Abstract
Rationale: Declining lung function in patients with interstitial lung disease is accompanied by epithelial remodeling and progressive scarring of the gas-exchange region. There is a need to better understand the contribution of basal cell hyperplasia and associated mucosecretory dysfunction to the development of idiopathic pulmonary fibrosis (IPF).
Objectives: We sought to decipher the transcriptome of freshly isolated epithelial cells from normal and IPF lungs to discern disease-dependent changes within basal stem cells.
Methods: Single-cell RNA sequencing was used to map epithelial cell types of the normal and IPF human airways. Organoid and air–liquid interface cultures were used to investigate functional properties of basal cell subtypes.
Measurements and Main Results: We found that basal cells included multipotent and secretory primed subsets in control adult lung tissue. Secretory primed basal cells include an overlapping molecular signature with basal cells obtained from the distal lung tissue of IPF lungs. We confirmed that NOTCH2 maintains undifferentiated basal cells and restricts basal-to-ciliated differentiation, and we present evidence that NOTCH3 functions to restrain secretory differentiation.
Conclusions: Basal cells are dynamically regulated in disease and are specifically biased toward the expansion of the secretory primed basal cell subset in IPF. Modulation of basal cell plasticity may represent a relevant target for therapeutic intervention in IPF.
Keywords: prematurity, pulmonary microvascular function, dynamic contrast enhanced magnetic resonance imaging, vascular simplification
At a Glance Commentary
Scientific Knowledge on the Subject
Basal cell expansion is a common feature of epithelial remodeling seen in idiopathic pulmonary fibrosis (IPF) and other chronic lung diseases. Basal cells serve as stem/progenitor cells that maintain the pseudostratified epithelium of the mammalian lung. However, little is known of their molecular and functional heterogeneity in the normal and diseased human lung and the roles they play in tissue remodeling.
What This Study Adds to the Field
We determined that basal cells of the normal human lung represent a heterogeneous population that include multipotent and secretory primed subsets. Secretory primed basal (SPB) cells share a molecular signature with basal cells recovered from fibrotic lung tissue obtained from patients undergoing transplant for end-stage IPF and are enriched in honeycomb regions of IPF lungs. Our data suggest that dynamic changes in the regional abundance and molecular phenotype of basal cells contribute to lung-tissue remodeling seen in end-stage IPF. Specifically, the identification of SPB cells at sites where MUC5B is expressed in honeycomb regions link our results to MUC5B allele–specific susceptibility to IPF and warrant for future investigations of the role of SPB cells in patient outcomes in IPF. Overall, these findings have the potential to help the identification of specific therapeutic targets for IPF.
Human airways are lined by a mucociliary epithelium that functions in the clearance of inhaled pathogens, particulates, and toxic gases. Major portions of this epithelium are pseudostratified, with basal cells located adjacent to the basement membrane and subtending specialized luminal cell types, including ciliated cells and various secretory cell types. Both basal and secretory cell types have proliferative capacity, but only basal cells have the capacity for the long-term self-renewal that is commonly associated with tissue stem cells (1, 2). Pathological remodeling of the airway epithelium is common in chronic lung disease (3, 4). Disease-related changes include hyperplasia of basal cells and altered differentiation of secretory cells, leading to overproduction of mucus (5, 6).
Idiopathic pulmonary fibrosis (IPF) is a progressive lung disease with a mean survival after initial diagnosis of approximately 3 years (7). Declining lung function results from scarring and remodeling of the interstitium surrounding the alveoli. Other pathological sequelae of IPF include the appearance of honeycomb cysts at sites of extensive parenchymal remodeling—structures that are composed of basal cells and mucin-producing airway secretory cells (5). These structural defects are considered to be secondary to epithelial progenitor cell dysfunction and defective epithelial–mesenchymal signaling (8, 9); furthermore, the detection of basal cells within BAL fluid of patients with IPF is associated with poor prognosis (10).
Herein, we comprehensively assess the transcriptome of single cells from normal human lungs and lung tissue of patients undergoing transplantation for end-stage IPF. We found that basal cells include multiple molecularly distinct states in the normal lung, including multipotent and secretory cell–primed, and present evidence suggesting that basal cell subsets form a hierarchy that is regulated by Notch signaling.
Methods
See also online supplement.
Study Population
Explant tissue was obtained from patients undergoing transplantation for end-stage IPF at either Duke University or Cedars-Sinai Medical Center in compliance with consent procedures accepted by the institutional review board of Cedars-Sinai Medical Center. Human lung specimens obtained through the International Institute for the Advancement of Medicine were obtained in compliance with consent procedures developed by International Institute for the Advancement of Medicine and approved by the Cedars-Sinai Medical Center internal review board.
Data Availability
All transcriptome data were deposited in Gene Expression Omnibus (GSE143706 and GSE143705). A GitHub repository with a markdown to show our quality control and data analysis is available at https://github.com/gc-github-bio/Normal-and-IPF-lung.
Histology
Lung tissue was formalin-fixed, paraffin-embedded, and sectioned at 4 μm. Cultures were fixed with 4% paraformaldehyde and washed with phosphate-buffered saline before whole-mount staining. Paraffin sections were deparaffinized in xylene and rehydrated through a gradient of ethanol. Before antibody staining, antigen retrieval was performed in 10-mM citrate buffer (pH 6.0) using a 2100-Retreiver (Aptum Biologics). Slides were washed with TRIS-buffered saline (TBS) and blocked for 1 hour at room temperature using blocking buffer containing 3% bovine serum albumin, 0.1% Triton X-100, and 10% serum (from secondary antibody species) in TBS. Primary antibodies MUC5B (HPA008246), MUC5AC (45M1; Thermofisher), FOXJ1 (14–9965–82; Biolegend), KRT5 (905904; Biolegend), and KRT8 (TROMA-1; Hybrodoma Bank) were diluted in 3% bovine serum albumin, 0.1% Triton X-100, and 1% serum in TBS, applied to tissue sections, and incubated overnight at 4°C. Nuclei were counterstained with 2-(4-Amidinophenyl)-1H-indole-6-carboxamidine (DAPI), and appropriate secondary antibodies conjugated to fluorochromes (Thermo Fisher Scientific) were applied for 1 hour at room temperature. Sections were mounted using Fluoromount G mounting medium (Thermo Fisher Scientific). Stained samples were imaged on a Zeiss LSM 780 Confocal Microscope and image-processed using Zen Blue software (Zeiss).
Cell Isolation
Human IPF explants were processed to obtain epithelial cells from fibrotic regions. Donor explants were processed to obtain airway epithelial cells. The tissue was processed as described previously (1), with the following modifications. Proximal tissue was enzymatically digested with Liberase followed by gentle scraping of epithelial cells off the basement membrane. IPF tissue was finely minced and washed in Ham’s F12 (Corning) at 4°C for 5 minutes with rocking, followed by centrifugation for 5 minutes at 600 g and 4°C. The minced cleaned tissue was then incubated in Dulbecco’s modified Eagle medium/F12 (Thermo Fisher Scientific) containing 1X Liberase (Sigma-Aldrich) incubated at 37°C with rocking for 45 minutes. Dissociated single-cell preparations were enriched for epithelial cells and depleted of erythrocytes, leukocytes, and endothelial cells using antibodies against the following molecules: EPCAM (CO17–1A, 369820), CD235a (HI264, 349106), CD45 (2D1, 368522), and CD31 (WM59, 303124) (Biolegend). For the basal cell enrichment experiment and subpopulation isolation, CD271 (ME20.4, 345106) and CD66 (ASL-32, 342306) (Biolegend) were also included. Labeled cells were washed in Hanks’ balanced salt solution with 2% fetal bovine serum and resuspended and placed on ice for fluorescence-activated cell sorting using a BD Influx cell sorter (Becton Dickinson). Viability was determined by staining cell preparations with either 7AAD (Biolegend), propidium iodide (Biolegend), or DAPI (Thermo Fisher Scientific) 15 minutes before cell sorting.
Results
Mapping the Molecular Phenotype of Normal Human Lung Airway Epithelial Cells
Single-cell RNA sequencing (scRNAseq) was performed on fractionated epithelial cells from six donors with no preexisting chronic lung disease and no or mild smoking history (Figure E1A in the online supplement). A single-cell transcriptome of the aggregated datasets containing 9,731 cells was generated after the initial filtering. We defined 37 clusters (Figure E2A) encompassing 12 cellular phenotypes, including four basal, five secretory, two cilated, and ionocyte cell types, that were visualized by Uniform Manifold Approximation and Projection (UMAP) and by their top differentially expressed genes (Figures 1A and 1B and Table E1). Clusters were first assigned to major cell populations using previously published gene lists (11) (Table E2) to create a signature score (12). Clustering and subclustering were than refined by multiple iterations of scoring based on differential gene expression (Figure E2C–E2L). Clusters 18, 28, and 31, initially uncharacterized, were recognized as secretory cell types after the first round of score refinement (Figure E2D) and were than manually annotated as mucous and serous cells on the basis of published markers for the submucosal gland (13) (Figures 1A, 1J, and 1K). Using the same approach, the following two separate clusters of ciliated cells were identified: a relatively immature (ciliated2) cluster that contains genes of early ciliogenesysis, such as LRRC6, DNAAF1, and DNAAF5 (Figures 1A, 1L, and 1M and E1J), and a mature ciliated1 cluster (Figures 1A, 1L, and 1M). Each cell type classification was represented by single cells from each donor (Figures E1B).
Figure 1.
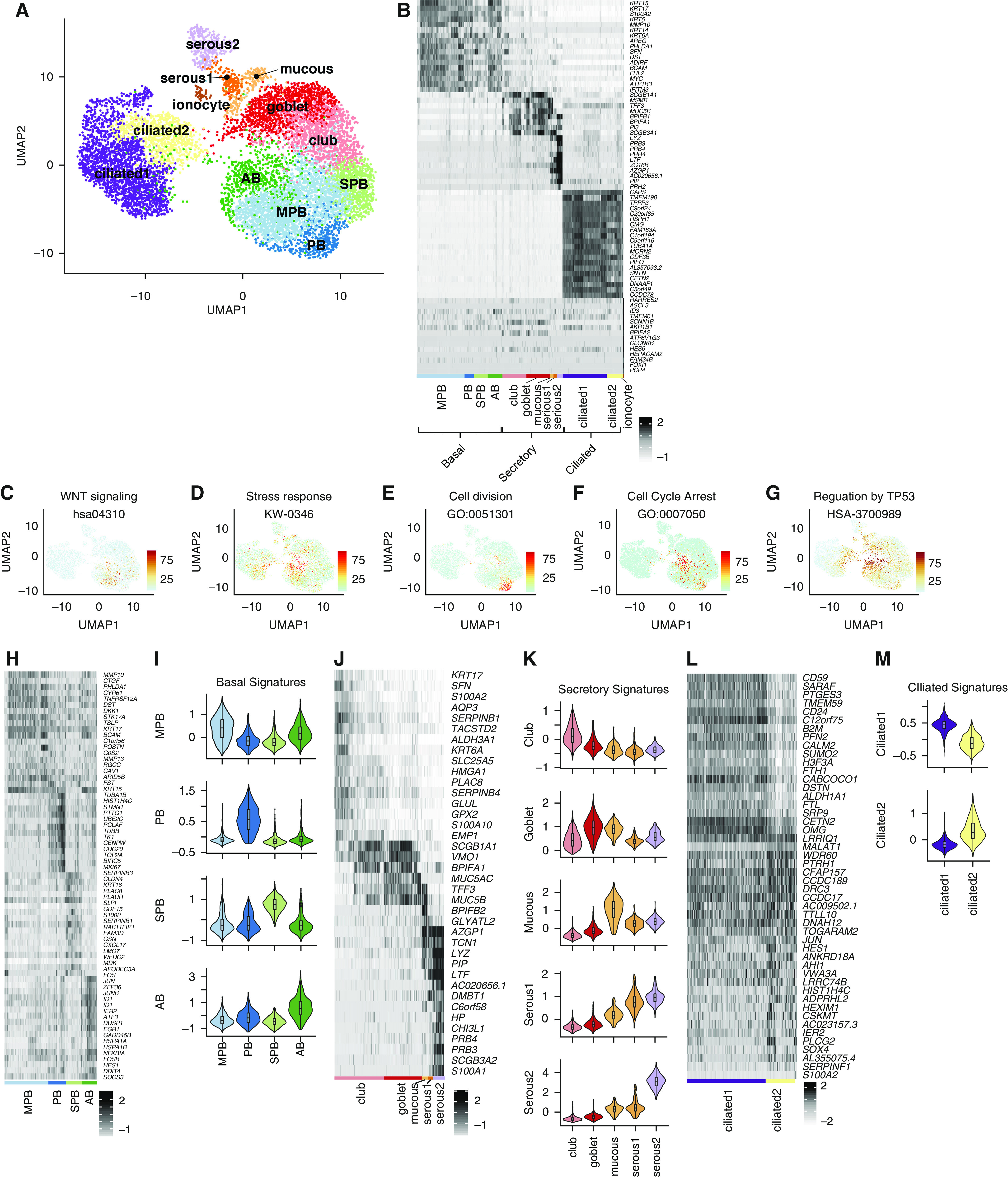
Classification of the human airway epithelium. (A) Uniform Manifold Approximation and Projection clustering of single-cell RNA-sequencing data generated for normal human lung airway epithelium. (B) Expression of differentially expressed genes that distinguish major cell types shown by heatmap with z-score values. Cell subtypes are shown within each major cell type. (C–G) Scoring derived respectively from Wnt signaling, stress response, cell division, cell-cycle arrest, and TP53-regulation gene signatures visualized by Uniform Manifold Approximation and Projection. (H) Expression of differentially expressed genes within basal cell subclusters shown by heatmap with z-score values. (I) Signatures of basal cell subclusters derived by scoring differentially expressed genes shown by violin plots. (J) Expression of differentially expressed genes within secretory cell subclusters shown by heatmap with z-score values. (K) Signatures of secretory cell subclusters derived by scoring differentially expressed genes shown by violin plots. (L) Expression of differentially expressed genes within ciliated cell subclusters shown by heatmap with z-score values. (M) Signatures of ciliated cell subclusters derived by scoring differentially expressed genes, shown by violin plots. AB = activated basal; MPB = multipotent basal; PB = proliferating basal; SPB = secretory primed basal; UMAP = Uniform Manifold Approximation and Projection.
Normal Human Airway Basal Cells Comprise Specialized Subpopulations
Basal cells were segregated into four clusters on the basis of their gene signatures (Figures 1A, 1B, 1H, and 1I and Table E3). The multipotent basal (MPB) cluster represents a “classical” MPB cell that was defined by a strong WNT-signaling signature (Figures 1A–1C). The proliferating basal (PB) cluster represents PB cells (Figures 1A, 1B, and E). Cells within the secretory primed basal (SPB) cluster express various members of the serpin family (Figures 1H and E1I) that regulate protein folding in the early secretory pathway (14). SPB cells also show enrichment of transcripts involved in O-glycan biosynthesis (Figure 2A), a pathway involved in the initial steps of mucin production by secretory cells (15). The activated basal (AB) cluster is characterized by a stress-response signature (Figures 1A, 1B, and 1D), including p38-SAPK MAP kinase cellular stress (Figure E1K). SAPK has been involved in centriolar satellite remodeling and ciliogenesis (16). Furthermore, the AB cluster is also characterized by the activation of the TP53 pathway (Figure 1G) and contains a cell-cycle arrest signature (Figure 1F).
Figure 2.
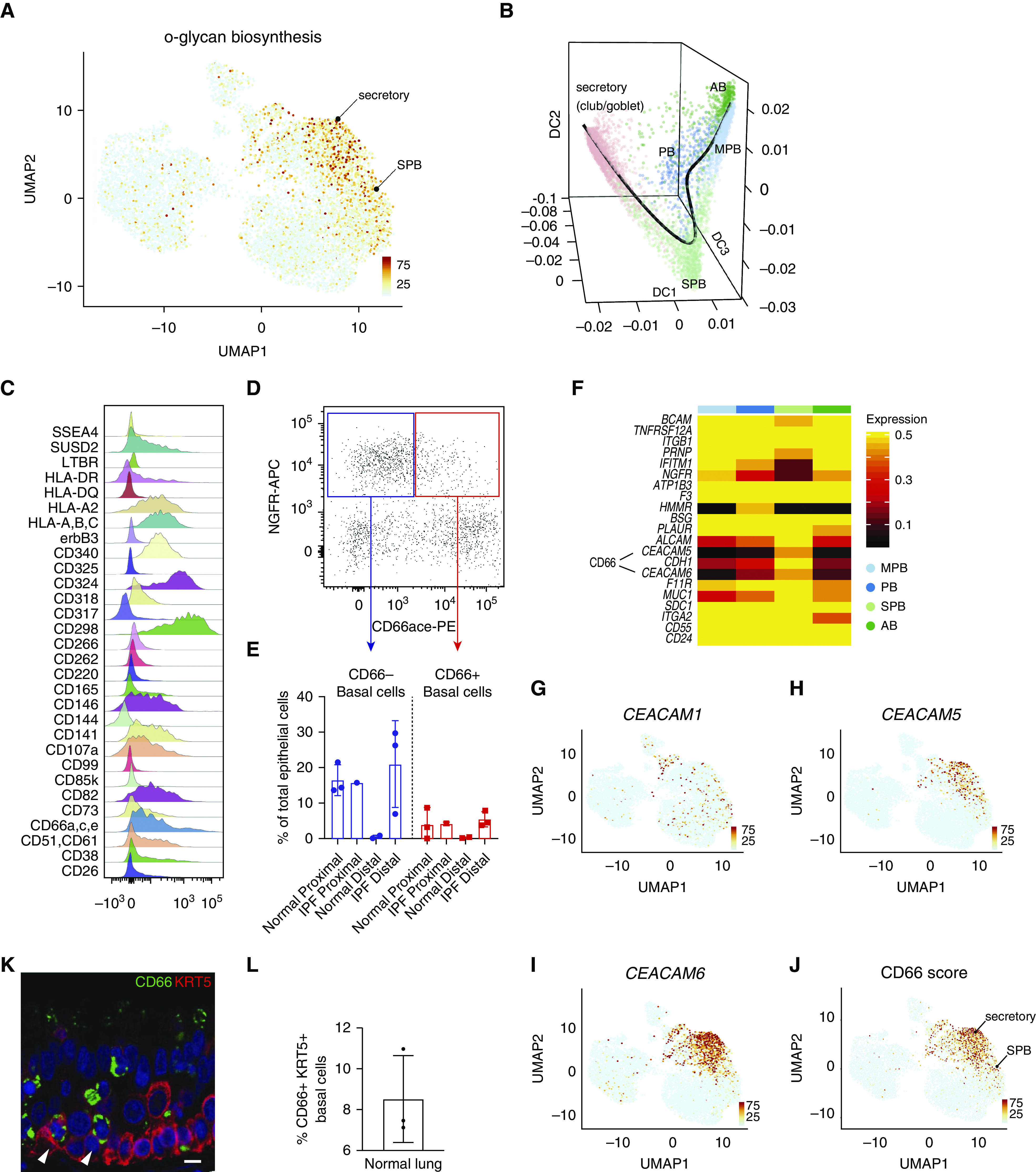
Characterization of secretory primed basal cells (secretory primed basal cluster). (A) Scoring of O-glycan biosynthesis pathway in the normal human airways visualized by Uniform Manifold Approximation and Projection. (B) Lineage reconstruction from basal to secretory cells (black line) was produced with “Slingshot” on the top of a diffusion map generated with “Destiny.” (C) Distribution analysis of CD markers discriminating basal cell subclusters identified by high-content flow cytometry. (D) Discrimination of basal cell subsets by NGFR and CD66 surface reactivity. (E) Percentage of basal subsets over total epithelial cells in proximal and distal compartments of normal and idiopathic pulmonary fibrosis lungs. (F) Differential expression of basal cell–specific CD markers from the single-cell RNA-sequencing data of the four basal cell subclusters. The entire repertoire of human CD molecules available from Human Genome Organization Gene Nomenclature Committee was screened. (G–J) Expression of CEACAM1, CEACAM5, and CEACAM6, and summarized signature score of CD66 in human normal airway epithelium visualized by Uniform Manifold Approximation and Projection. (K) Normal lung cross-section shows immunoreactivity of basal cell subpopulations for CD66 (arrowheads). Scale bar, 15 μm. (L) Quantification of CD66+ basal cells in normal lung, n = 3. AB = activated basal; DC = diffusion component; IPF = idiopathic pulmonary fibrosis; MPB = multipotent basal; NGFR-APC = NGFR allophycocyanin; PB = proliferating basal; PE = phycoerythrin; SPB = secretory primed basal; UMAP = Uniform Manifold Approximation and Projection.
In addition to scRNAseq, we used a high-content flow cytometry screening to detect basal cell heterogeneity based on surface-marker expression. This discovery screen was performed on pooled samples of epithelial cells isolated from the bronchi and the bronchiolar and distal lung regions from a single patient with IPF. The requirement for very high cell numbers for this type of high-throughput screen restricted our capacity to perform this screen on separate spatially localized samples. A total of 332 individual surface markers (plus isotype controls) were screened in combination with a viability dye and antibodies against CD31 (endothelial cells), CD45 (hematopoietic cells), EPCAM (epithelial cells), and NGFR (basal cells). The analysis for the distribution of cells in the CD31− CD45− EpCAM+ NGFR+ basal cell fraction revealed 30 surface markers that could be used for basal cell subclustering (Figures 2C and E3). Of these, CD66 recognition of CEACAM1,5,6 isoforms was one of the markers that was identified to correlate with the selective expression of CEACAM 5 and 6 by the SPB cluster in the scRNAseq data (Figure 2F and Table E4) and was thus chosen for further validation in normal (n = 3) and IPF (n = 3) lungs (Figures 2D and 2E). This confirmed differential surface reactivity of CD66 among basal cells in the normal proximal lung and showed a trend for an elevated proportion of CD66+ basal cells in IPF. Immunofluorescent staining of normal lung airways showed that CD66 immunoreactivity was observed at the plasma membrane in a rare population of basal cells in addition to previously described subpopulations of luminal transitional and secretory cells (Figures 2K and 2L and E4C) (17). The expression of CEACAM 5 and 6 isoforms that contribute to the CD66 immunoreactivity of SPB and secretory cells is also observable at the transcriptional level by UMAP (Figures 2G–2J). Interestingly, dimensional reduction by diffusion map with “Destiny” and lineage visualization with “Slingshot” show that SPB cells have a molecular phenotype that is transitional between basal and secretory cells (Figure 2B). However, our analysis does not allow determination of whether this population represents a transitory or phenotypically stable subset of basal cells.
SPB Cells Have Limited Capacity for Self-Renewal and Are Primed to Assume Secretory Cell Fates
We next sought to test the functional potential of SPB cells and validate the computational predictions from the scRNAseq data. Total basal cells were sorted for either EPCAM+, NGFR +, and CD66+ (CD66+ cells), or EPCAM+, NGFR+, and CD66− (referred to as CD66− cells). Mesenchyme-free three-dimensional in vitro cultures were used to assess the capacity of basal cell subsets to form clonal organoids and to evaluate differentiation potential. Both CD66+ and CD66− populations generated clonal organoids in the absence of mesenchymal support. However, CD66+ organoids displayed a reduced capacity for self-renewal starting at passage 2 compared with their CD66− counterparts (Figure 3A). We performed bulk RNA sequencing on either freshly isolated or cultured basal cell fractions. Top differentially expressed genes (log2-fold change; adjusted P value < 0.05) for CD66+ and CD66− are shown (Figures 3C and 3D and Table E5). Top differentially expressed genes (log2-fold change; adjusted P value < 0.05) that were also differentially expressed in SPB and MPB cells (scRNAseq data) are shown (Figures E4A and E4B and Table E5). Principal component analysis showed that CD66+ and CD66− cells diverged significantly 7 days after organoid culture, reflecting their disparate fates (Figure 3B). Immunofluorescence showed marked upregulation of secretory cell markers MUC5AC and MUC5B (Figures 3E and 3G and E5). When seeded at high density on Transwell cultures and grown at the air–liquid interface (ALI), CD66+ basal cells also demonstrated a more pronounced propensity for secretory cell differentiation compared with their CD66− counterparts (Figures 3F and 3H). These data are consistent with the notion that SPB CD66+ basal cells are primed to assume secretory cell fates.
Figure 3.
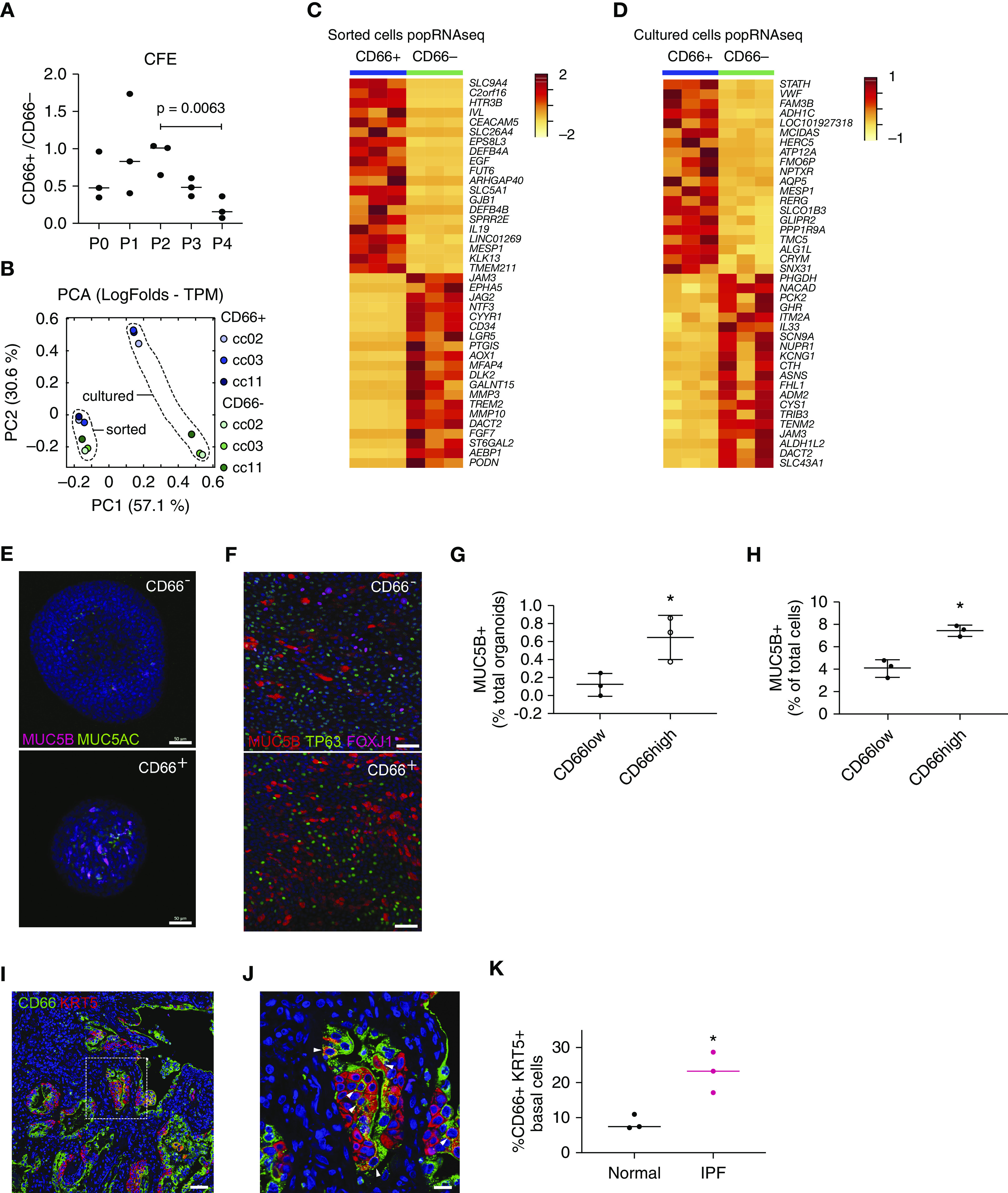
Functional properties of secretory primed basal cells. (A) Colony-forming efficiency of CD66+ and CD66− basal cells grown in mesenchyme-free three-dimensional cultures (n = 3; P = 0.0063; ANOVA). (B) Principal component analysis of popRNAseq data from freshly isolated (fluorescence-activated cell-sorting) or cultured CD66+ and CD66− basal cells. (C and D) Transcriptomic differences between freshly isolated and cultured CD66+ and CD66− basal cells visualized by heatmap. Top 20 differentially expressed genes (log2-fold change) with adjusted P value < 0.05 for CD66+ and CD66− basal cells are shown. (E and G) Immunofluorescence detection and quantification of mucins MUC5B and MUC5AC in organoid cultures of CD66+ and CD66− basal cells. Scale bars, 50 μm. (F and H) Immunofluorescence detection of basal (TP63), secretory (MUC5B), and ciliated (FOXJ1) markers for CD66+ and CD66− basal cells grown in air–liquid interface. Scale bars, 50 μm. Quantification of the percentage of MUC5B+ cells is shown. (I and J) Honeycomb regions of idiopathic pulmonary fibrosis lung showing enrichment of CD66+ basal cells. Scale bars, 50 μm for I and 20 μm for J. Arrowheads identify cells that are double positive for KRT5 and CD66. (K) Quantification of difference in proportion of CD66+ basal cells between normal and idiopathic pulmonary fibrosis lung (n = 3; *P < 0.1; Mann-Whitney test ). CFE = colony-forming efficiency; IPF = idiopathic pulmonary fibrosis; PCA = principal component analysis; popRNAseq = population RNA sequencing; TPM = transcript count per million.
IPF Distal Lung Contains an Expanded Pool of SPB Cells
We previously showed that decreases in the abundance of alveolar type 2 cells within explant tissue of patients undergoing transplant for end-stage IPF was accompanied by a corresponding increase in the abundance of NGFR+ basal cells (18). These findings were of particular interest in light of genome-wide association study data linking MUC5B-promoter polymorphism and susceptibility to IPF, which suggest potential roles for altered airway secretory cells in disease progression (19). Immunofluorescent staining to localize CD66+ KRT5+ basal cells in end-stage IPF explant tissue revealed an increase in SPB cells (Figures 3I and 3K). To better define changes to basal cells in the IPF lung, we evaluated scRNAseq data from IPF distal lung epithelium. Single-cell transcriptomes of seven IPF samples (Figures E1A, E1C, and E1G) were generated, and the initial quality control and filtering were performed as for control subjects. IPF datasets were classified by the gene signatures obtained for each of the major epithelial cell types defined in the normal lung (Figure 4A). Shared transcripts between IPF and control datasets were evaluated as integrated data, showing similarity among major cell types (Figure 4B). IPF datasets contained representative cells for AB, MPB, and SPB cells identified in the normal lung. Control and IPF cells showed an incomplete signature overlap. A specific proliferative cluster of IPF basal cells was not identified. Instead, proliferating cells were present in all IPF datasets (Figure E1H). Furthermore, two secretory cell clusters specifically enriched in IPF were named on the basis of their specificity for selected secretoglobins as follows: SCGB3A2+ and SCGB3A2+/SCGB3A1+ (Figure 4A). IPF datasets were enriched in CD66 expression, in SPB, and in all the secretory clusters (Figures 4C–4F and Table E6). IPF SPB cells were specifically enriched in CEACAM6 expression (Figure 4E) and could be distinguished from normal SPB cells by their gene signature (Figure 4G and Table E6). Dimensional reduction by diffusion map with “Destiny” and lineage visualization with “Slingshot” show that in IPF datasets, basal and secretory cells also form a continuum, with SPB cells being the connection to secretory cells (Figure 4H).
Figure 4.
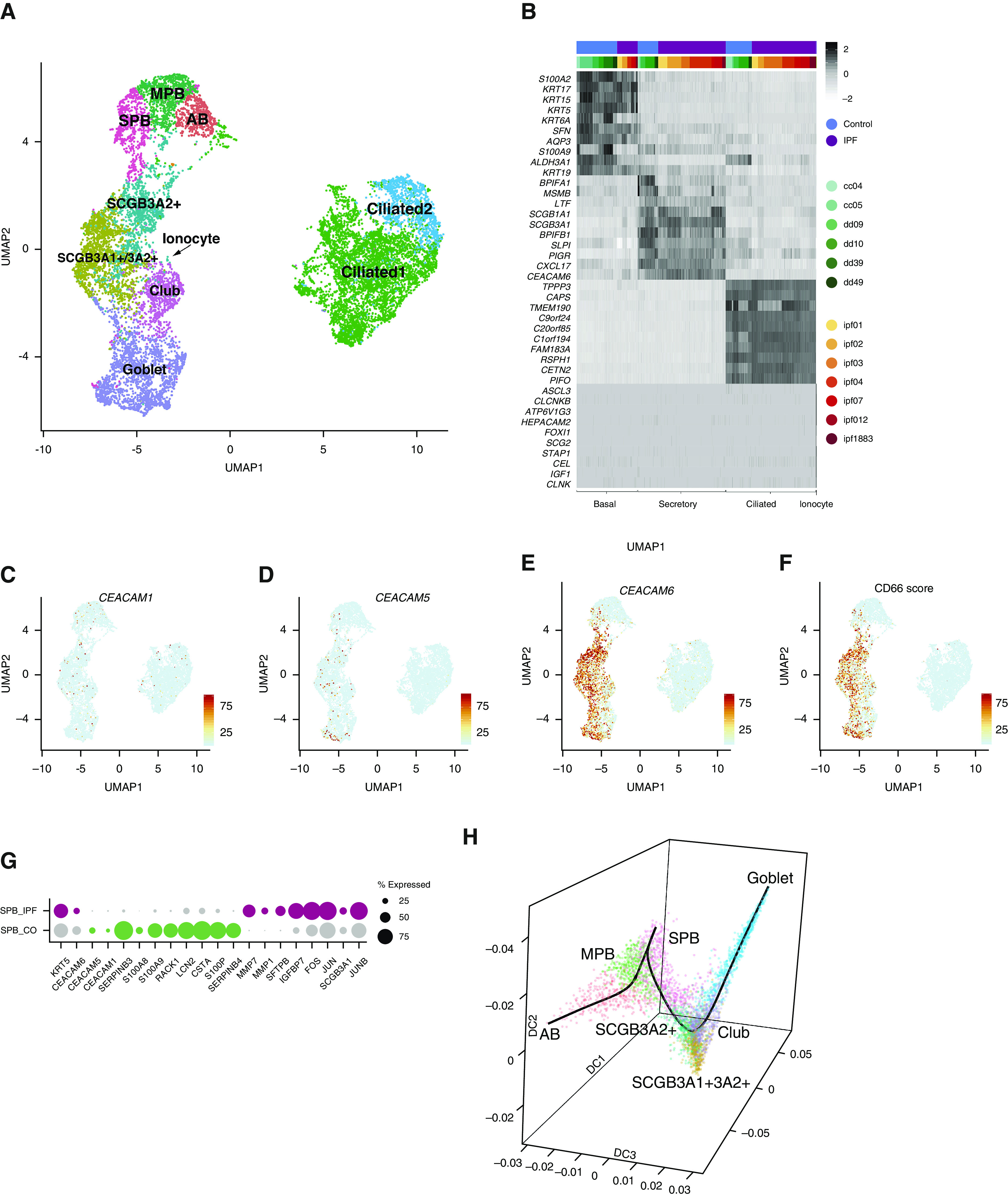
Analysis of idiopathic pulmonary fibrosis (IPF) datasets. (A) Clustering of human IPF datasets visualized by Uniform Manifold Approximation and Projection. (B) Visualization of conserved genes for major cell categories from integrated data of control and IPF datasets visualized by heatmap with z-score values. (C–F) Expression of CEACAM1, CEACAM5, and CEACAM6 and summarized signature score of CD66 in human IPF epithelium visualized by Uniform Manifold Approximation and Projection. (G) Dot plot visualization of top differentially expressed genes for secretory primed basal cells between control and IPF lung. (H) Diffusion map generated with “Destiny” from basal and secretory IPF cells shows a continuous transition of states. The superimposed black line was generated with “Slingshot” and describes the trajectory between cells. AB = activated basal; CO = control; DC = diffusion component; MPB = multipotent basal; SPB = secretory primed basal; UMAP = Uniform Manifold Approximation and Projection.
Notch Signaling Regulates the Maintenance of SPB Cells
Given the importance of Notch signaling in regulating lung airway maintenance and differentiation, we interrogated Notch pathway gene expression between basal cell subsets of control lung. Analysis of Notch ligand–receptor interactions suggests that both signaling and receiving cells were present within the total basal cell population (Figure 5). Interestingly, the SPB cluster possesses the largest amount of both autocrine and juxtacrine Notch signaling (Figure 5A). NOTCH1 was the most broadly involved receptor and was predicted to play a significant role in the regulation of all basal cell subsets, with the exception of MPB cells (Figure 5B). In contrast, signaling by NOTCH3 was the most restricted, showing significant regulation of only the SPB subset (Figures 5B and 5R–5T). The PB cells showed the least signaling activity and were defined predominantly as Notch receiving cells.
Figure 5.
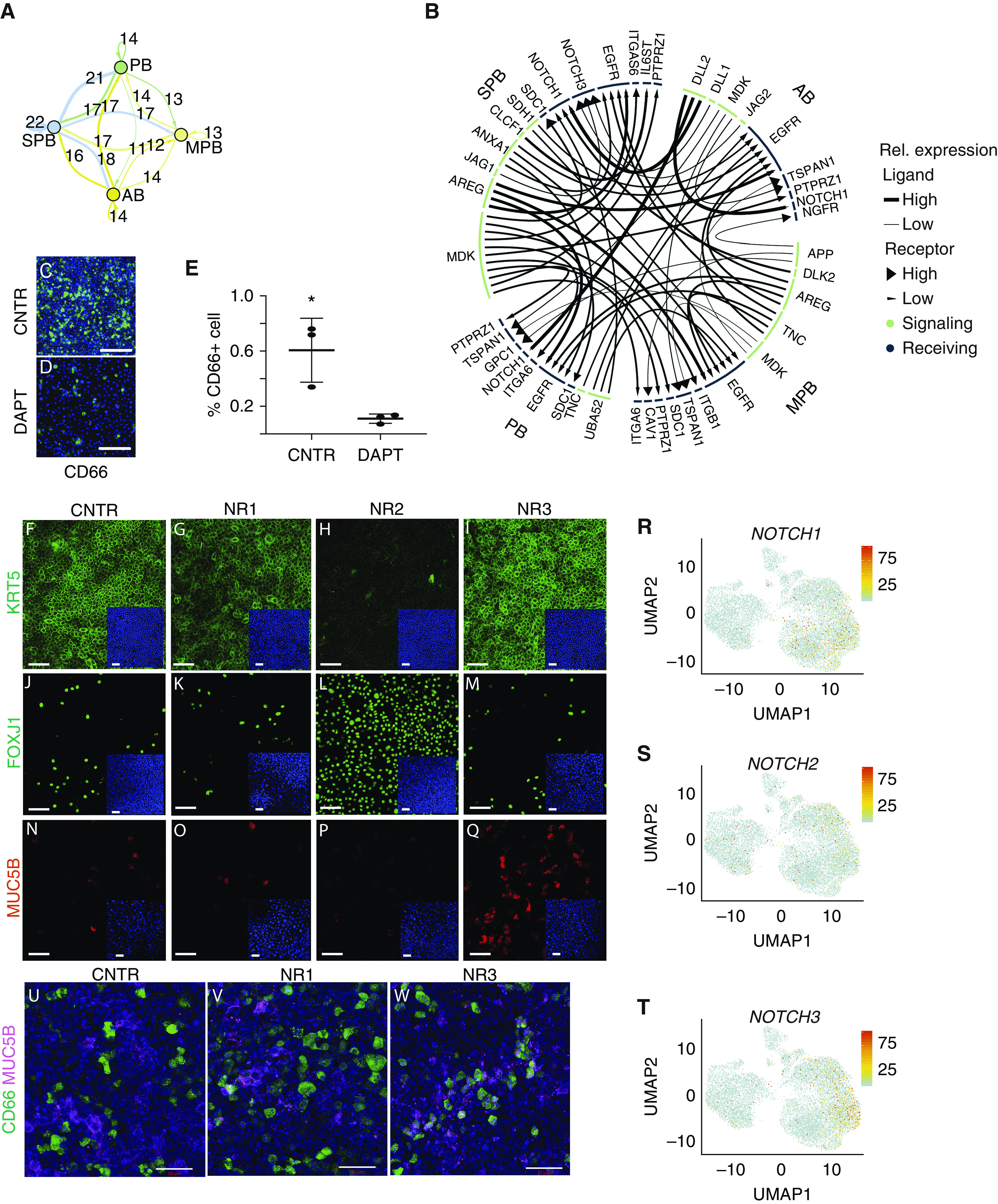
Analysis of Notch receptor specificity in human basal cells. (A) Summary of predicted ligand–receptor interactions for Notch signaling between pairs of basal cell subtypes and for autocrine Notch signaling visualized by network plot. Each arrow is color coded according to basal cell type, and the thickness of the arrow represents the number of ligand–receptor interactions. Arrows that go back to the cell of origin indicate paracrine signaling within the same cell type. Numbers above each arrow indicate how many ligand–receptor interactions were identified. (B) Circus plot showing ligands in green and receptors in blue, with arrows connecting signaling and receiving cells. Each arrow connects signaling and receiving cells, with the tip of the arrow pointing to the receiving cell. The size of the segment underneath each ligand or receptor is proportional to the interactions that were detected. (C–E) Air–liquid interface (ALI) coculture of CD66+ and CD66− basal cells showing the effect of DAPT treatment on CD66+ basal cells. Scale bars, 100 μm. *P < 0.1; Mann-Whitney test. (F–Q) CD66+ and CD66− basal cell ALI coculture treated with specific Notch-blocking antibody for NOTCH1 (NR1), NOTCH2 (NR2), and NOTCH3 (NR3). Effect on basal cell (KRT5+) maintenance, ciliated cell (FOXJ1+) differentiation at Day 14, and secretory cell (MUC5B+) differentiation at Day 8 is shown by immunoreactivity with cell type–specific antibodies. Scale bars, 50 μm. (R–T) Expression of NOTCH1, NOTCH2, and NOTCH3 in normal airway epithelial cells visualized by Uniform Manifold Approximation and Projection. (U–W) Coexpression of CD66 and MUC5B in CD66+ and CD66− cells at Day 14 of ALI coculture treated with specific Notch-blocking antibodies. Scale bars, 50 μm. AB = activated basal; CNTR = control; DAPT = N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester; MPB = multipotent basal; PB = proliferating basal; Rel. expression = relative expression; SPB = secretory primed basal; UMAP = Uniform Manifold Approximation and Projection.
To test the importance of Notch signaling in SPB cell maintenance, we established an ALI culture with a 1:1 proportion of CD66+ (SPB) and CD66− cells. Treatment with DAPT, a γ-secretase inhibitor that blocks release of the NOTCH intracellular domain, dramatically reduced the abundance of CD66+ SPB cells 48 hours after treatment, suggesting a functional role for Notch signaling in the maintenance of SPB cells (Figures 5C–5E). To specifically interrogate the significance of the three Notch-signaling receptors for the maintenance and differentiation of basal cells, the fate of basal cells in primary cultures of human tracheal epithelium was evaluated after treatment with specific blocking antibodies for NOTCH1 (NR1), NOTCH2 (NR2), and NOTCH3 (NR3). Cells grown in ALI culture were harvested after 24 hours, 8 days, and 14 days of treatment. From Day 8 to Day 14, NR2-treated ALI culture showed a dramatic reduction in KRT5+ cells compared with control culture (Figures 5F–5I). Interestingly, neither NR1 nor NR3 treatment impacted the abundance of KRT5+ cells, suggesting that NOTCH2 is the main ligand involved in basal cell maintenance. Furthermore, NOTCH2 inhibition also promoted the expansion of ciliated cell differentiation, as shown by an increase in FOXJ1-immunoreactive cells (Figures 5J–5M). These data are consistent with the previously reported effects of γ-secretase inhibitors, which promote commitment of secretory progenitors to multiciliated cell fates (20), and suggest that NOTCH2 is the relevant receptor that mediates these effects. Even though the inhibition of NOTCH3 had no impact on the abundance of KRT5+ basal or FOXJ1+ ciliated cells in culture, NR3 treatment led to an increase in the proportion of mucin-secreting cells at 8 days (Figures 5N–5Q) and 14 days (Figures 5U–5W) in culture. These data suggest that NOTCH3 may specifically restrain the differentiation of SPB cells, whereas NOTCH2 plays a more global role in regulating basal cell maintenance and restrains ciliated cell differentiation.
Discussion
Airway basal cells function as stem cells that maintain the pseudostratified epithelium of the mouse, and presumably also the human, lung. Even though basal cell heterogeneity has been implied from studies of the pseudostratified epithelium lining the mouse trachea, little is known of basal cell heterogeneity in the normal human lung and how this changes in the setting of the pathological tissue remodeling that accompanies disease. By employing single-cell transcriptomics of freshly isolated human airway epithelial cells, we were able to discern four broad classes of epithelial cells that included basal, secretory, ciliated, and ionocyte. Even though heterogeneity was observed within each of the abundant epithelial cell types, significant heterogeneity was observed among basal cells. Basal cell subsets included a Wnt-signaling multipotent cluster, PB cells, a cluster that showed evidence of secretory priming, and a cluster with activation of stress-response genes. SPB cells may either represent a transitory state of basal cells that are in the process of secretory cell differentiation or a phenotypically stable subset. Diffusion mapping pseudotime analysis of differentiation trajectory shows a continuum of differentiation, suggesting a transitional state. However, immunofluorescence analysis reveals rare basal cells that are positive for the SPB marker CD66 in control adult lung tissue, and we also found that these cells can be clonally expanded in vitro for multiple passages in the absence of mesenchymal support. Further work is therefore necessary to exhaustively determine the features of these cells and their contribution to homeostasis and disease. A recent study performing single-cell transcriptomic analysis of epithelial cells isolated from ALI cultures of nasal epithelium (21) described a KRT5+ cell type, termed “suprabasal,” that may represent a precursor of secretory cells. Suprabasal cells were characterized by the expression of KRT13, a transcript observed in SPB cells in our study. Furthermore, suprabasal cells share an additional similarity with SPB cells in their expression of NOTCH3. However, differences between transcriptomes of suprabasal and SPB cells were observed, such as the expression of KRT4 in suprabasal cells but the absence of this cytokeratin in SPB cells. These may represent cell-type differences between epithelia of the upper and lower respiratory tracts or represent differences in cell states associated with isolation from fresh tissue versus cultures.
Studies in mice have established the existence of subsets of quiescent basal cells in the pseudostratified epithelium that lines the mouse trachea (22). Notch2-ICD expression was shown to define a subpopulation of basal cells that preferentially generate luminal secretory cells after injury. In contrast, c-Myb expression defined basal cells that were primed to undergo multiciliogenesis after injury (23). Our analysis of basal cells in the human lung suggests the conservation of Notch signaling as a regulator of basal cell fate. In cultures of freshly isolated human basal cells, we found that either γ-secretase inhibitors or isoform-specific Notch-blocking antibodies biased basal cell differentiation toward secretory or ciliated phenotypes. We specifically found that NOTCH3 restrains SPB cells from assuming mature secretory cell fates and that it is a global regulator of basal cell maintenance. These findings are consistent with studies in mice showing that NOTCH3 signaling regulates the pool of epithelial progenitors that are competent to respond to NOTCH receptors 1 and 2 (24). In basal cells of the human airway, NOTCH2 is required for their differentiation (25). This is consistent with our observation that blocking NOTCH2 with anti-NOTCH2 antibody blocks secretory cell differentiation and enhances ciliated cell differentiation. Our results with anti-NOTCH3 differ from those of Danahay and colleagues (23) in that we see an increase in secretory cell differentiation, an effect that was not observed in the Danahay study. One explanation for this discrepancy could be that we coculture CD66+ and CD66− basal cells, hence increasing the proportion of CD66+ basal cells in culture and allowing more precise evaluation of this basal cell subset. JAG-blocking antibodies were reported to produce a loss of club cells and a gain in ciliated cells (26). This result is consistent with our observed effect of anti-NOTCH2 on ciliated cell expansion.
We provide evidence that basal cells in fibrotic explant lung tissue obtained from patients undergoing transplant for end-stage IPF include SPB cells that are found in distal airways and honeycomb regions of IPF lungs that are the site of excessive MUC5B expression (5). Even though the pathophysiological significance of honeycombing and excessive mucus secretion is not fully understood, Seibold and colleagues showed that a polymorphism of the MUC5B gene is associated with increased susceptibility to IPF, but surprisingly patients with IPF that carry this polymorphism show better outcomes compared with those with the wild-type MUC5B allele (19). We found that IPF SPB cells express genes previously identified as serum biomarkers for a higher risk of mortality (27) or proposed to be involved in disease development (28), such as MUC1, MMP7, and ICAM1.
This work defines a basal cell hierarchy that is dynamically regulated between health and disease, presenting new therapeutic targets for the modulation of normal tissue maintenance and remodeling.
Supplementary Material
Acknowledgments
Acknowledgment
The authors thank Adrianne Kurkciyan and Stephen Beil for technical assistance, Dr. Edo Israely for assistance with fluorescence-activated cell sorting, and Cedars-Sinai Medical Center Flow Cytometry, Genomics Cores, and Biobank. They also thank Dr. Allon M. Klein and Caleb Weinreb for helpful discussions.
Footnotes
Supported by grants from the California Institute for Regenerative Medicine (LA1-06915), the Cystic Fibrosis Foundation (CARRAR19G0 and STRIPP15XX0), Celgene, the NHLBI (R01-HL135163, HL138540, P01-HL108793, and T32-HL134637) and the NIDDK (P30 DK065988).
Author Contributions: G.C. and B.R.S. designed the project and wrote the manuscript. G.C. performed single-cell RNA-sequencing data analysis. A.M. designed and performed experiments for Notch signaling. T.M. designed and performed organoid experiments and population RNA sequencing. B.K. and M.P. performed tissue processing and immunostainings. C.Y. performed single-cell RNA-sequencing experiments and population RNA-sequencing data processing. M.P. performed single-cell RNA-sequencing experiments. J.L.M. performed high-throughput fluorescence-activated cell-sorting screening experiments and contributed to the project design. D.L., J.R.A., and S.H.R. contributed to data interpretation. C.M.H., P.C., D.J., and P.W.N. contributed with comments on the manuscript. All authors read and reviewed the manuscript.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1164/rccm.201904-0792OC on July 21, 2020
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Wells JM, Watt FM. Diverse mechanisms for endogenous regeneration and repair in mammalian organs. Nature. 2018;557:322–328. doi: 10.1038/s41586-018-0073-7. [DOI] [PubMed] [Google Scholar]
- 2.Teixeira VH, Nadarajan P, Graham TA, Pipinikas CP, Brown JM, Falzon M, et al. Stochastic homeostasis in human airway epithelium is achieved by neutral competition of basal cell progenitors. eLife. 2013;2:e00966. doi: 10.7554/eLife.00966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crystal RG, Randell SH, Engelhardt JF, Voynow J, Sunday ME. Airway epithelial cells: current concepts and challenges. Proc Am Thorac Soc. 2008;5:772–777. doi: 10.1513/pats.200805-041HR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Noble PW, Barkauskas CE, Jiang D. Pulmonary fibrosis: patterns and perpetrators. J Clin Invest. 2012;122:2756–2762. doi: 10.1172/JCI60323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seibold MA, Smith RW, Urbanek C, Groshong SD, Cosgrove GP, Brown KK, et al. The idiopathic pulmonary fibrosis honeycomb cyst contains a mucocilary pseudostratified epithelium. PLoS One. 2013;8:e58658. doi: 10.1371/journal.pone.0058658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rock JR, Randell SH, Hogan BL. Airway basal stem cells: a perspective on their roles in epithelial homeostasis and remodeling. Dis Model Mech. 2010;3:545–556. doi: 10.1242/dmm.006031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.George PM, Patterson CM, Reed AK, Thillai M. Lung transplantation for idiopathic pulmonary fibrosis. Lancet Respir Med. 2019;7:271–282. doi: 10.1016/S2213-2600(18)30502-2. [DOI] [PubMed] [Google Scholar]
- 8.Bagnato G, Harari S. Cellular interactions in the pathogenesis of interstitial lung diseases. Eur Respir Rev. 2015;24:102–114. doi: 10.1183/09059180.00003214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knudsen L, Ruppert C, Ochs M. Tissue remodelling in pulmonary fibrosis. Cell Tissue Res. 2017;367:607–626. doi: 10.1007/s00441-016-2543-2. [DOI] [PubMed] [Google Scholar]
- 10.Prasse A, Binder H, Schupp JC, Kayser G, Bargagli E, Jaeger B, et al. BAL cell gene expression is indicative of outcome and airway basal cell involvement in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2019;199:622–630. doi: 10.1164/rccm.201712-2551OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Plasschaert LW, Žilionis R, Choo-Wing R, Savova V, Knehr J, Roma G, et al. A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte. Nature. 2018;560:377–381. doi: 10.1038/s41586-018-0394-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tirosh I, Venteicher AS, Hebert C, Escalante LE, Patel AP, Yizhak K, et al. Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature. 2016;539:309–313. doi: 10.1038/nature20123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Widdicombe JH, Wine JJ. Airway gland structure and function. Physiol Rev. 2015;95:1241–1319. doi: 10.1152/physrev.00039.2014. [DOI] [PubMed] [Google Scholar]
- 14.Pan S, Iannotti MJ, Sifers RN. Analysis of serpin secretion, misfolding, and surveillance in the endoplasmic reticulum. Methods Enzymol. 2011;499:1–16. doi: 10.1016/B978-0-12-386471-0.00001-8. [DOI] [PubMed] [Google Scholar]
- 15.Strous GJ, Dekker J. Mucin-type glycoproteins. Crit Rev Biochem Mol Biol. 1992;27:57–92. doi: 10.3109/10409239209082559. [DOI] [PubMed] [Google Scholar]
- 16.Plotnikova OV, Golemis EA, Pugacheva EN. Cell cycle-dependent ciliogenesis and cancer. Cancer Res. 2008;68:2058–2061. doi: 10.1158/0008-5472.CAN-07-5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rock JR, Gao X, Xue Y, Randell SH, Kong YY, Hogan BL. Notch-dependent differentiation of adult airway basal stem cells. Cell Stem Cell. 2011;8:639–648. doi: 10.1016/j.stem.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu Y, Mizuno T, Sridharan A, Du Y, Guo M, Tang J, et al. Single-cell RNA sequencing identifies diverse roles of epithelial cells in idiopathic pulmonary fibrosis. JCI Insight. 2016;1:e90558. doi: 10.1172/jci.insight.90558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seibold MA, Wise AL, Speer MC, Steele MP, Brown KK, Loyd JE, et al. A common MUC5B promoter polymorphism and pulmonary fibrosis. N Engl J Med. 2011;364:1503–1512. doi: 10.1056/NEJMoa1013660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsao PN, Vasconcelos M, Izvolsky KI, Qian J, Lu J, Cardoso WV. Notch signaling controls the balance of ciliated and secretory cell fates in developing airways. Development. 2009;136:2297–2307. doi: 10.1242/dev.034884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruiz García S, Deprez M, Lebrigand K, Cavard A, Paquet A, Arguel MJ, et al. Novel dynamics of human mucociliary differentiation revealed by single-cell RNA sequencing of nasal epithelial cultures. Development. 2019;146:dev177428. doi: 10.1242/dev.177428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carraro G, Stripp BR. A new notch for lung stem cells. Cell Stem Cell. 2015;16:107–109. doi: 10.1016/j.stem.2015.01.012. [DOI] [PubMed] [Google Scholar]
- 23.Pardo-Saganta A, Law BM, Tata PR, Villoria J, Saez B, Mou H, et al. Injury induces direct lineage segregation of functionally distinct airway basal stem/progenitor cell subpopulations. Cell Stem Cell. 2015;16:184–197. doi: 10.1016/j.stem.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mori M, Mahoney JE, Stupnikov MR, Paez-Cortez JR, Szymaniak AD, Varelas X, et al. Notch3-Jagged signaling controls the pool of undifferentiated airway progenitors. Development. 2015;142:258–267. doi: 10.1242/dev.116855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Danahay H, Pessotti AD, Coote J, Montgomery BE, Xia D, Wilson A, et al. Notch2 is required for inflammatory cytokine-driven goblet cell metaplasia in the lung. Cell Rep. 2015;10:239–252. doi: 10.1016/j.celrep.2014.12.017. [DOI] [PubMed] [Google Scholar]
- 26.Lafkas D, Shelton A, Chiu C, de Leon Boenig G, Chen Y, Stawicki SS, et al. Therapeutic antibodies reveal Notch control of transdifferentiation in the adult lung. Nature. 2015;528:127–131. doi: 10.1038/nature15715. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y, Kaminski N. Biomarkers in idiopathic pulmonary fibrosis. Curr Opin Pulm Med. 2012;18:441–446. doi: 10.1097/MCP.0b013e328356d03c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ballester B, Milara J, Cortijo J. Idiopathic pulmonary fibrosis and lung cancer: mechanisms and molecular targets. Int J Mol Sci. 2019;20:593. doi: 10.3390/ijms20030593. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All transcriptome data were deposited in Gene Expression Omnibus (GSE143706 and GSE143705). A GitHub repository with a markdown to show our quality control and data analysis is available at https://github.com/gc-github-bio/Normal-and-IPF-lung.


