Abstract
The chaperonin-containing T-complex protein 1 (CCT) subunits participate in diverse diseases. However, little is known about their expression and prognostic values in human head and neck squamous cancer (HNSC). This article aims to evaluate the effects of CCT subunits regarding their prognostic values for HNSC. We mined the transcriptional and survival data of CCTs in HNSC patients from online databases. A protein–protein interaction network was constructed and a functional enrichment analysis of target genes was performed. We observed that the mRNA expression levels of CCT1/2/3/4/5/6/7/8 were higher in HNSC tissues than in normal tissues. Survival analysis revealed that the high mRNA transcriptional levels of CCT3/4/5/6/7/8 were associated with a low overall survival. The expression levels of CCT4/7 were correlated with advanced tumor stage. And the overexpression of CCT4 was associated with higher N stage of patients. Validation of CCTs’ differential expression and prognostic values was achieved by the Human Protein Atlas and GEO datasets. Mechanistic exploration of CCT subunits by the functional enrichment analysis suggests that these genes may influence the HNSC prognosis by regulating PI3K-Akt and other pathways. This study implies that CCT3/4/6/7/8 are promising biomarkers for the prognosis of HNSC.
Keywords: chaperonin-containing TCP-1, TRiC, head and neck squamous cancer, prognosis, Kaplan–Meier plot
1. Introduction
Head and neck squamous cancer (HNSC) is the sixth leading malignancy worldwide, with an annual incidence of more than 6,00,000 cases [1]. Despite considerable advancements in diagnostic and treatment methods, the 5 year overall survival (OS) rate of HNSC remains less than 50% [2]. Hence, prognostic markers and potential drug targets should be identified to enhance the prognosis and individualized treatments.
Molecular chaperones are key molecular complexes in the process of correct folding of proteins to produce energetically stable and functionally competent protein conformations. They are essential in maintaining protein homeostasis and proteome integrity, and in cell growth and survival [3]. They can be classified into two mechanistic classes – chaperones such as HSP70 and HSP90, and chaperonins. The eukaryotic chaperonin family includes the type I chaperonin, HSP60, and the type II hetero-oligomeric chaperonin, TRiC (T-complex protein-1 ring complex, also known as CCT). CCT is a multiprotein complex that functions to assist polypeptides in achieving a functional three-dimensional configuration. CCT is estimated to interact with 5–10% of proteome [4] and is the obligate chaperone for both actin and tubulin [5,6,7].
CCT features a cylindrical architecture composed of two rings stacked opposite one another. Each ring contains eight different subunits, namely CCT1–CCT8 [8,9], which recognize different motifs within substrate proteins. The specific arrangement of these subunits provides the ability to fold certain proteins and let them gain expected functions, leading to protein homeostasis. Disrupted protein homeostasis underlies various diseases and conditions including cancers. Researches have shown that CCTs mediate the folding of a number of proteins implicated in oncogenesis such as prooncogenic proteins signal transducer and activator of transcription 3 (STAT3), p53, cell cycle regulatory proteins cell division cycle protein 20 (CDC20), and tumor suppressor Von Hippel–Lindau (VHL) [10,11,12,13].
It is evident that CCT subunits promote the development of several tumor types, including breast cancer [14,15,16], colorectal cancer [17,18,19], uterine sarcoma [20], lung carcinoma [19], ovarian cancer [21], hepatocellular carcinoma (HCC) [22], and multitudinous tumor cells [23,24]. And HSP70, another member of the chaperone family, proves to be a biomarker of head and neck cancer [25]. However, the involvement of CCT subunits in head and neck cancer remains unknown.
To the best of our knowledge, bioinformatics analysis is yet to be applied to explore the role of CCT subunits in HNSC. RNA and DNA research, an essential component of the biological and biomedical studies, has been revolutionized with the development of microarray technology [26]. On the basis of the analyses of thousands of gene expressions or variations in copy numbers published online, we analyzed in detail the expression and mutations of different CCTs in patients with HNSC to determine the expression patterns, potential functions, and distinct prognostic values.
2. Materials and methods
2.1. mRNA transcriptional levels of CCT subunits
The mRNA transcriptional levels of CCT subunits in cancers were analyzed by the ONCOMINE database (www.oncomine.org). ONCOMINE is currently the world’s largest oncogene chip database and integrated data mining platform [27]. The mRNA expressions of CCTs in clinical cancer specimens were compared with those in normal controls, using a Student’s t-test to generate a P value. The cutoff of P value and fold change were defined as 0.0001 and 2, respectively. The mRNA transcriptional levels of CCTs in HNSC were also analyzed. CCTs expression was assessed in a HNSC tissue relative to its expression in a normal tissue, and the differences associated with P ˂ 0.01 were considered significant. Transcriptional levels of CCTs between head and neck cancer tissues and normal samples were also analyzed by Gene Expression Profiling Interactive Analysis (GEPIA) dataset (http://gepia.cancer-pku.cn/). GEPIA is an interactive web server for analyzing the RNA sequencing expression data from The Cancer Genome Atlas (TCGA) project, using a standard processing pipeline [28]. The unmatched normal and tumor tissues were compared. The raw data were filtered based on the cutoffs |log2FC| > 1 and P < 0.01. The log2(TPM + 1) transformed expression data were used for plotting and one-way ANOVA for differential analysis.
2.2. Relationship between the mRNA levels of CCTs and clinicopathological parameters
To identify the comparability between the tumor group and the normal group of TCGA HNSC, we downloaded the level-3 RNA sequence (RNA Seq) data (fragments per kilobase of transcript per million mapped reads upper quartile data) of HNSC and the corresponding normal tissue samples from the TCGA database (https://www.cancer.gov/tcga). Clinical characteristics of patients from the tumor group and the normal group were compared. The Fisher exact test and t-test were used, respectively, to determine the differences in gender and age between patients in different groups. Furthermore, the impact of age and gender on each CCT mRNA expression was calculated by the LinkedOmics database. The LinkedOmics database (http://www.linkedomics.org/login.php) is a web-based platform for analyzing TCGA datasets [29]. The statistical methods were Pearson correlation test and t-test for age and gender, respectively.
The expression of CCTs with tumor stage for HNSC was also analyzed using GEPIA databases. The statistical method for differential gene expression analysis was one-way ANOVA. Relationship between the mRNA expression levels and patients’ N stage was analyzed by the LinkedOmics database. The method for differential gene expression analysis is Jonckheere’s trend test.
2.3. Survival analysis
The prognostic value of CCTs mRNA expression was evaluated using the LinkedOmics database. The HNSC patient samples were split into two groups by median expression (high vs low expression) and assessed by Kaplan–Meier survival plots and log-rank test. In cases when two survival curves crossed each other – a clear violation of the assumption of proportional hazard rates for log-rank test [30], a two-stage procedure was used. The two-stage procedure includes the log-rank test as the first stage and a proposed procedure for addressing the crossing hazard rates as the second stage [31]. In brief, we set the significance level of the two-stage procedure at 0.05. If the P value in the first stage (P1) was less than 0.0253, then the total P value = P1; otherwise, the P value in the second stage (P2) was calculated, and the total P value = 0.0253 + (1 − 0.0253) × P2. This procedure was conducted using the R package ComparisonSurv.
2.4. Validation of CCTs’ expression level and prognostic value
IHC images of CCTs protein expression in clinical specimens of patients with HNSC and normal tissues were obtained from the Human Protein Atlas database (https://www.proteinatlas.org/) [32]. The HNSC datasets of GSE30784 and GSE41613 were obtained from the NCBI Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/). GSE30784 consists of 167 HNSC samples, 17 dysplasia samples, and 45 normal samples. This dataset was used to validate the expression level of CCTs between tumor and normal samples. GSE41613 consists of 97 HNSC patients with follow-up information. The R packages affy and annotate were used to process the raw data, make expression matrix, and match the probe to their gene symbol. Patient samples were split into two groups by median expression (high vs low expression) and assessed by the log-rank test and two-stage procedure, as mentioned above. Survival curves and the expression level of CCTs were visualized by GraphPad® PRISM version 8.0 (Graph Pad Software, Inc., La Jolla, CA, USA) with P values.
2.5. TCGA data and cBioPortal
TCGA had both sequencing and pathological data on 30 different cancers. The head and neck squamous cell carcinoma (TCGA, Firehose Legacy) dataset including data from 530 cases with pathology reports was selected for further analyses of CCTs using cBioPortal (www.cbioportal.org) [33,34]. The genomic profiles included mutations, copy-number variance from GISTIC, mRNA expression z-scores (RNA Seq V2 RSEM), and protein expression z-scores (reverse-phase protein arrays). Pearson correlation coefficient of co-expression was calculated according to the cBioPortal’s online instruction and visualized by R environment. For the other comparisons of correlations between CCTs mRNA transcriptional levels, the co-expression heatmaps with Pearson correlation coefficients were calculated and visualized by R packages – ggcorrplot and ggthemes.
2.6. Pathway commons and STRING
Pathway commons (https://www.pathwaycommons.org) is an integrated resource of publicly available information about biological pathways [35]. The Search Tool for the Retrieval of Interacting Genes (STRING) is an online database used to predict PPIs, which are essential for interpreting the molecular mechanisms of key cellular activities in carcinogenesis. In this study, Pathway Commons was used to identify the 50 most frequently altered neighbor genes of the CCTs family and the STRING database was used to build a PPI network of these genes mentioned above. Cytoscape (version 3.7.1), an open source bioinformatics software platform, was used to visualize molecular interaction networks [36].
2.7. Functional and pathway enrichment analysis
The Database for Annotation, Visualization, and Integrated Discovery (DAVID, https://david.ncifcrf.gov/) was used to perform the gene ontology (GO) enrichment analysis. DAVID is an online tool for systematic and integrative annotation and enrichment analysis that reveals the biological meaning related to large gene lists [37]. GO analysis for the cellular component (CC), biological process (BP), and molecular function (MF) categories [38] and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis [39] were performed for the selected genes (CCTs and the 50 most frequently altered neighbor genes) using the DAVID. A P value < 0.05 was considered statistically significant. The results were visualized using R packages.
3. Results
3.1. mRNA transcriptional levels of CCTs in patients with HNSC
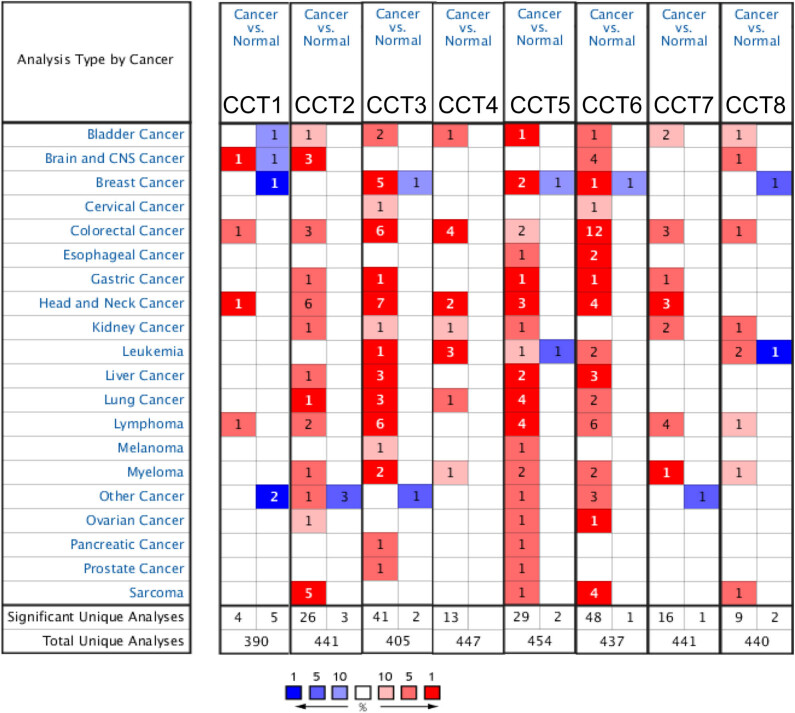
Eight CCT subunits have been identified in mammalian cells. We compared the transcriptional levels of CCTs in cancers with those in normal samples by using ONCOMINE databases (Figure 1). Red cell with a number in it indicated the number of datasets with a statistically significant (P < 0.0001) mRNA overexpression of CCTs in different types of cancers vs the corresponding normal tissue. There are 13 datasets regarding HNSC in total on ONCOMINE. Statistical details of Figure 1 are presented in Table 1. For instance, the mRNA expression levels of CCT3 were significantly upregulated in patients with HNSC in seven datasets. According to Pyeon’s study [40], CCT3 was overexpressed, compared with those in the normal samples in all of the HNSC types: tonsillar carcinoma with a fold change of 2.534, oral cavity carcinoma with a fold change of 3.115, floor of the mouth carcinoma with a fold change of 3.68, oropharyngeal carcinoma with a fold change of 3.001, and tongue carcinoma with a fold change of 2.529 (Table 1). In Estilo’s study [41] and Talbot’s study [42], CCT3 was also overexpressed in tongue squamous cell carcinoma with fold changes of 2.143 and 2.117, respectively. Similarly, the comparisons of other CCTs mRNA expression levels in HNSC and normal tissues are listed in Table 1. No significant difference was found between the mRNA transcriptional levels of CCT8 in head and neck cancer tissues and normal samples.
Figure 1.
The mRNA transcriptional levels of CCTs in different types of cancers (ONCOMINE). This graphic was generated from ONCOMINE, indicating the numbers of datasets with statistically significant (P < 0.0001) mRNA overexpression (red) or downexpression (blue) of CCTs (different types of cancers vs corresponding normal tissue). Cell color was determined by the best gene rank percentile for the analyses within the cell, and the gene rank was analyzed by the percentile of target gene in the top of all genes measured in each research.
Table 1.
The significant changes in CCTs expression in mRNA transcriptional level between different types of head and neck squamous cancers and normal tissues (ONCOMINE database)
| Types of head and neck squamous cancers vs normal tissue | Fold change | P value | t-Test | Ref. | |
|---|---|---|---|---|---|
| CCT1 | Oral cavity carcinoma | 2.505 | 7.21 × 10−6 | 5.987 | Pyeon [40] |
| CCT2 | Tongue squamous cell carcinoma | 2.104 | 5.29 × 10−9 | 6.749 | Estilo[41] |
| Tongue squamous cell carcinoma | 2.161 | 1.45 × 10−9 | 7.065 | Talbot [42] | |
| Oropharyngeal carcinoma | 3.137 | 2.43 × 10−5 | 5.405 | Pyeon [40] | |
| Floor of the mouth carcinoma | 3.146 | 9.32 × 10−6 | 5.823 | Pyeon [40] | |
| Tongue carcinoma | 2.122 | 7.05 × 10−5 | 4.437 | Pyeon [40] | |
| CCT3 | Tonsillar carcinoma | 2.534 | 6.24 × 10−5 | 4.52 | Pyeon [40] |
| Oral cavity carcinoma | 3.115 | 8.2 × 10−6 | 5.543 | Pyeon [40] | |
| Floor of the mouth carcinoma | 3.68 | 9.87 × 10−7 | 6.287 | Pyeon [40] | |
| Oropharyngeal carcinoma | 3.001 | 6.02 × 10−5 | 4.755 | Pyeon [40] | |
| Tongue carcinoma | 2.529 | 2.44 × 10−5 | 4.795 | Pyeon [40] | |
| Tongue squamous cell carcinoma | 2.143 | 1.94 × 10−8 | 6.605 | Estilo [41] | |
| Tongue squamous cell carcinoma | 2.117 | 1.00 × 10−7 | 6.051 | Talbot [42] | |
| CCT4 | Floor of the mouth carcinoma | 3.035 | 9.47 × 10−8 | 7.405 | Pyeon [40] |
| Oropharyngeal carcinoma | 2.76 | 2.89 × 10−5 | 5.516 | Pyeon [40] | |
| CCT5 | Tongue squamous cell carcinoma | 3.009 | 1.64 × 10−12 | 8.836 | Talbot [42] |
| Tongue squamous cell carcinoma | 2.965 | 1.38 × 10−11 | 8.354 | Estilo [41] | |
| Floor of the mouth carcinoma | 2.142 | 6.40 × 10−7 | 9.206 | Pyeon [40] | |
| CCT6 | Nasopharyngeal carcinoma | 2.287 | 5.59 × 10−6 | 6.21 | Sengupta [73] |
| Floor of the mouth carcinoma | 2.845 | 2.31 × 10−5 | 6.025 | Pyeon [40] | |
| Tongue carcinoma | 2.362 | 8.23 × 10−6 | 5.027 | Pyeon [40] | |
| CCT7 | Floor of the mouth carcinoma | 2.872 | 8.37 × 10−8 | 7.159 | Pyeon [40] |
| Oral cavity carcinoma | 2.113 | 2.95 × 10−5 | 4.922 | Pyeon [40] | |
| Oropharyngeal carcinoma | 2.273 | 5.85 × 10−5 | 4.731 | Pyeon [40] | |
| CCT8 | NA | NA | NA | NA | NA |
3.2. Relationship between the mRNA levels of CCTs and the clinicopathological parameters of patients with HNSC
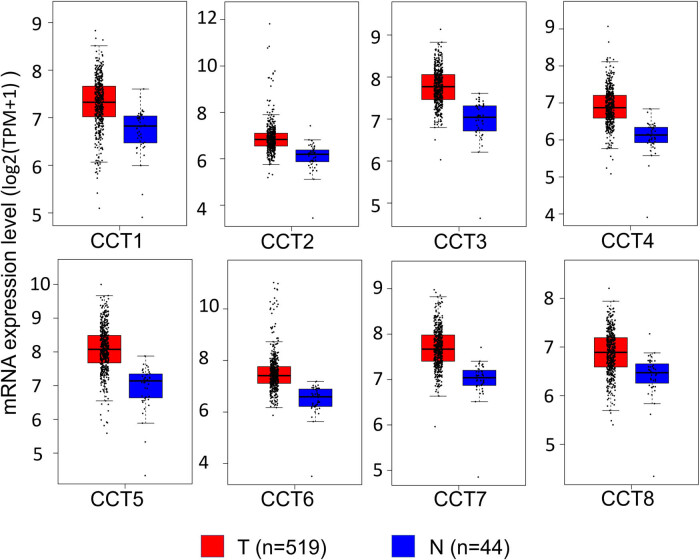
Using GEPIA dataset (http://gepia.cancer-pku.cn/), we compared the mRNA expression of CCTs between head and neck cancer tissues and normal samples. The results indicated that the expression levels of CCTs were higher in head and neck cancer tissues than in normal tissues (Figure 2). To identify the comparability between the tumor group and the normal group of TCGA HNSC, clinical characteristics of patients from the tumor group and the normal group were compared. There were no differences in gender or age between patients in the two groups (Table S1, ESI). Furthermore, the impact of age and gender on each CCT mRNA expression in TCGA HNSC samples was calculated. The results showed that CCT4 mRNA expression was related to gender statistically, with a higher expression level in male than in female (Table S2).
Figure 2.
The expression level of CCTs in HNSC (GEPIA). Boxplots showing the relative expression of eight CCTs in HNSC and normal samples. The red and blue boxes represent cancer and normal tissues, respectively.
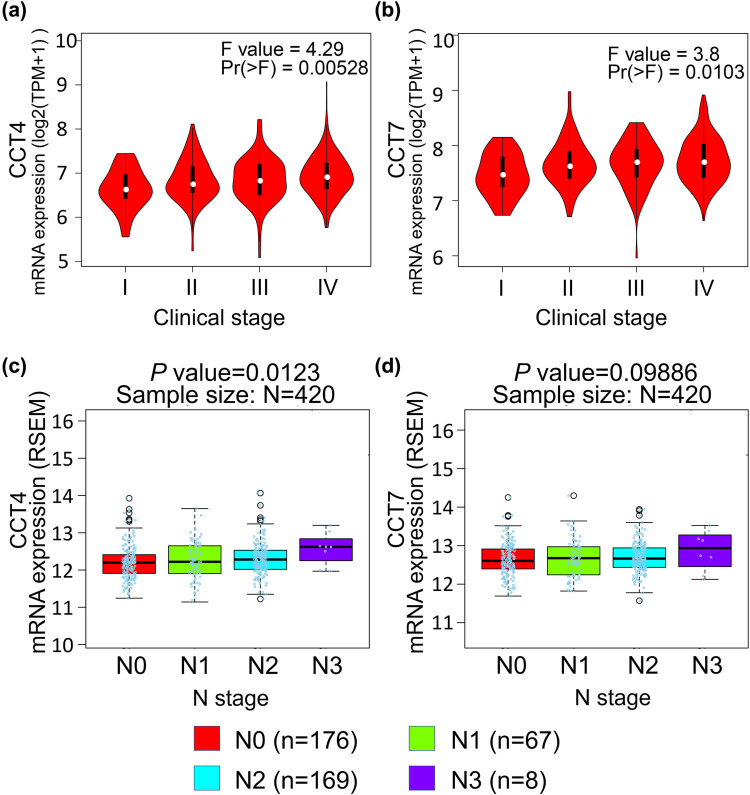
The expression of CCTs with tumor stage for HNSC was also analyzed. CCT4 and CCT7 groups significantly varied, whereas the other groups did not significantly differ (Figure 3a and b and Supplement Figure 1). Furthermore, there were differences for CCT4 (P = 0.0123) and CCT7 (P = 0.09886) in the mRNA expression levels and patients’ N stage, a staging system developed by the American Joint Committee on Cancer to classify patients by the involvement of regional lymph nodes (Figure 3c and d).
Figure 3.
Correlation between the CCT4/7 expression and tumor stage in HNSC patients (GEPIA and LinkedOmics). (a and b) Violin plots showing the relative expression of CCT4/7 in HNSC patients in stage I, II, III, or IV. (c and d) Boxplots indicating the relative expression of CCT4/7 in HNSC patients with N stage of 0, 1, 2, or 3.
3.3. Association of the decreased mRNA expression of CCTs with the improved prognosis of patients with HNSC
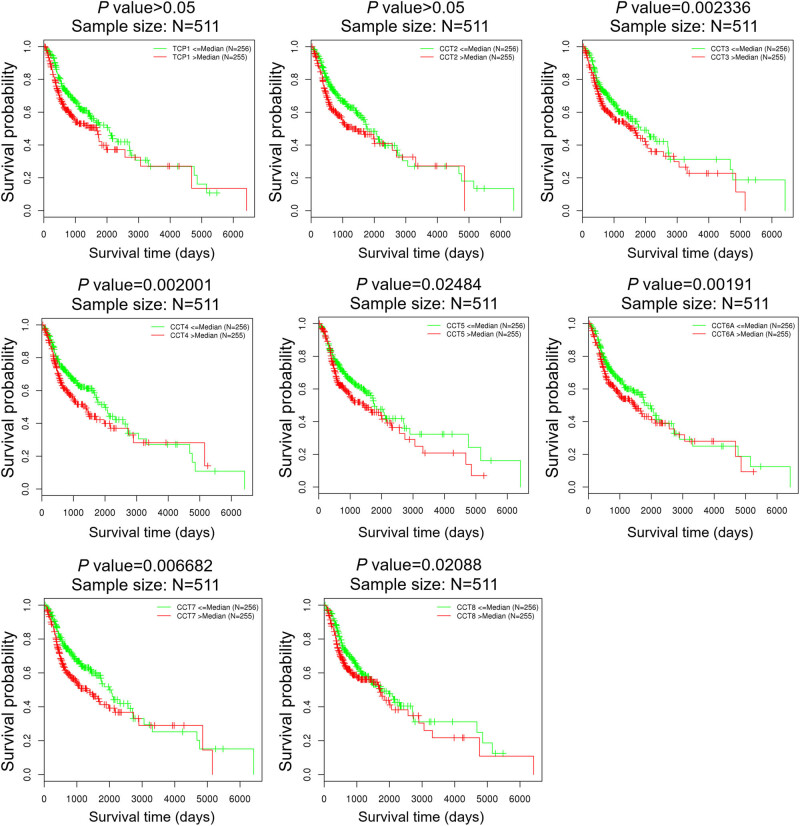
We further explored the critical efficiency of CCTs in the survival of patients with HNSC using the LinkedOmics database. The log-rank test and two-stage procedure analyses for Kaplan–Meier curves from LinkedOmics revealed that the decreased CCT3/4/5/6/7/8 mRNA levels were significantly associated with the OS (Figure 4).
Figure 4.
The prognosis value of CCTs in HNSC patients (LinkedOmics). The prognosis value of mRNA expression levels of CCTs in HNSC patients, analyzed by LinkedOmics.
3.4. Validation of the expression status and prognosis value of CCTs
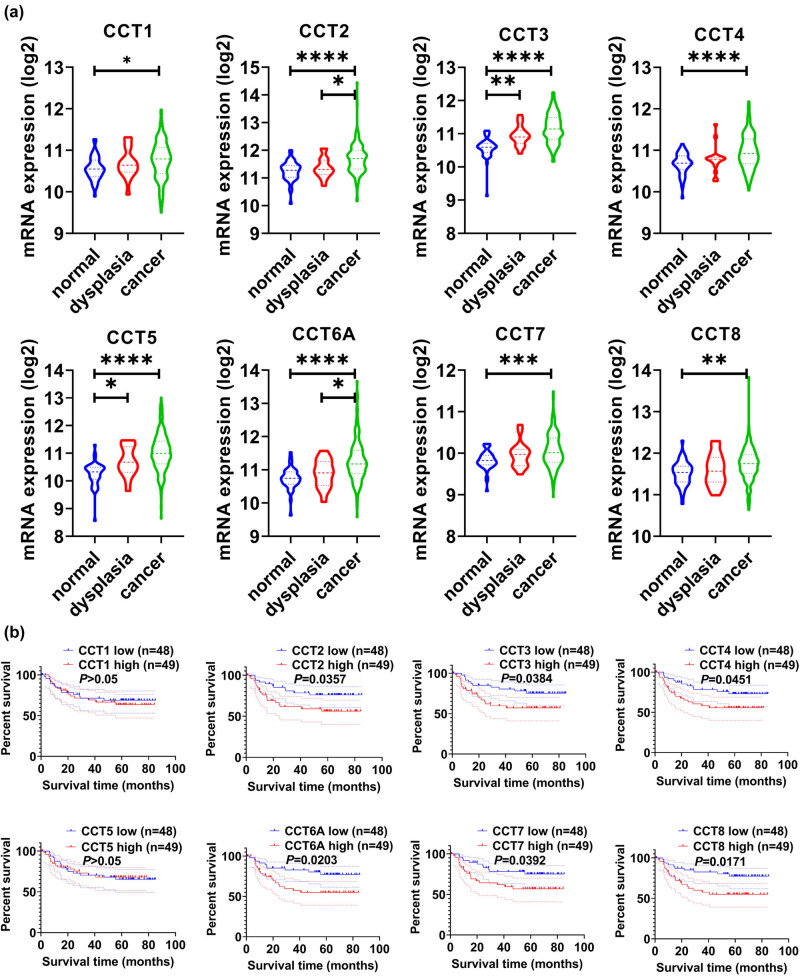
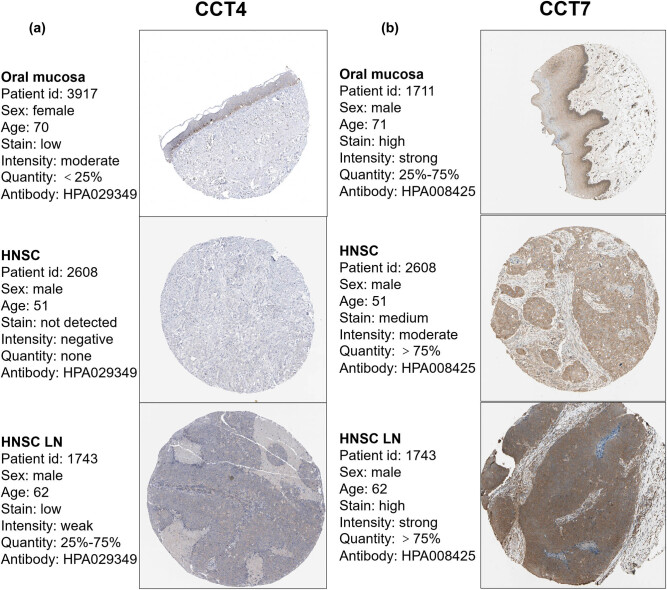
We used multiple datasets from the GEO database to validate the expression status and prognosis value of CCTs. Figure 5a demonstrates the expression status of CCT subunits in normal, dysplasia, and OSCC samples of GSE30784. As shown in the figure, the expression status of CCTs was positively correlated with the disease status. Furthermore, GSE41613 was used to validate CCTs’ prognosis value. This dataset contained not only gene transcriptional data but also follow-up information, including 97 patients of HNSC. Patient samples were split into two groups by median expression (high vs low expression) and assessed by the log-rank test and two-stage procedure, as mentioned in the Materials and methods section. Survival analysis was conducted and significant differences were observed between high CCTs and low CCTs groups (Figure 5b). Moreover, immunohistochemistry (IHC) staining obtained from The Human Protein Atlas database also demonstrated the expression status of CCTs family genes and the patient data, as shown in Figure 6 and Supplement Figure 2. We found that CCT2, CCT5, CCT6, and CCT8 proteins were more highly expressed in the HNSC tissues than in the normal tissues (Supplement Figure 2). Notably, the expressions of CCT4 and CCT7 were higher in HNSC lymph node metastasis than in the primary site or normal tissue (Figure 7), which indicated their potential role in tumor metastasis process. These results confirmed that CCTs were closely related to HNSC and were of prognosis value.
Figure 5.
Validation of CCTs’ differential expression and prognostic values using GEO datasets. (a) Validation of CCTs expression level in GSE30784. The expression status of eight CCTs was positively correlated with the disease status. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. (b) Validation of CCTs’ prognostic value in GSE41613. High CCTs expression groups had significantly lower OS than low CCTs expression groups. Shown is the log-rank P value.
Figure 6.
Validation of CCT4/7 in the translational level. IHC staining for CCTs in HNSC and normal tissue from the Human Protein Atlas Portal. (a and b) Validation of CCT4 and CCT7 in HNSC primary site, lymph node metastasis, and normal tissue by The Human Protein Atlas database (IHC).
Figure 7.
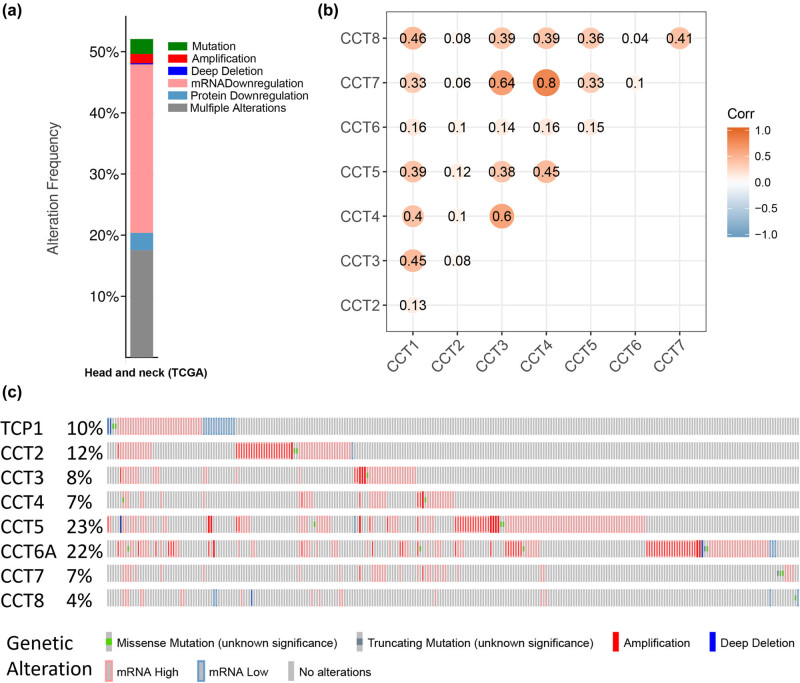
Genetic expression and alteration analysis associated with CCTs in TCGA HNSC (cBioPortal). (a) The total alteration frequency of eight CCT genes in TCGA HNSC is illustrated. (b) Co-expression heatmap of CCT genes in TCGA HNSC, visualized by the R environment. (c) Oncoprint in the cBioPortal represented the proportion and distribution of samples with alterations in CCTs.
3.5. Predicted functions and pathways of the changes in CCTs and their frequently altered neighbor genes in patients with HNSC
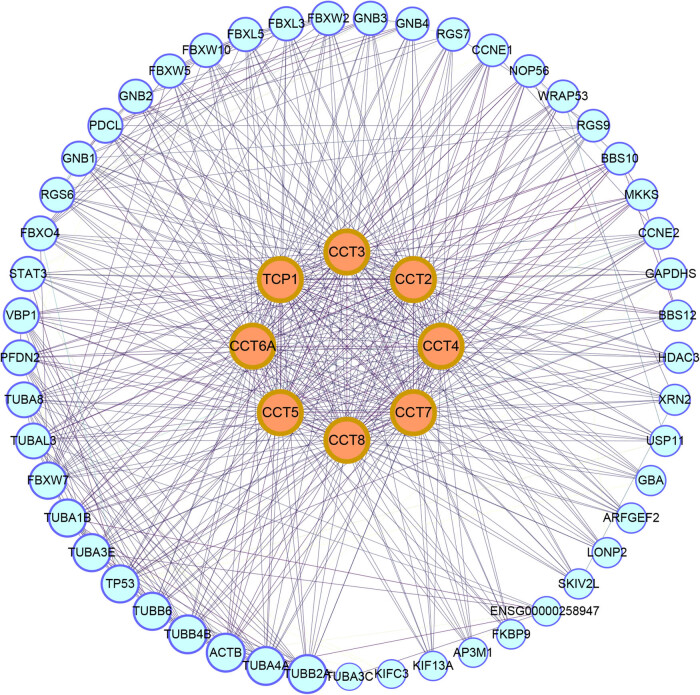
We analyzed the CCTs alterations by using the cBioPortal online tool for head and neck squamous cell carcinoma (TCGA, Firehose Legacy). CCTs were altered in 276 samples out of 530 patients with HNSC (52%) (Figure 7a). Two or more alterations were detected in almost one third of the samples (93 samples) (Figure 7c). We also calculated the correlations of CCTs with each other by analyzing their mRNA expression (RNA Seq V2 RSEM) via the cBioPortal online tool for head and neck squamous cell carcinoma (TCGA, Firehose Legacy), and Pearson’s correlation was included (Figure 7b). The results indicated significant and positive correlations in the following CCTs: CCT1 with CCT3, CCT4, and CCT8; CCT3 with CCT1, CCT4, and CCT7; CCT4 with CCT1, CCT3, CCT5, and CCT7; CCT7 with CCT3, CCT4, and CCT8; and CCT8 with CCT1 and CCT7 (Figure 7b). More correlation heatmaps are presented in Figure S3, which calculated and compared CCT subunits’ correlations in head and neck normal tissues (Figure S3A), in another dataset GSE41613 (Figure S3B), and at different cancer stages (Figure S3C and D). We then constructed the network for CCTs and the 50 most relevant genes on Pathway Commons (https://www.pathwaycommons.org) [35]. In detail, we input the eight CCT subunits to Pathway Commons and chose HNSC as the cancer type of interest. Seventy-seven relevant genes were calculated and presented by the website ranking by relevance. We chose 50 most relevant genes for further analysis. And a matched functional protein association network of the 50 genes was visualized on The Search Tool for the Retrieval of Interacting Genes (STRING) (Figure 8). The results showed that cancer pathway-related genes, such as TP53, STAT3, CCNE1, and CCNE2, were closely associated with CCTs alterations, just as previously reported by others [43].
Figure 8.
Protein–protein interaction (PPI) network. The PPI network of CCTs and 50 most frequently altered neighbor genes, which includes 58 nodes and 591 edges (PPI enrichment P value < 1.0 × 10−16), visualized by Cytoscape software.
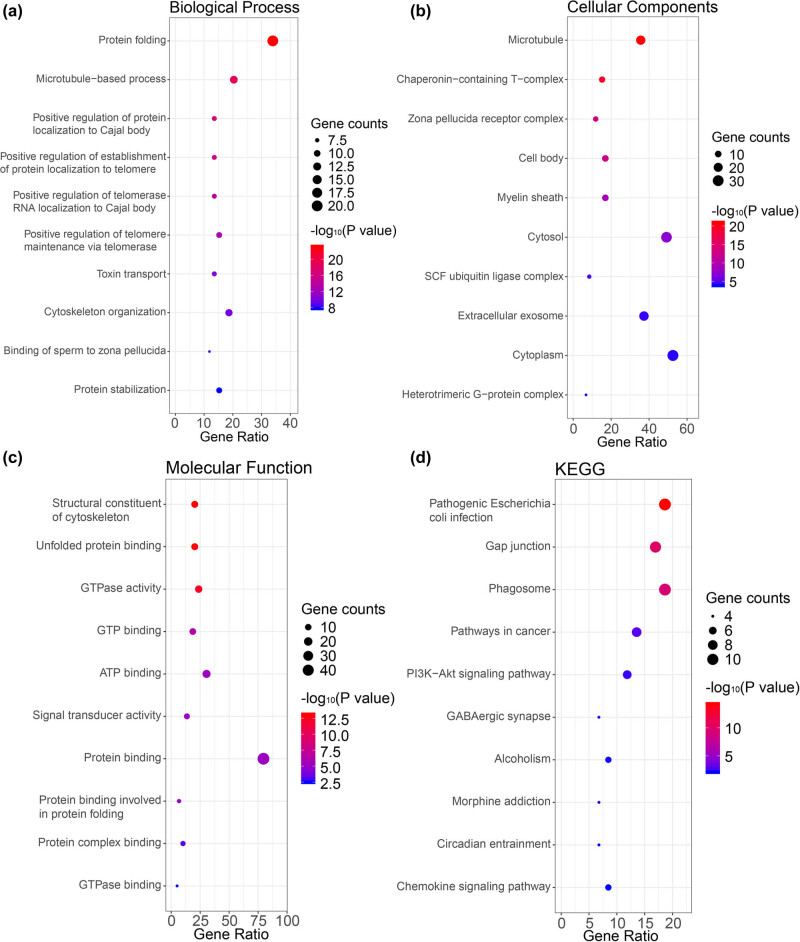
The functions of CCTs and the genes significantly associated with their alterations were predicted by analyzing GO and KEGG in the DAVID. The GO enrichment analysis predicted the functional roles of target genes on the basis of three aspects, including BPs, CCs, and MFs. We found that protein folding, microtubule-based process, and cytoskeleton organization were significantly regulated by the CCTs alterations in HNSC (Figure 9a and Table S3). These genes were largely related to microtubule and extracellular exosome, and were mostly located in cytoplasm or cytosol (Figure 9b and Table S4). Their MF was significantly related to protein binding and ATP binding (Figure 9c and Table S5). The predicted functions and locations were consistent with CCTs’.
Figure 9.
Functional enrichment analysis of CCTs and neighbor genes in HNSC. The bubble diagrams display the enrichment results of CCTs and top 50 genes altered in the CCTs neighborhood in HNSC. (a) Biological processes. (b) Cellular components. (c) Molecular functions. (d) KEGG pathways.
KEGG analysis can define the pathways related to the functions of CCTs alterations and the frequently altered neighbor genes. Ten pathways related to the functions of CCTs alterations in HNSC were found through the KEGG analysis (Figure 9d and Table S6). Among these pathways, gap junction, pathways in cancer, phagosome, and PI3K-Akt signaling pathway were closely related to the tumorigenesis and pathogenesis of cancer.
4. Discussion
CCTs dysregulation has been reported in many cancers. To our knowledge, the present study is the first to explore the mRNA expression and prognostic values of different CCT genes in head and neck squamous cancer. We hope that our findings will contribute to the available knowledge, improve treatment designs, and enhance the accuracy of prognosis for patients with HNSC.
In the present study, we applied ONCOMINE and TCGA databases to identify the mRNA expression differences of CCTs in HNSC samples and normal tissues. GEPIA and LinkedOmics contributed to comparing the relationship between the mRNA levels of CCTs and the clinicopathological parameters and prognosis. Multiple GEO datasets were used to validate the expression status and prognosis value of CCTs. IHC staining obtained from the HPA database was used to validate the expression difference on a protein level. Functional enrichment analysis of CCTs and most frequently altered genes predicted by Pathway Commons was performed by DAVID.
ONCOMINE consisted of multiple datasets, including DNA-seq data of TCGA, some data from GEO, and many other datasets from literature [44]. It is difficult for us to make comparisons of its comprehensive data. GEPIA and LinkedOmics were both based on the TCGA RNA-seq data [28,29], so the results from these two databases were merely different. Each of these interactive web servers had advantages and disadvantages in terms of application. For example, LinkedOmics could be used to compare detailed clinical information such as age, gender, N stage, etc., while GEPIA could be used to compare the mRNA expression level between the tumor tissue and the normal tissue. These webs simplified the work of downloading and reprocessing the raw data from TCGA and could serve as good assistants for researchers. Distinguishing from the TCGA data, GEO datasets were used for validation. The comparability between the tumor group and the normal group of TCGA HNSC was identified by comparing patients’ gender and age. The impact of age and gender on each CCT mRNA expression in TCGA HNSC samples was also calculated. The results showed that only the CCT4 mRNA expression was related to gender statistically, while others were not related to gender or age. However, the age distribution and gender composition of the patients were similar among different groups. Hence, gender or age has no impact on the CCTs mRNA expression between groups.
In this study, transcriptional levels of CCTs in cancers with those in normal samples indicated that CCTs’ significant overexpression was observed in various types of cancers, including liver cancer, lung cancer, breast cancer, bladder cancer, and so on. This was consistent with the result of literature review, suggesting that CCTs overexpression was not a tissue-specific phenomenon [15,18,45]. According to GEPIA, the expression level of CCTs in HNSC was all higher than normal tissues, but none of them showed significant differences. The expression levels of CCT4 and CCT7 demonstrated to be significantly associated with the tumor stage of HNSC, and only CCT4 was related to patients’ N stage. Interestingly, most of the CCTs were significantly associated with patients’ OS, as confirmed by GSE41613.
Validation of expression status and prognosis value of CCTs was done using datasets from the GEO database and IHC pictures from the HPA. As shown in Figure 5a, there were significant differences in CCTs expression level among normal, dysplasia, and cancer tissue. The result reflected that the expression status of CCTs was positively correlated with the disease status. On a translational level, the IHC showed that CCT4 and CCT7 were less expressed in HNSC samples than in oral mucosa, but their expressions in lymph node metastasis samples were significantly higher than that of a normal tissue, which further indicated their potential role in cancer migration and metastasis. Furthermore, survival analysis between high and low CCTs groups from the GEO database definitely showed their prognostic values in HNSC.
We cannot ignore the fact that the IHC staining for some CCTs was not higher in HNSC tissues than in normal tissues, despite their mRNA transcription being higher in HNSC tissues. The IHC data were obtained from The Human Protein Atlas database. Although the mRNA expression data of CCTs were from large sample sizes, the IHC data of each CCT were from one or two patients’ tumor tissue. The heterogeneity of biological samples in studying RNA and protein expression levels might be one of the causes. In addition, the translation process from mRNA to proteins might be hampered by mRNA modifications in cancer [46]. The phenomenon of inconsistency of RNA and protein expression levels did exist [47], and further experiments were needed to unveil the possible reasons. Genetic expression and alteration analysis associated with CCTs in TCGA HNSC showed that more than half of the patients possessed at least one CCT subunit alteration. Correlations between CCTs mRNA transcriptional levels in head and neck normal tissues revealed a strong correlation among the eight CCT subunits, which was consistent with their biological function of a multiprotein complex [8]. While the correlation heatmaps in HNSC, from TCGA or GSE41613, differed from those in normal tissues, some CCT subunits barely correlated with each other, suggesting an abnormal function of CCTs in tumor. Such correlations were similar at different cancer clinical stages. Pathway analysis showed that pathways related to the functions of CCTs and neighbor genes included pathways in cancer, PI3K-Akt signaling pathway, gap junction, and phagosome, other than expected pathways such as protein folding and microtubule-based process.
PI3K-Akt signaling pathway was one of the most important pathways in HNSC, which was a critical regulatory axis for cell growth, survival, motility, and metabolism in both normal physiology and cancer. TCGA and other studies revealed that the majority of HNSCCs possess alterations in the PI3K/Akt/mTOR pathway [48,49]. In addition, the members of the PI3K/Akt/mTOR axis interacted with and contributed to the regulation of several other signaling molecules in HNSC, including tumor suppressor, TP53, NF-κB, and MAPK/ERK. Since CCTs were predicted to interact with the PI3K-Akt signaling pathway by a functional enrichment analysis, we presumed that the potential mechanism underlying CCTs’ differential expression and prognostic values in HNSC was a cross talk with the PI3K-Akt pathway. Further experiments were needed to better confirm the findings of this study.
The molecular chaperone network (including chaperons and chaperonins) played a central role in maintaining protein homeostasis and proteome integrity [50,51]. The main substrates for CCT seemed to be involved in the folding of cytoskeletal proteins, such as actins and tubulins [5,6,7], and other intracellular proteins, such as protein phosphatase PP2A regulatory subunit B [52], histone deacetylase [53], and cyclin E1 [54]. In addition, CCT was involved in the regulation of cell cycle progression and cytoskeletal organization [55]. The transcriptions of CCT subunits were apparently increased from the G1/S phase transition to the early phase in the process of cell cycle in murine and human cell lines [13,56]. Reduction of the CCT or CCT subunit could lead to a growth arrest, with a great change in cell morphology and motility [57]. Scientists also deemed that CCT’s function in assisting the folding of actin and tubulin could enhance cell migration related to cancer metastasis [58,59]. Moreover, CCT may be overexpressed in cancer cells [24]. Inevitably, members of the molecular chaperone pathway have been implicated in the development of cancers [60,61,62]. Researches have shown that prooncogenic proteins, such as STAT3, p53, CDC20, and tumor suppressor VHL [10,11,12,13], were mediated by CCTs, which partly explained their role in oncogenesis.
The CCTs are essential for cell survival, correct folding and for the function of diverse proteins. Each CCT subunit is the product of an individual gene [63]. However, at present, it is still unclear whether or not the eight subunits form a single particle and cover all functions, and whether they exist as individual subunits or smaller complexes. For example, CCT4 and CCT5 homo-oligomers have been found to form 8-fold double rings without the other subunits. And the CCT4 or CCT5 oligomer rings exhibited activities of ATP hydrolysis and protein folding, comparable to the TRiC ring [64]. CCT4 associates with the plasma membrane and alters the cell shape [65], whereas CCT5 regulates actin expression via the serum response factor pathway [66]. As increasing information on the individual roles played by CCTs components is being accumulated, the possibility of subunit-specific roles for these proteins in cell growth and tumorigenesis also needs to be considered [67].
CCT subunits are also irregulated in different types of cancers. For instance, CCT1 and CCT2 are upregulated in patients with HCC and colonic cancer and correlate with tumor proliferation and poor prognosis [56]. CCT3 is widely studied in different cancers. CCT3 is overexpressed in gastric cancer, and CCT3 knockdown can also suppress the proliferation and induce cell apoptosis in gastric cancer [68] and papillary thyroid carcinoma (PTC) [45]. CCT4 participates in protein CCT6A as a potential prognostic biomarker in glioblastoma [69]. CCT7 effectively regulates VHL proteostasis, which is responsible for sporadic renal cell carcinoma [70]. CCT8 can promote the migration and invasion of esophageal squamous carcinoma by regulating actin and tubulin [71]. And CCT8 has been reported to be upregulated in colon cancer and HCC [56,72]. These studies concluded, as we do, that CCT subunit expression could be a marker of cancer.
We acknowledge that there were some limitations and shortcomings in this study. First, head and neck squamous cancer consists of various types of cancers derived from different anatomic sites, such as pharynx, oral cavity, tongue, larynx, upper esophagus, etc. They have the same histological type of squamous cell carcinoma, but also have different prognoses and responses to treatment due to anatomic differences. This study was mainly focused on the available data of ONCOMINE and TCGA, whose HNSCs were mostly comprised of cancer from the oral cavity and tongue. Hence, a data bias of the cancer type was difficult to be avoided. Second, this study was mainly conducted by the data mining of an online public database. The results were analyzed by the methodology but not validated fully by experiments. Third, more datasets should be involved to obtain a solid result. The datasets for validation, GSE30784 and GSE41613, were from the same departments and using the same array in analysis. Although they had different sets of data and different roles for validation, datasets with demographic diversity were of better reliability.
Here, we systemically analyzed the expression and prognostic values of CCTs in HNSC and provided a better understanding of the heterogeneity and complexity of the molecular biological properties of HNSC. Our results implied that CCT3/4/6/7/8 were promising prognostic biomarkers for the improvement of HNSC survival and prognostic accuracy.
Abbreviations
- CCT
Chaperonin-containing T-complex protein 1
- TRiC
T-complex protein-1 ring complex
- HNSC
Head and neck squamous cancer
- DAVID
Database for annotation, visualization, and integrated discovery
- PPI
Protein–protein interaction
- GEPIA
Gene expression profiling interactive analysis
- STRING
Search tool for the retrieval of interacting genes
- OS
Overall survival
- STAT3
Signal transducer and activator of transcription 3
- CDC20
Cell cycle regulatory proteins cell division cycle protein 20
- VHL
Von Hippel–Lindau
- HCC
Hepatocellular carcinoma
- TCGA
The cancer genome atlas
- LN
Lymph node
- IHC
Immunohistochemistry
- HPA
Human protein atlas
- GO
gene ontology
- CC
Cellular component
- BP
Biological process
- MF
Molecular function
- KEGG
Kyoto encyclopedia of genes and genomes
- HSP
Heat shock protein
- PI3K
Phosphoinositide 3-kinase
- Akt
Protein kinase B
- mTOR
Mammalian target of rapamycin
- NF-κB
Nuclear factor-kappa B
- MAPK
Mitogen-activated protein kinase
- ERK
Extracellular signal-regulated kinase
Acknowledgments
This study was supported by the Research and Development Project of Scientific Research Instruments and Equipment of the Chinese Academy of Sciences – major instruments project (YJKYYQ20180039) and the Digestive Medical Coordinated Development Center of Beijing Municipal Administration of Hospitals (No. XXZ0604).
Footnotes
Conflict of interest: The authors declare that there are no competing interests.
Author contributions: YD and LL conceived and designed the research. YD and ZW acquired and analyzed the data. YD, ZW, and SL interpreted the results. YD and SL prepared figures and tables. YD and LL drafted, edited, and revised manuscript. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work is appropriately investigated and resolved.
Availability of data and materials: The datasets generated and/or analyzed during the current study are available in the ONCOMINE (www.oncomine.org), the Human Protein Atlas (http://www.proteinatlas.org), TCGA (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga), and GEO (https://www.ncbi.nlm.nih.gov/gds/) repositories.
Ethics approval and consent to participate: Approval by a local Ethics Committee was not needed because all data used in this study were publicly available and open-access. All the datasets were retrieved from the publishing literature, so it was confirmed that all written informed consent was obtained.
Patient consent for publication: Not applicable.
References
- [1].Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA A Cancer J Clinic. 2018;68(1):7–30. [DOI] [PubMed]; Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA A Cancer J Clinic. 2018;68(1):7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- [2].Bose P, Brockton NT, Dort JC. Head and neck cancer: from anatomy to biology. Int J Cancer. 2013;133(9):2013–23. [DOI] [PubMed]; Bose P, Brockton NT, Dort JC. Head and neck cancer: from anatomy to biology. Int J Cancer. 2013;133(9):2013–23. doi: 10.1002/ijc.28112. [DOI] [PubMed] [Google Scholar]
- [3].Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475(7356):324–32. [DOI] [PubMed]; Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475(7356):324–32. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- [4].Yam AY, Xia Y, Lin H-TJ, Burlingame A, Gerstein M, Frydman J. Defining the TRiC/CCT interactome links chaperonin function to stabilization of newly made proteins with complex topologies. Nat Struct Mol Biol. 2008;15(12):1255–62. [DOI] [PMC free article] [PubMed]; Yam AY, Xia Y, Lin H-TJ, Burlingame A, Gerstein M, Frydman J. Defining the TRiC/CCT interactome links chaperonin function to stabilization of newly made proteins with complex topologies. Nat Struct Mol Biol. 2008;15(12):1255–62. doi: 10.1038/nsmb.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Gao Y, Thomas JO, Chow RL, Lee GH, Cowan NJ. A cytoplasmic chaperonin that catalyzes beta-actin folding. Cell. 1992;69(6):1043–50. [DOI] [PubMed]; Gao Y, Thomas JO, Chow RL, Lee GH, Cowan NJ. A cytoplasmic chaperonin that catalyzes beta-actin folding. Cell. 1992;69(6):1043–50. doi: 10.1016/0092-8674(92)90622-j. [DOI] [PubMed] [Google Scholar]
- [6].Frydman J, Nimmesgern E, Erdjument-Bromage H, Wall JS, Tempst P, Hartl FU. Function in protein folding of TRiC, a cytosolic ring complex containing TCP-1 and structurally related subunits. EMBO J. 1992;11:4767–78. [DOI] [PMC free article] [PubMed]; Frydman J, Nimmesgern E, Erdjument-Bromage H, Wall JS, Tempst P, Hartl FU. Function in protein folding of TRiC, a cytosolic ring complex containing TCP-1 and structurally related subunits. EMBO J. 1992;11:4767–78. doi: 10.1002/j.1460-2075.1992.tb05582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Ursic D, Culbertson MR. The yeast homolog to mouse Tcp-1 affects microtubule-mediated processes. Mol Cell Biol. 1991;11(5):2629–40. [DOI] [PMC free article] [PubMed]; Ursic D, Culbertson MR. The yeast homolog to mouse Tcp-1 affects microtubule-mediated processes. Mol Cell Biol. 1991;11(5):2629–40. doi: 10.1128/mcb.11.5.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Spiess C, Meyer AS, Reissmann S, Frydman J. Mechanism of the eukaryotic chaperonin: protein folding in the chamber of secrets. Trends Cell Biol. 2004;14:598–604. [DOI] [PMC free article] [PubMed]; Spiess C, Meyer AS, Reissmann S, Frydman J. Mechanism of the eukaryotic chaperonin: protein folding in the chamber of secrets. Trends Cell Biol. 2004;14:598–604. doi: 10.1016/j.tcb.2004.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Carrascosa JL, Llorca O, Valpuesta JM. Structural comparison of prokaryotic and eukaryotic chaperonins. Micron. 2001;32(1):43–50. [DOI] [PubMed]; Carrascosa JL, Llorca O, Valpuesta JM. Structural comparison of prokaryotic and eukaryotic chaperonins. Micron. 2001;32(1):43–50. doi: 10.1016/s0968-4328(00)00027-5. [DOI] [PubMed] [Google Scholar]
- [10].McClellan AJ, Scott MD, Frydman J. Folding and quality control of the VHL tumor suppressor proceed through distinct chaperone pathways. Cell. 2005;121(5):739–48. [DOI] [PubMed]; McClellan AJ, Scott MD, Frydman J. Folding and quality control of the VHL tumor suppressor proceed through distinct chaperone pathways. Cell. 2005;121(5):739–48. doi: 10.1016/j.cell.2005.03.024. [DOI] [PubMed] [Google Scholar]
- [11].Trinidad AG, Muller PAJ, Cuellar J, Klejnot M, Nobis M, Valpuesta JM, et al. Interaction of p53 with the CCT complex promotes protein folding and wild-type p53 activity. Mol Cell. 2013;50(6):805–17. [DOI] [PMC free article] [PubMed]; Trinidad AG, Muller PAJ, Cuellar J, Klejnot M, Nobis M, Valpuesta JM. et al. Interaction of p53 with the CCT complex promotes protein folding and wild-type p53 activity. Mol Cell. 2013;50(6):805–17. doi: 10.1016/j.molcel.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kasembeli M, Lau WCY, Roh S-H, Eckols TK, Frydman J, Chiu W, et al. Modulation of STAT3 folding and function by TRiC/CCT chaperonin. PLoS Biol. 2014;12:e1001844. [DOI] [PMC free article] [PubMed]; Kasembeli M, Lau WCY, Roh S-H, Eckols TK, Frydman J, Chiu W. et al. Modulation of STAT3 folding and function by TRiC/CCT chaperonin. PLoS Biol. 2014;12:e1001844. doi: 10.1371/journal.pbio.1001844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Yokota S, Yanagi H, Yura T, Kubota H. Cytosolic chaperonin is up-regulated during cell growth. Preferential expression and binding to tubulin at G(1)/S transition through early S phase. J Biol Chem. 1999;52:37070–8. [DOI] [PubMed]; Yokota S, Yanagi H, Yura T, Kubota H. Cytosolic chaperonin is up-regulated during cell growth. Preferential expression and binding to tubulin at G(1)/S transition through early S phase. J Biol Chem. 1999;52:37070–8. doi: 10.1074/jbc.274.52.37070. [DOI] [PubMed] [Google Scholar]
- [14].Chen L, Zhang Z, Qiu J. Chaperonin CCT-mediated AIB1 folding promotes the growth of ER alpha-positive breast cancer cells on hard substrates. PLoS One. 2014;9(5):e96085. [DOI] [PMC free article] [PubMed] [Retracted]; Chen L, Zhang Z, Qiu J. Chaperonin CCT-mediated AIB1 folding promotes the growth of ER alpha-positive breast cancer cells on hard substrates. PLoS One. 2014;9(5):e96085. doi: 10.1371/journal.pone.0096085. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [15].Guest ST, Kratche ZR, Bollig-Fischer A, Haddad R, Ethier SP. Two members of the TRiC chaperonin complex, CCT2 and TCP1 are essential for survival of breast cancer cells and are linked to driving oncogenes. Exp Cell Res. 2015;332(2):223–35. [DOI] [PubMed]; Guest ST, Kratche ZR, Bollig-Fischer A, Haddad R, Ethier SP. Two members of the TRiC chaperonin complex, CCT2 and TCP1 are essential for survival of breast cancer cells and are linked to driving oncogenes. Exp Cell Res. 2015;332(2):223–35. doi: 10.1016/j.yexcr.2015.02.005. [DOI] [PubMed] [Google Scholar]
- [16].Bassiouni R, Nemec KN, A, Iketani, Flores O, Showalter A, et al. Chaperonin containing TCP-1 protein level in breast cancer cells predicts therapeutic application of a cytotoxic peptide. Clin Cancer Res. 2016;22(17):4366–79. [DOI] [PMC free article] [PubMed]; Bassiouni R, Nemec KN, Iketani A, Flores O, Showalter A. et al. Chaperonin containing TCP-1 protein level in breast cancer cells predicts therapeutic application of a cytotoxic peptide. Clin Cancer Res. 2016;22(17):4366–79. doi: 10.1158/1078-0432.CCR-15-2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Seiden-Long IM, Brown KR, Shih W, Wigle DA, Radulovich N, Jurisica I, et al. Transcriptional targets of hepatocyte growth factor signaling and Ki-ras oncogene activation in colorectal cancer. Oncogene. 2006;25(1):91–102. [DOI] [PubMed]; Seiden-Long IM, Brown KR, Shih W, Wigle DA, Radulovich N, Jurisica I. et al. Transcriptional targets of hepatocyte growth factor signaling and Ki-ras oncogene activation in colorectal cancer. Oncogene. 2006;25(1):91–102. doi: 10.1038/sj.onc.1209005. [DOI] [PubMed] [Google Scholar]
- [18].Coghlin C, Carpenter B, Dundas SR, Lawrie LC, Telfer C, Murray GI. Characterization and over-expression of chaperonin t-complex proteins in colorectal cancer. J Pathol. 2006;210:351–7. [DOI] [PubMed]; Coghlin C, Carpenter B, Dundas SR, Lawrie LC, Telfer C, Murray GI. Characterization and over-expression of chaperonin t-complex proteins in colorectal cancer. J Pathol. 2006;210:351–7. doi: 10.1002/path.2056. [DOI] [PubMed] [Google Scholar]
- [19].LLeonart ME, Vidal F, Gallardo D, Diaz-Fuertes M, Rojo F, Cuatrecasas M, et al. New p53 related genes in human tumors: significant downregulation in colon and lung carcinomas. Oncol Rep. 2006;16:603–8. [DOI] [PubMed]; LLeonart ME, Vidal F, Gallardo D, Diaz-Fuertes M, Rojo F, Cuatrecasas M. et al. New p53 related genes in human tumors: significant downregulation in colon and lung carcinomas. Oncol Rep. 2006;16:603–8. doi: 10.3892/or.16.3.603. [DOI] [PubMed] [Google Scholar]
- [20].Lin Y-F, Tsai W-P, Liu H-G, Liang P-H. Intracellular β-tubulin/chaperonin containing TCP1-β complex serves as a novel chemotherapeutic target against drug-resistant tumors. Cancer Res. 2009;69(17):6879–88. [DOI] [PubMed]; Lin Y-F, Tsai W-P, Liu H-G, Liang P-H. Intracellular β-tubulin/chaperonin containing TCP1-β complex serves as a novel chemotherapeutic target against drug-resistant tumors. Cancer Res. 2009;69(17):6879–88. doi: 10.1158/0008-5472.CAN-08-4700. [DOI] [PubMed] [Google Scholar]
- [21].Macleod K, Mullen P, Sewell J, Rabiasz G, Lawrie S, Miller E, et al. Altered ErbB receptor signaling and gene expression in cisplatin-resistant ovarian cancer. Cancer Res. 2005;(15):6789–800. [DOI] [PubMed]; Macleod K, Mullen P, Sewell J, Rabiasz G, Lawrie S, Miller E. et al. Altered ErbB receptor signaling and gene expression in cisplatin-resistant ovarian cancer. Cancer Res. 2005;(15):6789–800. doi: 10.1158/0008-5472.CAN-04-2684. [DOI] [PubMed] [Google Scholar]
- [22].Zhang Y, Wang Y, Wei Y, Wu J, Zhang P, Shen S, et al. Molecular chaperone CCT3 supports proper mitotic progression and cell proliferation in hepatocellular carcinoma cells. Cancer Lett. 2016;372(1):101–9. [DOI] [PubMed]; Zhang Y, Wang Y, Wei Y, Wu J, Zhang P, Shen S. et al. Molecular chaperone CCT3 supports proper mitotic progression and cell proliferation in hepatocellular carcinoma cells. Cancer Lett. 2016;372(1):101–9. doi: 10.1016/j.canlet.2015.12.029. [DOI] [PubMed] [Google Scholar]
- [23].Myung J-K, Afjehi-Sadat L, Felizardo-Cabatic M, Slavc I, Lubec G. Expressional patterns of chaperones in ten human tumor cell lines. Proteome Sci. 2004;2:8. [DOI] [PMC free article] [PubMed]; Myung J-K, Afjehi-Sadat L, Felizardo-Cabatic M, Slavc I, Lubec G. Expressional patterns of chaperones in ten human tumor cell lines. Proteome Sci. 2004;2:8. doi: 10.1186/1477-5956-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Boudiaf-Benmammar C, Cresteil T, Melki R. The cytosolic chaperonin CCT/TRiC and cancer cell proliferation. PLoS One. 2013;8(4):e60895. [DOI] [PMC free article] [PubMed]; Boudiaf-Benmammar C, Cresteil T, Melki R. The cytosolic chaperonin CCT/TRiC and cancer cell proliferation. PLoS One. 2013;8(4):e60895. doi: 10.1371/journal.pone.0060895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Gehrmann M, Specht HM, Bayer C. Hsp70 – a biomarker for tumor detection and monitoring of outcome of radiation therapy in patients with squamous cell carcinoma of the head and neck. Radiat Oncol. 2014;9:131. [DOI] [PMC free article] [PubMed]; Gehrmann M, Specht HM, Bayer C. Hsp70 – a biomarker for tumor detection and monitoring of outcome of radiation therapy in patients with squamous cell carcinoma of the head and neck. Radiat Oncol. 2014;9:131. doi: 10.1186/1748-717X-9-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Sealfon SC, Chu TT. RNA and DNA microarrays. Methods Mol Biol. 2011;671:3–34. [DOI] [PubMed]; Sealfon SC, Chu TT. RNA and DNA microarrays. Methods Mol Biol. 2011;671:3–34. doi: 10.1007/978-1-59745-551-0_1. [DOI] [PubMed] [Google Scholar]
- [27].Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9(2):166–80. [DOI] [PMC free article] [PubMed]; Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB. et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9(2):166–80. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. [DOI] [PMC free article] [PubMed]; Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Vasaikar S, Straub P, Wang J, Zhang B. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018;46(D1):D956–63. [DOI] [PMC free article] [PubMed]; Vasaikar S, Straub P, Wang J, Zhang B. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018;46(D1):D956–63. doi: 10.1093/nar/gkx1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Bouliotis G, Billingham L. Crossing survival curves: alternatives to the logrank test. Trials. 2011;12:A137.; Bouliotis G, Billingham L. Crossing survival curves: alternatives to the logrank test. Trials. 2011;12:A137. [Google Scholar]
- [31].Li H, Han D, Hou Y, Chen H, Chen Z. Statistical inference methods for two crossing survival curves: a comparison of methods. PLoS One. 2015;10(1):e0116774. [DOI] [PMC free article] [PubMed]; Li H, Han D, Hou Y, Chen H, Chen Z. Statistical inference methods for two crossing survival curves: a comparison of methods. PLoS One. 2015;10(1):e0116774. doi: 10.1371/journal.pone.0116774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ponten F, Jirstrom K, Uhlen M. The Human Protein Atlas – a tool for pathology. J Pathol. 2008;216(4):387–93. [DOI] [PubMed]; Ponten F, Jirstrom K, Uhlen M. The Human Protein Atlas – a tool for pathology. J Pathol. 2008;216(4):387–93. doi: 10.1002/path.2440. [DOI] [PubMed] [Google Scholar]
- [33].Gao J, Aksoy B, Dogrusoz U, Dresdner G, Gross B, Sumer S, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. [DOI] [PMC free article] [PubMed]; Gao J, Aksoy B, Dogrusoz U, Dresdner G, Gross B, Sumer S. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discovery. 2012;2(5):401–4. [DOI] [PMC free article] [PubMed]; Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discovery. 2012;2(5):401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Rodchenkov I, Babur O, Luna A, Aksoy BA, Wong JV, Fong D, et al. Pathway commons 2019 update: integration, analysis and exploration of pathway data. Nucleic Acids Res. 2019;48(D1):D489–97. [DOI] [PMC free article] [PubMed]; Rodchenkov I, Babur O, Luna A, Aksoy BA, Wong JV, Fong D. et al. Pathway commons 2019 update: integration, analysis and exploration of pathway data. Nucleic Acids Res. 2019;48(D1):D489–97. doi: 10.1093/nar/gkz946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Smoot ME, Ono K, Ruscheinski J, Wang P-L, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2010;27(3):431–2. [DOI] [PMC free article] [PubMed]; Smoot ME, Ono K, Ruscheinski J, Wang P-L, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2010;27(3):431–2. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. [DOI] [PubMed]; Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- [38].Gene Ontology Consortium. The gene ontology (GO) project in 2006. Nucleic Acids Res. 2006;34:D322–6. [DOI] [PMC free article] [PubMed]; Gene Ontology Consortium. The gene ontology (GO) project in 2006. Nucleic Acids Res. 2006;34:D322–6. doi: 10.1093/nar/gkj021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kanehisa M, Sato Y, Kawashima M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44(D1):D457–62. [DOI] [PMC free article] [PubMed]; Kanehisa M, Sato Y, Kawashima M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44(D1):D457–62. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Pyeon D, Newton MA, Lambert PF, den Boon JA, Sengupta S, Marsit CJ, et al. Fundamental differences in cell cycle deregulation in human papillomavirus-positive and human papillomavirus-negative head/neck and cervical cancers. Cancer Res. 2007;67(10):4605–19. [DOI] [PMC free article] [PubMed]; Pyeon D, Newton MA, Lambert PF, den Boon JA, Sengupta S, Marsit CJ. et al. Fundamental differences in cell cycle deregulation in human papillomavirus-positive and human papillomavirus-negative head/neck and cervical cancers. Cancer Res. 2007;67(10):4605–19. doi: 10.1158/0008-5472.CAN-06-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Estilo CL, Oc P, Talbot S, Socci ND, Carlson DL, Ghossein R, et al. Oral tongue cancer gene expression profiling: identification of novel potential prognosticators by oligonucleotide microarray analysis. BMC Cancer. 2009;9:11. [DOI] [PMC free article] [PubMed]; Estilo CL, Oc P, Talbot S, Socci ND, Carlson DL, Ghossein R. et al. Oral tongue cancer gene expression profiling: identification of novel potential prognosticators by oligonucleotide microarray analysis. BMC Cancer. 2009;9:11. doi: 10.1186/1471-2407-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Talbot SG, Estilo C, Maghami E, Sarkaria IS, Pham DK, Oc P, et al. Gene expression profiling allows distinction between primary and metastatic squamous cell carcinomas in the lung. Cancer Res. 2005;65(8):3063–71. [DOI] [PubMed]; Talbot SG, Estilo C, Maghami E, Sarkaria IS, Pham DK, Oc P. et al. Gene expression profiling allows distinction between primary and metastatic squamous cell carcinomas in the lung. Cancer Res. 2005;65(8):3063–71. doi: 10.1158/0008-5472.CAN-04-1985. [DOI] [PubMed] [Google Scholar]
- [43].Roh S-H, Kasembeli M, Bakthavatsalam D, Chiu W, Tweardy DJ. Contribution of the type II chaperonin, TRiC/CCT, to oncogenesis. Int J Mol Sci. 2015;(11):26706–20. [DOI] [PMC free article] [PubMed]; Roh S-H, Kasembeli M, Bakthavatsalam D, Chiu W, Tweardy DJ. Contribution of the type II chaperonin, TRiC/CCT, to oncogenesis. Int J Mol Sci. 2015;(11):26706–20. doi: 10.3390/ijms161125975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. [DOI] [PMC free article] [PubMed]; Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D. et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Shi X, Cheng S, Wang W. Suppression of CCT3 inhibits malignant proliferation of human papillary thyroid carcinoma cell. Oncol Lett. 2018;15(6):9202–8. [DOI] [PMC free article] [PubMed]; Shi X, Cheng S, Wang W. Suppression of CCT3 inhibits malignant proliferation of human papillary thyroid carcinoma cell. Oncol Lett. 2018;15(6):9202–8. doi: 10.3892/ol.2018.8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Frye M, Harada BT, Behm M, He C. RNA modifications modulate gene expression during development. Science. 2018;361(6409):1346–9. [DOI] [PMC free article] [PubMed]; Frye M, Harada BT, Behm M, He C. RNA modifications modulate gene expression during development. Science. 2018;361(6409):1346–9. doi: 10.1126/science.aau1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Klooster R, Straasheijm K, Shah B, Sowden J, Frants R, Thornton C, et al. Comprehensive expression analysis of FSHD candidate genes at the mRNA and protein level. Eur J Hum Genet EJHG. 2009;17(12):1615–24. [DOI] [PMC free article] [PubMed]; Klooster R, Straasheijm K, Shah B, Sowden J, Frants R, Thornton C. et al. Comprehensive expression analysis of FSHD candidate genes at the mRNA and protein level. Eur J Hum Genet EJHG. 2009;17(12):1615–24. doi: 10.1038/ejhg.2009.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Hayes DN, Grandis J, El-Naggar AK. Comprehensive genomic characterization of squamous cell carcinoma of the head and neck in the Cancer Genome Atlas. Cancer Res. 2013;73(Suppl 8):1117.; Hayes DN, Grandis J, El-Naggar AK. Comprehensive genomic characterization of squamous cell carcinoma of the head and neck in the Cancer Genome Atlas. Cancer Res. 2013;73(Suppl 8):1117. [Google Scholar]
- [49].Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517(7536):576–82. [DOI] [PMC free article] [PubMed]; Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;82;517(7536):576. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–9. [DOI] [PubMed]; Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–9. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- [51].Morimoto RI. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev. 2008;22(11):1427–38. [DOI] [PMC free article] [PubMed]; Morimoto RI. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev. 2008;22(11):1427–38. doi: 10.1101/gad.1657108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Siegers K, Bölter B, Schwarz JP, Böttcher UM, Guha S, Hartl FU. TRiC/CCT cooperates with different upstream chaperones in the folding of distinct protein classes. EMBO J. 2003;22(19):5230–40. [DOI] [PMC free article] [PubMed] [Retracted]; Siegers K, Bölter B, Schwarz JP, Böttcher UM, Guha S, Hartl FU. TRiC/CCT cooperates with different upstream chaperones in the folding of distinct protein classes. EMBO J. 2003;22(19):5230–40. doi: 10.1093/emboj/cdg483. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [53].Guenther MG, Yu J, Kao GD, Yen TJ, Lazar MA. Assembly of the SMRT-histone deacetylase 3 repression complex requires the TCP-1 ring complex. Genes Dev. 2002;16(24):3130–5. [DOI] [PMC free article] [PubMed]; Guenther MG, Yu J, Kao GD, Yen TJ, Lazar MA. Assembly of the SMRT-histone deacetylase 3 repression complex requires the TCP-1 ring complex. Genes Dev. 2002;16(24):3130–5. doi: 10.1101/gad.1037502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Won KA, Schumacher RJ, Farr GW, Horwich AL, Reed SI. Maturation of human cyclin E requires the function of eukaryotic chaperonin CCT. Mol Cell Biol. 1998;18(12):7584–9. [DOI] [PMC free article] [PubMed]; Won KA, Schumacher RJ, Farr GW, Horwich AL, Reed SI. Maturation of human cyclin E requires the function of eukaryotic chaperonin CCT. Mol Cell Biol. 1998;18(12):7584–9. doi: 10.1128/mcb.18.12.7584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Thulasiraman V, Yang CF, Frydman J. In vivo newly translated polypeptides are sequestered in a protected folding environment. EMBO J. 1999;18(1):85–95. [DOI] [PMC free article] [PubMed]; Thulasiraman V, Yang CF, Frydman J. In vivo newly translated polypeptides are sequestered in a protected folding environment. EMBO J. 1999;18(1):85–95. doi: 10.1093/emboj/18.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Yokota S, Yamamoto Y, Shimizu K, Momoi H, Kamikawa T, Yamaoka Y, et al. Increased expression of cytosolic chaperonin CCT in human hepatocellular and colonic carcinoma. Cell Stress Chaperones. 2001;6(4):345–50. [DOI] [PMC free article] [PubMed]; Yokota S, Yamamoto Y, Shimizu K, Momoi H, Kamikawa T, Yamaoka Y. et al. Increased expression of cytosolic chaperonin CCT in human hepatocellular and colonic carcinoma. Cell Stress Chaperones. 2001;6(4):345–50. doi: 10.1379/1466-1268(2001)006<0345:ieoccc>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Grantham J, Brackley KI, Willison KR. Substantial CCT activity is required for cell cycle progression and cytoskeletal organization in mammalian cells. Exp Cell Res. 2006;12:2309–24. [DOI] [PubMed]; Grantham J, Brackley KI, Willison KR. Substantial CCT activity is required for cell cycle progression and cytoskeletal organization in mammalian cells. Exp Cell Res. 2006;12:2309–24. doi: 10.1016/j.yexcr.2006.03.028. [DOI] [PubMed] [Google Scholar]
- [58].Poon TCW, Wong N, Lai PBS, Rattray M, Johnson PJ, Sung JJY. A tumor progression model for hepatocellular carcinoma: bioinformatic analysis of genomic data. Gastroenterology. 2007;131(4):1262–70. [DOI] [PubMed]; Poon TCW, Wong N, Lai PBS, Rattray M, Johnson PJ, Sung JJY. A tumor progression model for hepatocellular carcinoma: bioinformatic analysis of genomic data. Gastroenterology. 2007;131(4):1262–70. doi: 10.1053/j.gastro.2006.08.014. [DOI] [PubMed] [Google Scholar]
- [59].Gu J, Xuan Z. Inferring the perturbed microRNA regulatory networks in cancer using hierarchical gene co-expression signatures. PLoS One. 2013;8(11):e81032. [DOI] [PMC free article] [PubMed]; Gu J, Xuan Z. Inferring the perturbed microRNA regulatory networks in cancer using hierarchical gene co-expression signatures. PLoS One. 2013;8(11):e81032. doi: 10.1371/journal.pone.0081032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Calderwood SK, Khaleque MA, Sawyer DB, Ciocca DR. Heat shock proteins in cancer: chaperones of tumorigenesis. Trends Biochemical Sci. 2006;31:164–72. [DOI] [PubMed]; Calderwood SK, Khaleque MA, Sawyer DB, Ciocca DR. Heat shock proteins in cancer: chaperones of tumorigenesis. Trends Biochemical Sci. 2006;31:164–72. doi: 10.1016/j.tibs.2006.01.006. [DOI] [PubMed] [Google Scholar]
- [61].Santagata S, Hu R, Lin NU, Mendillo ML, Collins LC, Hankinson SE, et al. High levels of nuclear heat-shock factor 1 (HSF1) are associated with poor prognosis in breast cancer. Proc Natl Acad Sci USA. 2011;108(45):18378. [DOI] [PMC free article] [PubMed]; Santagata S, Hu R, Lin NU, Mendillo ML, Collins LC, Hankinson SE. et al. High levels of nuclear heat-shock factor 1 (HSF1) are associated with poor prognosis in breast cancer. Proc Natl Acad Sci USA. 2011;108(45):18378. doi: 10.1073/pnas.1115031108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nat Rev Cancer. 2005;5(10):761–72. [DOI] [PubMed]; Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nat Rev Cancer. 2005;5(10):761–72. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- [63].Kubota H, Hynes G, Carne A, Ashworth A, Willison K. Identification of six Tcp-1-related genes encoding divergent subunits of the TCP-1-containing chaperonin. Curr Biol. 1994;4(2):89–99. [DOI] [PubMed]; Kubota H, Hynes G, Carne A, Ashworth A, Willison K. Identification of six Tcp-1-related genes encoding divergent subunits of the TCP-1-containing chaperonin. Curr Biol. 1994;4(2):89–99. doi: 10.1016/s0960-9822(94)00024-2. [DOI] [PubMed] [Google Scholar]
- [64].Sergeeva OA, Chen B, Haase-Pettingell C, Ludtke SJ, Chiu W, King JA. Human CCT4 and CCT5 chaperonin subunits expressed in Escherichia coli form biologically active homo-oligomers. J Biol Chem. 2013;288(24):17734–44. [DOI] [PMC free article] [PubMed]; Sergeeva OA, Chen B, Haase-Pettingell C, Ludtke SJ, Chiu W, King JA. Human CCT4 and CCT5 chaperonin subunits expressed in Escherichia coli form biologically active homo-oligomers. J Biol Chem. 2013;288(24):17734–44. doi: 10.1074/jbc.M112.443929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Spiess M, Echbarthi M, Svanström A, Karlsson R, Grantham J. Over-expression analysis of all eight subunits of the molecular chaperone CCT in mammalian cells reveals a novel function for CCT delta. J Mol Biol. 2015;427(17):2757–64. [DOI] [PubMed]; Spiess M, Echbarthi M, Svanström A, Karlsson R, Grantham J. Over-expression analysis of all eight subunits of the molecular chaperone CCT in mammalian cells reveals a novel function for CCT delta. J Mol Biol. 2015;427(17):2757–64. doi: 10.1016/j.jmb.2015.06.007. [DOI] [PubMed] [Google Scholar]
- [66].Elliott KL, Svanstrom A, Spiess M, Karlsson R, Grantham J. A novel function of the monomeric CCT epsilon subunit connects the serum response factor pathway to chaperone-mediated actin folding. Mol Biol Cell. 2015;26(15):2801–9. [DOI] [PMC free article] [PubMed]; Elliott KL, Svanstrom A, Spiess M, Karlsson R, Grantham J. A novel function of the monomeric CCT epsilon subunit connects the serum response factor pathway to chaperone-mediated actin folding. Mol Biol Cell. 2015;26(15):2801–9. doi: 10.1091/mbc.E15-01-0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Lawrie LC, Fothergill JE, Murray GI. Spot the differences: proteomics in cancer research. Lancet Oncol. 2001;2(5):270–7. [DOI] [PubMed]; Lawrie LC, Fothergill JE, Murray GI. Spot the differences: proteomics in cancer research. Lancet Oncol. 2001;2(5):270–7. doi: 10.1016/S1470-2045(00)00321-1. [DOI] [PubMed] [Google Scholar]
- [68].Li L-J, Zhang L-S, Han Z-J, He Z-Y, Chen H, Li Y-M. Chaperonin containing TCP-1 subunit 3 is critical for gastric cancer growth. Oncotarget. 2017;(67):111470–81. [DOI] [PMC free article] [PubMed]; Li L-J, Zhang L-S, Han Z-J, He Z-Y, Chen H, Li Y-M. Chaperonin containing TCP-1 subunit 3 is critical for gastric cancer growth. Oncotarget. 2017;(67):111470–81. doi: 10.18632/oncotarget.22838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Liu Y, Yu L, Wu X, Chen Y, Ge J, Li Q, et al. Silencing CCT6A suppresses cell migration and invasion in glioblastoma in vitro. Int J Clin Exp Med. 2017;10(9):13263–71.; Liu Y, Yu L, Wu X, Chen Y, Ge J, Li Q. et al. Silencing CCT6A suppresses cell migration and invasion in glioblastoma in vitro . Int J Clin Exp Med. 2017;10(9):13263–71. [Google Scholar]
- [70].Ding Z, German P, Bai S, Sun M, Liu X, Jonasch E. Modulation of TRiC/CCT and proteasome regulates VHL proteostasis and functionality. BJU Int. 2012;17–8.; Ding Z, German P, Bai S, Sun M, Liu X, Jonasch E. Modulation of TRiC/CCT and proteasome regulates VHL proteostasis and functionality. BJU Int. 2012:17–8. [Google Scholar]
- [71].Yang X, Ren H, Shao Y. Chaperonin-containing T-complex protein 1 subunit 8 promotes cell migration and invasion in human esophageal squamous cell carcinoma by regulating alpha-actin and beta-tubulin expression. Int J Oncol. 2018;52(6):2021–30. [DOI] [PubMed]; Yang X, Ren H, Shao Y. Chaperonin-containing T-complex protein 1 subunit 8 promotes cell migration and invasion in human esophageal squamous cell carcinoma by regulating alpha-actin and beta-tubulin expression. Int J Oncol. 2018;52(6):2021–30. doi: 10.3892/ijo.2018.4335. [DOI] [PubMed] [Google Scholar]
- [72].Huang X, Wang X, Cheng C, Cai J, He S, Wang H, et al. Chaperonin containing TCP1, subunit 8 (CCT8) is upregulated in hepatocellular carcinoma and promotes HCC proliferation. APMIS Acta Pathol Microbiol Immunol Scand; 2014. [DOI] [PubMed]; Huang X, Wang X, Cheng C, Cai J, He S, Wang H. et al. Chaperonin containing TCP1, subunit 8 (CCT8) is upregulated in hepatocellular carcinoma and promotes HCC proliferation. APMIS Acta Pathol Microbiol Immunol Scand. 2014 doi: 10.1111/apm.12258. [DOI] [PubMed] [Google Scholar]
- [73].Sengupta S, den Boon JA, Chen IH, Newton MA, Dahl DB, Chen M, et al. Genome-wide expression profiling reveals EBV-associated inhibition of MHC class I expression in nasopharyngeal carcinoma. Cancer Res. 2006;66(16):7999–6. [DOI] [PubMed]; Sengupta S, den Boon JA, Chen IH, Newton MA, Dahl DB, Chen M. et al. Genome-wide expression profiling reveals EBV-associated inhibition of MHC class I expression in nasopharyngeal carcinoma. Cancer Res. 2006;66(16):7999–6. doi: 10.1158/0008-5472.CAN-05-4399. [DOI] [PubMed] [Google Scholar]