1073/pnas.1005776107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aron A. T.; Reeves A. G.; Chang C. J. Activity-Based Sensing Fluorescent Probes for Iron in Biological Systems. Curr. Opin. Chem. Biol. 2018, 43, 113–118. 10.1016/j.cbpa.2017.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo C.; Balci H.; Ishitsuka Y.; Buranachai C.; Ha T. Advances in Single-Molecule Fluorescence Methods for Molecular Biology. Annu. Rev. Biochem. 2008, 77 (1), 51–76. 10.1146/annurev.biochem.77.070606.101543. [DOI] [PubMed] [Google Scholar]
- Zlatanova J.; van Holde K. Single-Molecule Biology: What Is It and How Does It Work?. Mol. Cell 2006, 24 (3), 317–329. 10.1016/j.molcel.2006.10.017. [DOI] [PubMed] [Google Scholar]
- Wasmeier C.; Hume A. N.; Bolasco G.; Seabra M. C. Melanosomes at a Glance. J. Cell Sci. 2008, 121 (24), 3995–3999. 10.1242/jcs.040667. [DOI] [PubMed] [Google Scholar]
- Cieutat A. M.; Lobel P.; August J. T.; Kjeldsen L.; Sengeløv H.; Borregaard N.; Bainton D. F. Azurophilic Granules of Human Neutrophilic Leukocytes Are Deficient in Lysosome-Associated Membrane Proteins but Retain the Mannose 6-Phosphate Recognition Marker. Blood 1998, 91 (3), 1044–1058. 10.1182/blood.V91.3.1044.1044_1044_1058. [DOI] [PubMed] [Google Scholar]
- Blott E. J.; Griffiths G. M. Secretory Lysosomes. Nat. Rev. Mol. Cell Biol. 2002, 3 (2), 122–131. 10.1038/nrm732. [DOI] [PubMed] [Google Scholar]
- Luzio J. P.; Pryor P. R.; Bright N. A. Lysosomes: Fusion and Function. Nat. Rev. Mol. Cell Biol. 2007, 8 (8), 622–632. 10.1038/nrm2217. [DOI] [PubMed] [Google Scholar]
- Dell’Angelica E. C.; Mullins C.; Caplan S.; Bonifacino J. S. Lysosome-Related Organelles. FASEB J. 2000, 14 (10), 1265–1278. 10.1096/fasebj.14.10.1265. [DOI] [PubMed] [Google Scholar]
- Issaq H. J.; Veenstra T. D. Two-Dimensional Polyacrylamide Gel Electrophoresis (2D-PAGE): Advances and Perspectives. BioTechniques 2008, 44 (5), 697–700. 10.2144/000112823. [DOI] [PubMed] [Google Scholar]
- Lu S.; Sung T.; Lin N.; Abraham R. T.; Jessen B. A. Lysosomal Adaptation: How Cells Respond to Lysosomotropic Compounds. PLoS One 2017, 12 (3), e0173771. 10.1371/journal.pone.0173771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu T.; Kathayat R. S.; Cao Y.; Beck M. W.; Dickinson B. C. A Fluorescent Probe with Improved Water Solubility Permits the Analysis of Protein S-Depalmitoylation Activity in Live Cells. Biochemistry 2018, 57 (2), 221–225. 10.1021/acs.biochem.7b00835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M.; Qu H. PathLocdb: A Comprehensive Database for the Subcellular Localization of Metabolic Pathways and Its Application to Multiple Localization Analysis. BMC Genomics 2010, 11 (S4), S13. 10.1186/1471-2164-11-S4-S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwald E. C.; Mehta S.; Zhang J. Genetically Encoded Fluorescent Biosensors Illuminate the Spatiotemporal Regulation of Signaling Networks. Chem. Rev. 2018, 118 (24), 11707–11794. 10.1021/acs.chemrev.8b00333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta S.; Zhang Y.; Roth R. H.; Zhang J.-F.; Mo A.; Tenner B.; Huganir R. L.; Zhang J. Single-Fluorophore Biosensors for Sensitive and Multiplexed Detection of Signalling Activities. Nat. Cell Biol. 2018, 20 (10), 1215–1225. 10.1038/s41556-018-0200-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulyani A.; Vitriol E.; Allen R.; Wu J.; Gremyachinskiy D.; Lewis S.; Dewar B.; Graves L. M.; Kay B. K.; Kuhlman B.; Elston T.; Hahn K. M. A Biosensor Generated via High Throughput Screening Quantifies Cell Edge Src Dynamics. Nat. Chem. Biol. 2011, 7 (7), 437–444. 10.1038/nchembio.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhuniya S.; Maiti S.; Kim E.-J.; Lee H.; Sessler J. L.; Hong K. S.; Kim J. S. An Activatable Theranostic for Targeted Cancer Therapy and Imaging. Angew. Chem., Int. Ed. 2014, 53 (17), 4469–4474. 10.1002/anie.201311133. [DOI] [PubMed] [Google Scholar]
- Lefevre C.; Kang H. C.; Haugland R. P.; Malekzadeh N.; Arttamangkul S.; Haugland R. P. Texas Red-X and Rhodamine Red-X, New Derivatives of Sulforhodamine 101 and Lissamine Rhodamine B with Improved Labeling and Fluorescence Properties. Bioconjugate Chem. 1996, 7 (4), 482–489. 10.1021/bc960034p. [DOI] [PubMed] [Google Scholar]
- Bredt D. S.; Snyder S. H. NITRIC OXIDE: A Physiologic Messenger Molecule. Annu. Rev. Biochem. 1994, 63 (1), 175–195. 10.1146/annurev.bi.63.070194.001135. [DOI] [PubMed] [Google Scholar]
- Thomas D. D.; Liu X.; Kantrow S. P.; Lancaster J. R. The Biological Lifetime of Nitric Oxide: Implications for the Perivascular Dynamics of NO and O2. Proc. Natl. Acad. Sci. U. S. A. 2001, 98 (1), 355–360. 10.1073/pnas.98.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förstermann U.; Sessa W. C. Nitric Oxide Synthases: Regulation and Function. Eur. Heart J. 2012, 33 (7), 829–837. 10.1093/eurheartj/ehr304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton D.; Fontana J.; Sowa G.; Gratton J.-P.; Lin M.; Li K.-X.; Michell B.; Kemp B. E.; Rodman D.; Sessa W. C. Localization of Endothelial Nitric-Oxide Synthase Phosphorylated on Serine 1179 and Nitric Oxide in Golgi and Plasma Membrane Defines the Existence of Two Pools of Active Enzyme. J. Biol. Chem. 2002, 277 (6), 4277–4284. 10.1074/jbc.M106302200. [DOI] [PubMed] [Google Scholar]
- Bagal S. K.; Brown A. D.; Cox P. J.; Omoto K.; Owen R. M.; Pryde D. C.; Sidders B.; Skerratt S. E.; Stevens E. B.; Storer R. I.; Swain N. A. Ion Channels as Therapeutic Targets: A Drug Discovery Perspective. J. Med. Chem. 2013, 56 (3), 593–624. 10.1021/jm3011433. [DOI] [PubMed] [Google Scholar]
- Shen D.; Wang X.; Li X.; Zhang X.; Yao Z.; Dibble S.; Dong X.; Yu T.; Lieberman A. P.; Showalter H. D.; Xu H. Lipid Storage Disorders Block Lysosomal Trafficking by Inhibiting a TRP Channel and Lysosomal Calcium Release. Nat. Commun. 2012, 3, 731. 10.1038/ncomms1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samie M.; Wang X.; Zhang X.; Goschka A.; Li X.; Cheng X.; Gregg E.; Azar M.; Zhuo Y.; Garrity A. G.; Gao Q.; Slaugenhaupt S.; Pickel J.; Zolov S. N.; Weisman L. S.; Lenk G. M.; Titus S.; Bryant-Genevier M.; Southall N.; Juan M.; Ferrer M.; Xu H. A TRP Channel in the Lysosome Regulates Large Particle Phagocytosis via Focal Exocytosis. Dev. Cell 2013, 26 (5), 511–524. 10.1016/j.devcel.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicoli E.-R.; Weston M. R.; Hackbarth M.; Becerril A.; Larson A.; Zein W. M.; Baker P. R.; Burke J. D.; Dorward H.; Davids M.; Huang Y.; Adams D. R.; Zerfas P. M.; Chen D.; Markello T. C.; Toro C.; Wood T.; Elliott G.; Vu M.; Zheng W.; Garrett L. J.; Tifft C. J.; Gahl W. A.; Day-Salvatore D. L.; Mindell J. A.; Malicdan M. C. V. Lysosomal Storage and Albinism Due to Effects of a De Novo CLCN7 Variant on Lysosomal Acidification. Am. J. Hum. Genet. 2019, 104 (6), 1127–1138. 10.1016/j.ajhg.2019.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellokumpu S. Golgi PH, Ion and Redox Homeostasis: How Much Do They Really Matter?. Front. Cell Dev. Biol. 2019, 7, 93. 10.3389/fcell.2019.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akira S.; Uematsu S.; Takeuchi O. Pathogen Recognition and Innate Immunity. Cell 2006, 124 (4), 783–801. 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- West A. P.; Koblansky A. A.; Ghosh S. Recognition and Signaling by Toll-like Receptors. Annu. Rev. Cell Dev. Biol. 2006, 22, 409–437. 10.1146/annurev.cellbio.21.122303.115827. [DOI] [PubMed] [Google Scholar]
- Reinink P.; Buter J.; Mishra V. K.; Ishikawa E.; Cheng T.-Y.; Willemsen P. T. J.; Porwollik S.; Brennan P. J.; Heinz E.; Mayfield J. A.; Dougan G.; van Els C. A.; Cerundolo V.; Napolitani G.; Yamasaki S.; Minnaard A. J.; McClelland M.; Moody D. B.; Van Rhijn I. Discovery of Salmonella Trehalose Phospholipids Reveals Functional Convergence with Mycobacteria. J. Exp. Med. 2019, 216 (4), 757–771. 10.1084/jem.20181812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga I.; Moody D. B. Mincle Is a Long Sought Receptor for Mycobacterial Cord Factor. J. Exp. Med. 2009, 206 (13), 2865–2868. 10.1084/jem.20092533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendiola A. S.; Ryu J. K.; Bardehle S.; Meyer-Franke A.; Ang K. K.-H.; Wilson C.; Baeten K. M.; Hanspers K.; Merlini M.; Thomas S.; Petersen M. A.; Williams A.; Thomas R.; Rafalski V. A.; Meza-Acevedo R.; Tognatta R.; Yan Z.; Pfaff S. J.; Machado M. R.; Bedard C.; Rios Coronado P. E.; Jiang X.; Wang J.; Pleiss M. A.; Green A. J.; Zamvil S. S.; Pico A. R.; Bruneau B. G.; Arkin M. R.; Akassoglou K. Transcriptional Profiling and Therapeutic Targeting of Oxidative Stress in Neuroinflammation. Nat. Immunol. 2020, 21 (5), 513–524. 10.1038/s41590-020-0654-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou G.-Y.; Storz P. Detecting Reactive Oxygen Species by Immunohistochemistry. Methods Mol. Biol. 2015, 1292, 97–104. 10.1007/978-1-4939-2522-3_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.; Ge C.; Zhu C.; Salaita K. DNA-Based Digital Tension Probes Reveal Integrin Forces during Early Cell Adhesion. Nat. Commun. 2014, 5, 5167. 10.1038/ncomms6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saminathan A.; Devany J.; Pillai K. S.; Veetil A. T.; Schwake M.; Krishnan Y.. A DNA-Based Voltmeter for Organelles. bioRxiv, 523019, 2019. https://www.biorxiv.org/content/10.1101/523019v1. [DOI] [PMC free article] [PubMed]
- Zhao B.; Li N.; Xie T.; Bagheri Y.; Liang C.; Keshri P.; Sun Y.; You M. Quantifying Tensile Forces at Cell-Cell Junctions with a DNA-Based Fluorescent Probe. Chem. Sci. 2020, 11, 8558. 10.1039/D0SC01455A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma R.; Mayor S. GPI-Anchored Proteins Are Organized in Submicron Domains at the Cell Surface. Nature 1998, 394 (6695), 798–801. 10.1038/29563. [DOI] [PubMed] [Google Scholar]
- Schueder F.; Lara-Gutiérrez J.; Beliveau B. J.; Saka S. K.; Sasaki H. M.; Woehrstein J. B.; Strauss M. T.; Grabmayr H.; Yin P.; Jungmann R. Multiplexed 3D Super-Resolution Imaging of Whole Cells Using Spinning Disk Confocal Microscopy and DNA-PAINT. Nat. Commun. 2017, 8 (1), 2090. 10.1038/s41467-017-02028-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B.-C.; Legant W. R.; Wang K.; Shao L.; Milkie D. E.; Davidson M. W.; Janetopoulos C.; Wu X. S.; Hammer J. A.; Liu Z.; English B. P.; Mimori-Kiyosue Y.; Romero D. P.; Ritter A. T.; Lippincott-Schwartz J.; Fritz-Laylin L.; Mullins R. D.; Mitchell D. M.; Bembenek J. N.; Reymann A.-C.; Böhme R.; Grill S. W.; Wang J. T.; Seydoux G.; Tulu U. S.; Kiehart D. P.; Betzig E. Lattice Light-Sheet Microscopy: Imaging Molecules to Embryos at High Spatiotemporal Resolution. Science 2014, 346 (6208), 1257998. 10.1126/science.1257998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmchen F.; Denk W. Deep Tissue Two-Photon Microscopy. Nat. Methods 2005, 2 (12), 932–940. 10.1038/nmeth818. [DOI] [PubMed] [Google Scholar]
- Jonstrup A. T.; Fredsøe J.; Andersen A. H. DNA Hairpins as Temperature Switches, Thermometers and Ionic Detectors. Sensors 2013, 13 (5), 5937–5944. 10.3390/s130505937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies D. R.; Gelinas A. D.; Zhang C.; Rohloff J. C.; Carter J. D.; O’Connell D.; Waugh S. M.; Wolk S. K.; Mayfield W. S.; Burgin A. B.; Edwards T. E.; Stewart L. J.; Gold L.; Janjic N.; Jarvis T. C. Unique Motifs and Hydrophobic Interactions Shape the Binding of Modified DNA Ligands to Protein Targets. Proc. Natl. Acad. Sci. U. S. A. 2012, 109 (49), 19971–19976. 10.1073/pnas.1213933109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levsky J. M.; Singer R. H. Fluorescence in Situ Hybridization: Past, Present and Future. J. Cell Sci. 2003, 116 (14), 2833–2838. 10.1242/jcs.00633. [DOI] [PubMed] [Google Scholar]
- Qing Z.; Xu J.; Hu J.; Zheng J.; He L.; Zou Z.; Yang S.; Tan W.; Yang R. In Situ Amplification-Based Imaging of RNA in Living Cells. Angew. Chem., Int. Ed. 2019, 58 (34), 11574–11585. 10.1002/anie.201812449. [DOI] [PubMed] [Google Scholar]
- Nieves D. J.; Gaus K.; Baker M. A. B. DNA-Based Super-Resolution Microscopy: DNA-PAINT. Genes 2018, 9 (12), 621. 10.3390/genes9120621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta D.; Ebrahimi S. B.; Kusmierz C. D.; Cheng H. F.; Mirkin C. A. Protein Spherical Nucleic Acids for Live-Cell Chemical Analysis. J. Am. Chem. Soc. 2020, 142, 13350. 10.1021/jacs.0c06866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahimi S. B.; Samanta D.; Mirkin C. A. DNA-Based Nanostructures for Live-Cell Analysis. J. Am. Chem. Soc. 2020, 142 (26), 11343–11356. 10.1021/jacs.0c04978. [DOI] [PubMed] [Google Scholar]
- Deo C.; Abdelfattah A. S.; Bhargava H. K.; Berro A. J.; Falco N.; Moeyaert B.; Chupanova M.; Lavis L. D.; Schreiter E. R.. Bright and Tunable Far-Red Chemigenetic Indicators. bioRxiv, 2020.01.08.898783, 2020. https://www.biorxiv.org/content/10.1101/2020.01.08.898783v1. [DOI] [PubMed]
- Ellington A. D.; Szostak J. W. In Vitro Selection of RNA Molecules That Bind Specific Ligands. Nature 1990, 346 (6287), 818–822. 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- Tuerk C.; Gold L. Systematic Evolution of Ligands by Exponential Enrichment: RNA Ligands to Bacteriophage T4 DNA Polymerase. Science 1990, 249 (4968), 505–510. 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- Paige J. S.; Wu K. Y.; Jaffrey S. R. RNA Mimics of Green Fluorescent Protein. Science 2011, 333 (6042), 642–646. 10.1126/science.1207339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal M.; Breaker R. R. Gene Regulation by Riboswitches. Nat. Rev. Mol. Cell Biol. 2004, 5 (6), 451–463. 10.1038/nrm1403. [DOI] [PubMed] [Google Scholar]
- Kellenberger C. A.; Wilson S. C.; Sales-Lee J.; Hammond M. C. RNA-Based Fluorescent Biosensors for Live Cell Imaging of Second Messengers Cyclic Di-GMP and Cyclic AMP-GMP. J. Am. Chem. Soc. 2013, 135 (13), 4906–4909. 10.1021/ja311960g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paige J. S.; Nguyen-Duc T.; Song W.; Jaffrey S. R. Fluorescence Imaging of Cellular Metabolites with RNA. Science 2012, 335 (6073), 1194–1194. 10.1126/science.1218298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCown P. J.; Corbino K. A.; Stav S.; Sherlock M. E.; Breaker R. R. Riboswitch Diversity and Distribution. RNA 2017, 23 (7), 995–1011. 10.1261/rna.061234.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S.; Zaveri A.; Visweswariah S.; Krishnan Y. A Fluorescent Nucleic Acid Nanodevice Quantitatively Images Elevated Cyclic Adenosine Monophosphate in Membrane-Bound Compartments. Small 2014, 10, 4276. 10.1002/smll.201400833. [DOI] [PubMed] [Google Scholar]
- Wu R.; Karunanayake Mudiyanselage A. P. K. K.; Ren K.; Sun Z.; Tian Q.; Zhao B.; Bagheri Y.; Lutati D.; Keshri P.; You M. Ratiometric Fluorogenic RNA-Based Sensors for Imaging Live-Cell Dynamics of Small Molecules. ACS Appl. Bio Mater. 2020, 3 (5), 2633–2642. 10.1021/acsabm.9b01237. [DOI] [PubMed] [Google Scholar]