Abstract
The ability of the liver to regenerate and restore mass limits the increasing mortality rate due to life‐threatening liver diseases. Successful liver regeneration is accomplished in multiple stages, of which the priming and proliferation phases are well studied. However, the regulatory pathways, specifically microRNA (miRNA)‐mediated posttranscriptional regulation, which prevent uncontrolled proliferation and mediate the termination of liver regeneration, are not well understood. We identified differentially regulated miRNAs during the termination phase after 2/3 partial hepatectomy (PH) in mice, which is a well‐established mouse model of liver regeneration. We further evaluated the function of differentially regulated miRNAs in primary mouse hepatocytes by using mimics and inhibitors and in vivo by using adeno‐associated virus (AAV) serotype 8. A candidate miRNA target was identified by messenger RNA array in silico analyses and validated in primary mouse and human hepatocytes. Using miRNA profiling, we discovered miR‐125b‐5p as a novel regulator of hepatocyte proliferation in the late phase of liver regeneration. AAV‐mediated miR‐125b‐5p delivery in mice enhanced the endogenous regenerative capacity and resulted in improved restoration of liver mass after 2/3 PH. Further, we found that ankyrin repeat and BTB/POZ domain containing protein 1 (Abtb1) is a direct target of miR‐125b‐5p in primary mouse and human hepatocytes and contributes to the pro‐proliferative activity of miR‐125b‐5p by forkhead box G1 (FOXG1) and the cyclin‐dependent kinase inhibitor 1A (p21) pathway. Conclusion: miR‐125b‐5p has an important role in regulating hepatocyte proliferation in the termination phase of liver regeneration and may serve as a potential therapeutic target in various liver diseases that often exhibit deregulated hepatocyte proliferation.
Abbreviations
- AAV8
adeno‐associated virus 8
- Abtb1
ankyrin repeat and BTB/POZ domain containing protein 1
- BrdU
bromodeoxyuridine
- CCNA2
cyclin A2
- DAPI
4′,6‐diamidino‐2‐phenylindole
- Foxg1
forkhead box G1
- FOXO
forkhead box protein O1
- miRNA/miR
microRNA
- mRNA
messenger RNA
- p21
cyclin‐dependent kinase inhibitor 1A
- PH
partial hepatectomy
- PHH3
phosphorylation of histone 3
- qRT‐PCR
quantitative reverse‐transcription polymerase chain reaction
- siRNA
small interfering RNA
- SMAD
small mothers against decapentaplegic
- TGFβ
transforming growth factor beta
- Ttr
transthyretin
- UTR
untranslated region
- VCL
vinculin
- WST‐1
water‐soluble tetrazolium salts 1
Liver regeneration is commonly classified into three different phases based on distinct functions and molecular pathways activated, namely, priming phase, proliferation phase, and termination phase.( 1 ) In rodents, the whole process of liver regeneration is completed in around 7 to 14 days, whereas in humans, it may take weeks to several months. In the process of mouse liver regeneration, the period of 24 hours after 2/3 partial hepatectomy (PH) is defined as the priming phase, in which hepatocytes quickly progress from quiescent G0 to G1 states. This is followed by a proliferation phase, which lasts for 48 hours and where expansion of the hepatocyte population takes place. Finally, the termination phase of liver regeneration follows, in which proliferation is suppressed and hepatic cells regain a quiescent state.( 2 ) The regulators in the termination of liver regeneration, collectively termed the hepatostat control system,( 3 ) remain poorly investigated when compared to the abundance of studies on regulators of early phases of liver regeneration, such as initiation and proliferation. Thus far, the best characterized signaling in the termination of liver regeneration is the transforming growth factor beta (TGFβ)–small mothers against decapentaplegic (SMAD) pathway.( 4 ) Further, cooperation between transcription factors, such as CCAAT‐enhancer‐binding proteins and chromatin remodeling proteins, have been shown to be essential for the proper termination of liver regeneration.( 5 )
We( 6 , 7 , 8 ) and others( 9 , 10 ) have reported identification and functional elucidation of microRNAs (miRNAs ), which act as posttranscriptional regulators of gene expression, during liver diseases and early phases of liver regeneration. However, identification and functional elucidation of miRNAs during the termination phase remains to be shown. Hence, we aimed to identify miRNAs that regulate the regenerative capacity of hepatocytes during the termination of liver regeneration. By miRNA profiling at different time points during the termination phase in a 2/3 PH mouse model, we identified miR‐125b‐5p as a key regulator of liver regeneration. miR‐125b‐5p overexpression in vivo significantly extends the regenerative capacity of hepatocytes during the termination phase. Furthermore, we demonstrate that miR‐125b‐5p directly targeted ankyrin repeat and BTB/POZ domain containing protein 1 (Abtb1),( 11 ) which regulates the expression levels of forkhead box G1 (Foxg1) and cyclin‐dependent kinase inhibitor 1A (p21), contributing to the proliferative activity of miR‐125b‐5p.
Materials and Methods
Mice
Animal experiments were performed according to the guidelines of the Hannover Medical School, Germany. For all experiments presented in this study, we used BALB/c mice purchased from Charles River Laboratories (Germany).
Partial Hepatectomy
We performed 2/3 PH on BALB/c mice as described.( 7 ) Briefly, the median (left and right) and left lateral liver lobes were surgically removed after mice were anesthetized by inhaling isoflurane mixed with oxygen.
Hepatocyte Transfection and Proliferation Assay
Freshly isolated primary human hepatocytes were provided by Regenerative Medicine and Experimental Surgery, Hannover Medical School, Hannover, as reported.( 12 ) Mouse primary hepatocytes were isolated from mouse liver as described.( 8 ) Briefly, mice were anesthetized, and their livers were perfused with Liberase (Roche). After perfusion, livers were disintegrated mechanically before collecting hepatocytes by low‐speed centrifugation. Nonparenchymal cells were removed by discarding the supernatant. For all in vitro transfection experiments, we used Percoll density gradient‐purified mouse hepatocytes to achieve high transfection efficiency. We seeded 100,000 primary hepatocytes per well of a collagen‐coated 12‐well plate (TPP). Twelve hours after seeding, hepatocytes were transfected with 25 nM or 50 nM miR‐125b‐5p mimic, miR‐125b‐5p inhibitor, control scramble (Qiagen), or 100 nM ABTB1 small interfering RNA (siRNA) (Qiagen) using Targefect reagent in the presence of virofect enhancer (Targeting Systems). Transfected hepatocytes were cultured in hepatocyte culture medium (Lonza) containing recombinant epidermal growth factor (an inducer of hepatocyte proliferation), transferrin, ascorbic acid, insulin, hydrocortisone, bovine serum albumin, and gentamicin sulfate‐amphotericin.
Adeno‐Associated Virus 8 Preparation
Adeno‐associated virus 8 (AAV8)‐transthyretin (Ttr)‐miR‐125b‐5p and control AAV vectors were prepared as described.( 8 ) The titer was determined by quantitative real‐time polymerase chain reaction (qRT‐PCR) using primers spanning the region of the Ttr promoter as published.( 8 )
Luciferase Reporter Assay
The 3′ untranslated region (UTR) of mouse Abtb1 messenger RNA (mRNA) was amplified by PCR from genomic DNA using the following primers: forward primer,
GGAAAGTTTAAACACAAGGAAGCCCCAAGATTT; reverse primer, GGAAATCTAGATATTGCCACACACCCCCTAC. The amplicon was cloned into a miRGLO vector (Promega). Luciferase reporter assay was performed, as described.( 8 )
Statistical Analysis
Significance was determined with the two‐tailed Student t test for comparison of two groups. Significance between multiple groups was determined by one‐way analysis of variance. P < 0.05 was considered significant.
Results
Identification of Regulatory miRNAs During the Termination Phase of Liver Regeneration
To identify candidate miRNAs that regulate the endogenous regenerative capacity of hepatocytes during the termination phase of liver regeneration, total RNA from mouse liver at 0 hours, 72 hours, 96 hours, 7 days, and 14 days after 2/3 PH were subjected to miRNA profiling (Fig. 1A). These time points were chosen based on the bromodeoxyuridine (BrdU) labeling index, which clearly demonstrated minimal proliferation between 72 hours and 14 days after 2/3 PH (Fig. 1B). The differentially expressed miRNAs in the above‐mentioned time points after 2/3 PH compared to day 0 (Fig. 1C) were further selected based on the following three criteria: (i) miRNA must be conserved between human and mouse; (ii) miRNA must be differentially expressed at least at a 2‐fold level at any of the time points; and (iii) miRNA must have high or at least modest expression in normal liver (>1,000 reads in the profiling assay).( 13 ) Based on these criteria, we identified five miRNAs: miR‐1224‐5p, miR‐494‐3p, miR‐451, miR‐125b‐5p, and miR‐99a (Fig. 1D). To validate the result of the miRNA profiling, we then determined the levels of these five miRNAs by qRT‐PCR. The miRNA expression levels for the five miRNAs were consistent with the results obtained by miRNA profiling (Fig. 1E).
FIG. 1.
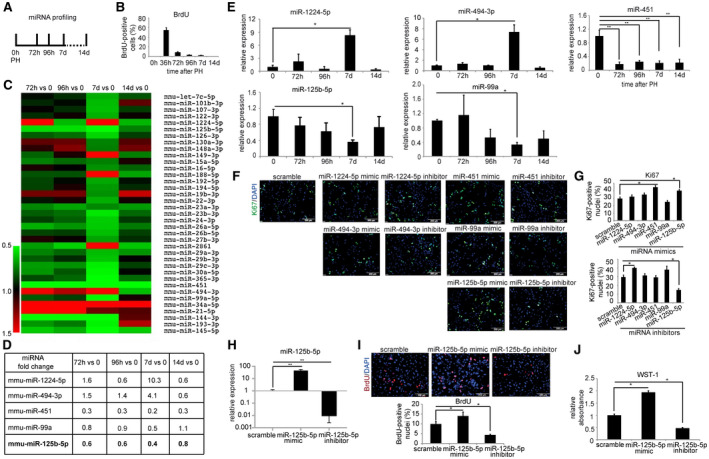
Setup and results of the miRNA profiling assay for miRNAs that impact the endogenous regenerative capacity of hepatocytes during the termination phase of liver regeneration. (A) Schematic outline of the miRNA profiling assay. Livers from mice subjected to 2/3 PH were analyzed by miRNA profiling at different time points. (B) BrdU labeling index at 0 hours, 36 hours, 72 hours, 96 hours, 7 days, and 14 days after 2/3 PH in wild‐type BALB/c mice. (C) Heat map of the differentially regulated miRNAs from the miRNA profiling assay. (D) List of miRNAs that showed at least 2‐fold change (up‐ or down‐regulation) at either of the indicated time points after 2/3 PH compared to the expression of the respective miRNA at 0 hours. (E) Validation of miRNA profiling by qRT‐PCR. (F) Representative photographs and (G) quantification of Ki67 immunostained primary hepatocytes (magnification ×200) transfected with 25 nM miRNA mimic, inhibitor, or scramble control. (H) qRT‐PCR analysis confirmed overexpression and down‐regulation of miR‐125b‐5p. (I) Representative photographs and quantification of BrdU‐immunostained primary hepatocytes (magnification ×200) transfected with miR‐125b‐5p mimic, inhibitor, or scramble control. (J) WST‐1 assay of hepatocytes transfected with miR‐125b‐5p mimic, inhibitor, or scramble control. *P < 0.05, **P < 0.01; data are presented as mean ± SEM (n = 4 per group). Abbreviations: d, days; h, hours.
As two candidate miRNAs, miR‐125b‐5p and miR‐99a, have been shown to cluster with let‐7c as a tricistron on human chromosome 21,( 14 ) we also examined the expression of let‐7c in mouse livers at different time points after PH. The qRT‐PCR analyses, similar to miRNA profiling, revealed that let‐7c expression was not significantly altered in the termination of liver regeneration (Supporting Fig. S1). Thus, we kept our short‐listed set of five miRNAs for further functional studies.
Overexpression of miR‐125b‐5p in Primary Mouse Hepatocytes Regulates Proliferation In Vitro
To investigate the effects of the five miRNAs on hepatocyte proliferation in vitro, freshly isolated and purified primary mouse hepatocytes were transfected with mimics and inhibitors of each miRNA. Of note, we cultured hepatocytes in a medium containing epidermal growth factor, a known inducer of hepatocyte proliferation.( 15 , 16 ) miRNA scramble was transfected as a control. Ki67 immunofluorescence staining was performed 48 hours after transfection to determine proliferation of hepatocytes. The result showed that mimics of miR‐451 and miR‐125b‐5p significantly enhanced hepatocyte proliferation whereas suppression of miR‐1224 and miR‐125b‐5p enhanced and decreased cell proliferation, respectively (Fig. 1F,G). In addition to these five miRNAs, we analyzed the effect of gain and loss of miR‐188‐5p and miR‐149‐3p on hepatocyte proliferation because they showed a robust up‐regulation in miRNA profiling (Supporting Fig. S2A) and subsequent qRT‐PCR analyses (Supporting Fig. S2B). Despite high expression of both miRNAs, both failed to exert a significant effect on proliferation of primary hepatocytes (Supporting Fig. S2C,D).
Because the gain as well as loss of only miR‐125b‐5p significantly affected proliferation of primary hepatocytes, we aimed to confirm these results by qRT‐PCR, BrdU staining, and water‐soluble tetrazolium salts 1 (WST‐1) analyses (Fig. 1H‐J). These analyses showed that miR‐125b‐5p mimic and inhibitor inversely regulate hepatocyte proliferation in vitro. Therefore, we selected miR‐125b‐5p for further in vivo studies.
Overexpression of miR‐125b‐5p In Vivo Enhances the Endogenous Regenerative Capacity During Termination of Liver Regeneration
To investigate the effect of miR‐125b‐5p during the termination of liver regeneration, miR‐125b‐5p was overexpressed in the mouse liver by an AAV8 (AAV8‐Ttr‐miR‐125b‐5p) vector that expresses miR‐125b‐5p under the transcriptional control of the Ttr promoter, a hepatocyte‐specific promoter. We injected 1 × 1010 viral particles of AAV8‐Ttr‐miR‐125b‐5p or control AAV into BALB/c mice through the tail vein (Fig. 2A). We performed 2/3 PH 7 days after AAV8 injection to induce liver regeneration, and the mice were killed for analysis after an additional 5 days. We first confirmed successful overexpression of miR‐125b‐5p at day 5 after 2/3 PH by qRT‐PCR in the livers of mice injected with AAV8‐Ttr‐miR‐125b‐5p (Fig. 2B). miR‐125b‐5p‐overexpressing mice showed a higher liver to body weight ratio (Fig. 2C) and more Ki67‐positive cells compared to control mice livers (Fig. 2D). Cyclin A2 (CCNA2) accumulates during the S‐phase and is also required for cell‐cycle progression through G2/M transition. Hence, we next examined CCNA2 expression by western blot. The result showed a strong accumulation of CCNA2 in miR‐125b‐5p‐overexpressing mice livers at day 5 after 2/3 PH (Fig. 2E). Phosphorylation of histone 3 (PHH3) is observed during the mitosis phase, whereas dephosphorylation of histone occurs as cells exit mitosis. Therefore, we investigated whether miR‐125b‐5p overexpression enhances mitosis by staining livers with PHH3 antibody. We observed PHH3‐positive cells in miR‐125b‐5p‐overexpressing mice livers at day 5 after PH while the control mice livers were devoid of PHH3‐positive cells (Fig. 2F). However, we did not find any significant difference in hepatocyte size in miR‐125b‐5p‐overexpressing mice at day 5 after PH (Fig. 2G). Thus, results of liver to body weight ratio, Ki67, CCNA2, and PHH3 together indicate that miR‐125b‐5p overexpression leads to enhanced proliferation during termination of liver regeneration.
FIG. 2.
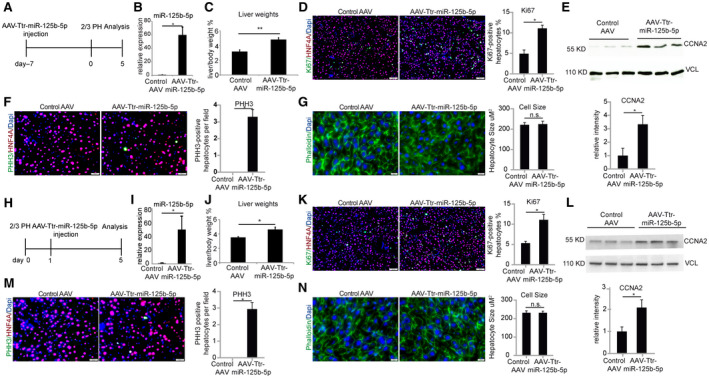
miR‐125b‐5p overexpression promotes mouse hepatocyte proliferation at day 5 after 2/3 PH. (A) Schematic representation of the experimental design. (B) The qRT‐PCR analysis confirmed overexpression of miR‐125b‐5p at day 5 of 2/3 PH in liver of mice injected with AAV8‐Ttr‐miR‐125b‐5p. (C) Liver to body weight at day 5 after 2/3 PH. (D) Representative photographs and quantification of Ki67‐ and HNF4A‐immunostained liver sections (magnification ×100) in miR‐125b‐5p‐overexpressing livers at day 5 after 2/3 PH compared to control mice livers with control AAV injection at the same time point. (E) Western blot analysis of CCNA from mouse livers injected with AAV8‐Ttr‐miR‐125b‐5p and control AAV. Vinculin was used as a loading control. (F) Representative photographs of PHH3 and HNF4A (magnification ×200) in miR‐125b‐5p‐overexpressing livers at day 5 after 2/3 PH compared to control mice livers with control AAV injection at the same time point (n = 4 per group). (G) Representative photographs of phalloidin staining and quantification of hepatocyte size in miR‐125b‐5p‐overexpressing livers. (H) Schematic representation of the experimental design. (I) qRT‐PCR analysis revealed overexpression of miR‐125b‐5p at day 5 after 2/3 PH in mice livers injected with AAV8‐Ttr‐miR‐125b‐5p. (J) Liver to body weight ratio. (K) Representative photographs and quantification of Ki67‐ and HNF4A‐immunostained liver sections (magnification ×100) in miR‐125b‐5p‐overexpressing livers at day 5 after 2/3 PH compared to control mice livers with control AAV injection at the same time point. (L) Western blot analysis of cyclin A from mouse livers that underwent 2/3 PH and were then injected with AAV8‐Ttr‐miR‐125b‐5p or control AAV. Vinculin was used as a loading control. (M) Representative photographs and quantification of PHH3‐ and HNF4A‐immunostained liver sections (magnification ×200) from mice injected with AAV8‐Ttr‐miR‐125b‐5p or control AAV at the same time point (n = 4 per group). (N) Representative photographs of phalloidin staining and quantification of hepatocyte size in miR‐125b‐5p‐overexpressing livers. *P < 0.05, **P < 0.01; data are presented as mean ± SEM. Abbreviation: HNF4A, hepatocyte nuclear factor 4A.
Administration of miR‐125b‐5p After PH Specifically Facilitates the Regenerative Capacity During Termination of Liver Regeneration
We next examined whether miR‐125b‐5p regulates the termination of liver regeneration when AAV8 is administered after 2/3 PH. To examine this, we injected AAV8 encoding for miR‐125b‐5p at 24 hours after 2/3 PH (Fig. 2H).
We first confirmed overexpression of miR‐125b‐5p at day 5 after 2/3 PH in livers of mice treated with AAV8‐Ttr‐miR‐125b‐5p (Fig. 2I). Liver to body weight ratio was higher in miR‐125b‐5p‐overexpressing mice compared to their respective controls (Fig. 2J). Moreover, we found a higher number of Ki67‐positive cells, elevated expression of CCNA2 protein, and presence of more PHH3‐positive cells in miR‐125b‐5p‐overexpressing mice without any significant difference in hepatocyte size (Fig. 2K‐N). Taken together, these data demonstrate that miR‐125b‐5p overexpression both before or after 2/3 PH enhances the endogenous regenerative capacity of hepatocytes during the termination phase of liver regeneration.
We then sought to investigate if miR‐125b‐5p modulation is able to enhance restoration of liver mass when liver regeneration is terminated. To this end, we administered AAV8‐Ttr‐miR‐125b‐5p at 24 hours after 2/3 PH and killed the mice at day 14 after 2/3 PH, a time point when liver regeneration has completed (Fig. 3A). We confirmed overexpression of miR‐125b‐5p at this time point (Fig. 3B), although levels of miR‐125b‐5p at day 14 were lower than those observed at day 5 (Fig. 2B.H). We found that the liver to body weight ratio remained significantly higher in miR‐125b‐5p‐overexpressing mice compared to the respective controls (Fig. 3C), indicating that increased liver mass during the termination phase remained stable. However, the pro‐proliferative effect diminished, as reflected by the absence of Ki67‐positive cells (Fig. 3D) and PHH3‐positive cells (Fig. 3E).
FIG. 3.
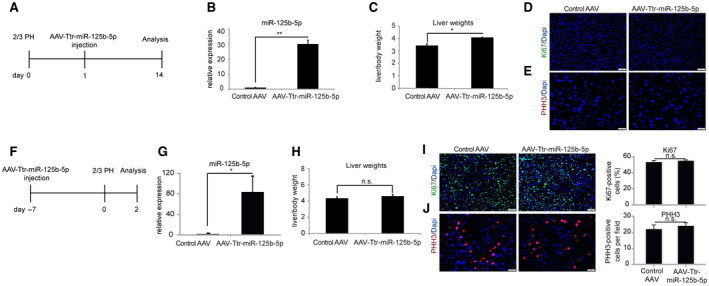
miR‐125b‐5p overexpression does not cause abnormal liver regeneration at day 14 and 48 hours after 2/3 PH. (A) Schematic representation of the experimental design. (B) qRT‐PCR analysis revealed overexpression of miR‐125b‐5p at day 14 after 2/3 PH in liver of mice injected with AAV8‐Ttr‐miR‐125b‐5p compared to mice injected with control AAV. (C) Liver to body weight ratio. (D,E) Representative photographs of (D) Ki67‐ and (E) PHH3‐immunostained liver sections in miR‐125b‐5p‐overexpressing livers and control mice at day 14 after 2/3 PH. (F) Schematic representation of the experimental design. (G) qRT‐PCR analysis revealed overexpression of miR‐125b‐5p at 2 days after 2/3 PH in liver of mice injected with AAV8‐Ttr‐miR‐125b‐5p compared to mice injected with control AAV. (H) Liver to body weight ratio at 2 days after 2/3 PH. (I,J) Representative photographs of (I) Ki67‐ and (J) PHH3‐immunostained liver sections in miR‐125b‐5p‐overexpressing livers and control mice at day 2 after 2/3 PH. *P < 0.05, **P < 0.01; data are presented as mean ± SEM (n = 4 per group). Abbreviation: n.s., not significant.
To address the relevance of miR‐125b‐5p during the proliferation phase, we injected mice with AAV8‐Ttr‐miR‐125b‐5p 7 days before 2/3 PH and analyzed livers at 48 hours after 2/3 PH (Fig. 3F,G). We did not find any significant differences in liver to body weight ratio (Fig. 3H) or in the expression of proliferation markers (Fig. 3I,J) between miR‐125b‐5p‐overexpressing mice and control mice, indicating that miR‐125b‐5p enhances hepatocyte proliferation specifically in the termination phase.
miR‐125b‐5p Enhances Hepatocyte Regeneration by Direct Down‐Regulation of Abtb1
miRNAs regulate gene expression at the posttranscriptional level, and up to 80% of genes are reported to show down‐regulation at mRNA levels in addition to their protein levels.( 17 ) Therefore, to elucidate the mechanism by which miR‐125b‐5p promotes hepatocyte proliferation during the termination phase, we carried out microarray‐based mRNA expression analysis covering 32,000 murine transcripts on RNA isolated from livers of mice injected either with AAV‐Ttr‐miR‐125b‐5p or control AAV (Fig. 4A). The results revealed that Abtb1 transcript, encoding ABTB1 protein with an ankyrin repeats region and two BTB/POZ domains, was consistently down‐regulated (<2‐fold) in mice treated with AAV8‐Ttr‐miR‐125b‐5p (Fig. 4B). Furthermore, Abtb1 was predicted as the conserved target between mouse and human by three miRNA target prediction tools: miRanda, PicTar, and TargetScan (Fig. 4C). Western blot analysis confirmed the reduced protein levels of ABTB1 in miR‐125b‐5p‐overexpressing mice livers (Fig. 4D).
FIG. 4.
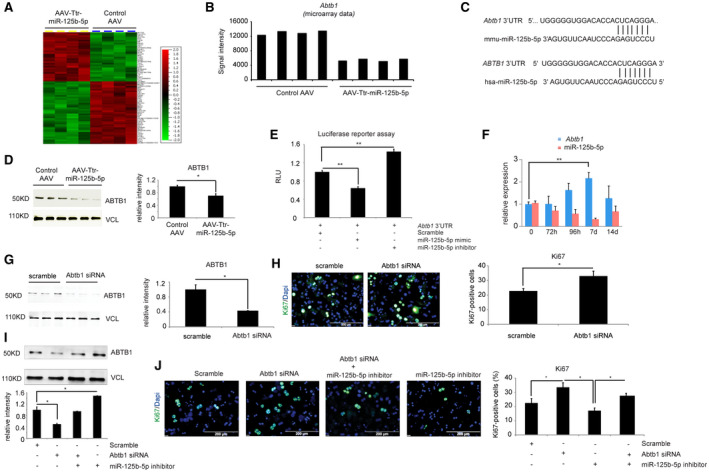
miR‐125b‐5p regulates Abtb1 expression in vivo. (A) The heat map shows differentially up‐ or down‐regulated genes (above 2‐fold change) obtained from microarray‐based mRNA expression analysis. (B) Microarray‐based mRNA expression analysis indicated that ABTB1 expression was down‐regulated in liver of mice injected with AAV8‐Ttr‐miR‐125b‐5p. (C) TargetScan‐based in silico analyses predicts Abtb1 as a target of miR‐125b‐5p in mouse and human. (D) Western blot analysis of ABTB1 in mice livers at day 5 after 2/3 PH. Vinculin was used as a loading control. (E) Luciferase reporter assay confirms the binding of miR‐125b‐5p with 3′ UTR of Abtb1. Primary hepatocytes transfected with miR‐125b‐5p mimic have lower RLUs, while hepatocytes transfected with miR‐125b‐5p inhibitor showed higher RLUs than the control miRNA scramble transfection. (F) qRT‐PCR analysis of ABTB1 expression and miR‐125b‐5p at the indicated time points during the termination phase, indicating an inverse correlation between the expression of ABTB1 and miR‐125b‐5p. (G) Determination of knockdown efficiency by western blot of primary mouse hepatocytes transfected with siABTB1 or scramble control. Vinculin was used as a loading control. Experiments were done in triplicates. Quantification of western blots is shown in the right panel. (H) Representative photographs and quantification of Ki67‐immunostained primary hepatocytes transfected with Abtb1 siRNA or scramble control. Quantification is shown in the right panel. (I) Western blot for ABTB1 and vinculin in primary mouse hepatocytes transfected with Abtb1 siRNA or miR‐125b‐5p inhibitor alone and together. (J) Ki67 immunofluorescence of primary mouse hepatocytes transfected with either Abtb1 siRNA or miR‐125b‐5p inhibitor alone and together. Scale bars, 200 μm. *P < 0.05, **P < 0.01; data are presented as mean ± SEM (n = 4 per group). Abbreviations: d, days; h, hours; hsa, human serum albumin; mmu, Mus musculus (house mouse); RLU, relative luciferase unit.
To assess whether Abtb1 is a direct target of miR‐125b‐5p, we generated luciferase reporter constructs in which the 3′ UTR of Abtb1 was cloned downstream of the luciferase gene. Luciferase reporter assay revealed decreased luciferase activity in the presence of miR‐125b‐5p mimic treatment and enhanced luciferase activity following inhibition of miR‐125b‐5p (Fig. 4E). These in vitro results were confirmed in vivo by analyzing Abtb1 and miR‐125b‐5p expression at different time points during the termination phase by qRT‐PCR; this revealed an inverse correlation between the expression pattern of Abtb1 and miR‐125b‐5p (Fig. 4F). Together, these data indicate that miR‐125b‐5p regulates Abtb1 at the posttranscriptional level.
To investigate the effect of ABTB1 loss, primary mouse hepatocytes were transfected with siRNA against Abtb1 mRNA. The transfection of Abtb1 siRNA led to efficient knockdown of ABTB1, as shown by western blot (Fig. 4G). ABTB1 loss led to increased primary hepatocyte proliferation, as indicated by Ki67 immunofluorescence staining (Fig. 4H). Furthermore, to study whether suppression of miR‐125b‐5p would rescue the effect caused by loss of ABTB1, we transfected primary mouse hepatocytes with Abtb1 siRNA alone or in combination with miR‐125b‐5p inhibitor. The cotransfection of Abtb1 siRNA and miR‐125b‐5p inhibitor led to a restoration of ABTB1 protein level (Fig. 4I). After confirming the restoration of ABTB1 protein levels, we analyzed proliferation by Ki67 immunofluorescence staining, which revealed a higher number of Ki67‐positive cells following cotransfection of Abtb1 siRNA and miR‐125b‐5p inhibitor than the number with miR‐125b‐5p inhibitor alone; however, this number remained less than that of siABTB1 transfection alone (Fig. 4J). Thus, our cotransfection experiments suggested that ABTB1 contributes, albeit in part, to the pro‐proliferative effect of miR‐125b‐5p on primary mouse hepatocytes.
Abtb1 Expression Correlates With The FOXG1/P21 Signaling Pathway
To identify the downstream signaling pathway of antiproliferative ABTB1,( 18 ) we transfected primary mouse hepatocytes with Abtb1 siRNA and performed microarray‐based mRNA expression analysis. The results revealed that ABTB1 loss led to significant up‐regulation of Foxg1 expression (Fig. 5A,B). Gene Ontology and Kyoto Encyclopedia of Genes and Genomes pathway enrichment analyses further indicated that these differentially expressed genes are involved in cell‐cycle arrest (Fig. 5C) and in enrichment of the forkhead box protein O1 (FOXO) signaling pathway (Fig. 5D), respectively. Foxg1, a member of the Forkhead box family, is shown to act as a transcription repression factor, which binds to the FOXO‐SMAD complex and inhibits p21 expression.( 19 ) Our western blot analysis confirmed higher protein levels of FOXG1 and lower protein levels of P21 (Fig. 5E), indicating Abtb1 regulates the FOXG1/p21 signaling pathway in primary hepatocytes. In concert with these in vitro results, qRT‐PCR results of Foxg1 expression at different time points during the termination phase also revealed an inverse correlation between the expression pattern of Abtb1 and Foxg1 in vivo (Figs. 4F and 5F). Importantly, higher expression levels of FOXG1 and lower levels of P21 in miR‐125b‐5p‐overexpressing mice at day 5 after 2/3 PH (Fig. 5G‐I) further highlighted the relevance of miR‐125b‐5p during the termination phase. Taken together, these results suggested that Abtb1 contributes to the pro‐proliferative effect of miR‐125b‐5p on mouse hepatocytes, at least in part through the regulation of the FOXG1/p21 pathway (Fig. 5J).
FIG. 5.
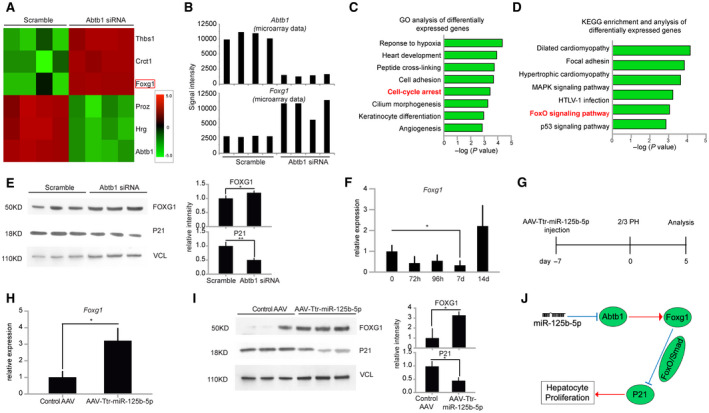
ABTB1 regulates hepatocytes proliferation by FOXG1/p21 pathway in vitro and in vivo. (A) The heat map shows top differentially up‐ or down‐regulated genes obtained from microarray data in siAbtb1‐transfected primary mouse hepatocytes. (B) Microarray‐based mRNA expression analysis shows knockdown of Abtb1 expression and up‐regulation of Foxg1 expression. (C) Gene ontology analysis of differentially expressed genes identified in siAbtb1‐transfected hepatocytes and their P values obtained using DAVID 6.8. (D) KEGG pathway enrichment and statistical analysis of differentially expressed genes identified in siAbtb1‐transfected hepatocytes. (E) Western blot analysis of FOXG1 and P21 in siAbtb1‐transfected hepatocytes and their quantification. Vinculin was used as a loading control. (F) qRT‐PCR analysis of Foxg1 expression at the indicated time points during the termination phase. (G) Schematic representation of the experimental design. (H) qRT‐PCR analysis of Foxg1 expression at day 5 after 2/3 PH in mice livers injected with AAV8‐Ttr‐miR‐125b‐5p. (I) Western blot analysis of FOXG1 and P21 at day 5 after 2/3 PH in mice livers injected with AAV8‐Ttr‐miR‐125b‐5p and their quantification. Vinculin was used as a loading control. (J) Schematic of miR‐125b‐5p‐mediated effect on hepatocyte proliferation through the ABTB1/FOXG1/p21 pathway. *P < 0.05, **P < 0.01; data are presented as mean ± SEM. Abbreviations: Crct1, cysteine rich C‐terminal 1; d, days; h, hours; Hrg, histidine rich glycoprotein; HTLV, human T‐lymphotropic virus; KEGG, Kyoto Encyclopedia of Genes and Genomes; MAPK, mitogen‐activated protein kinase; Proz, pritein Z; Thbs1, thrombospondin 1.
miR‐125b‐5p Also Regulates ABTB1 at the Posttranscriptional Level in Primary Human Hepatocytes
To examine whether miR‐125b‐5p also regulates human ABTB1, we transfected primary human hepatocytes with miR‐125b‐5p mimic and inhibitor. The results indicate that transfection of cells with miR‐125b‐5p mimic led to decreased levels of human ABTB1, both at mRNA (Fig. 6A) and protein levels (Fig. 6B). Likewise, cells transfected with miR‐125b‐5p inhibitor possess higher levels of human ABTB1 mRNA (Fig. 6A) and ABTB1 protein (Fig. 6B). Of note, the change in ABTB1 expression in the presence of miR‐125b‐5p or miR‐125b‐5p inhibitor treatment is modest but significant. Furthermore, luciferase reporter assay demonstrated that miR‐125b‐5p binds to the 3′ UTR of ABTB1 (Fig. 6C, S3). WST‐1 assay (Fig. 6D, Supporting Fig. S3) and Ki67 immunofluorescence staining (Fig. 6E) revealed enhanced proliferation of human hepatocytes following transfection with miR‐125b‐5p mimic and reduced proliferation following transfection with miR‐125b‐5p inhibitor.
FIG. 6.
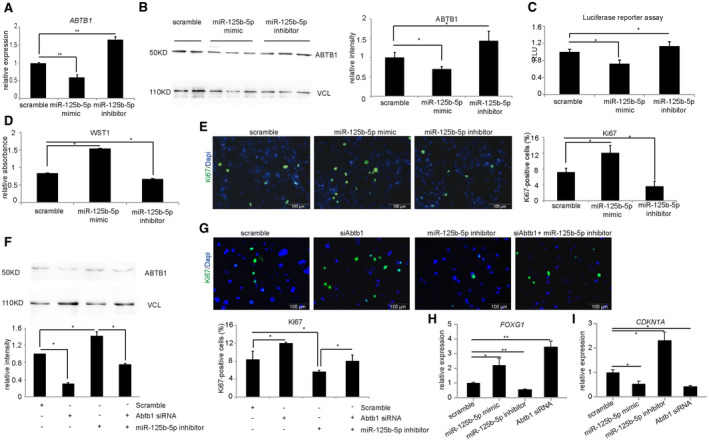
miR‐125b‐5p enhances proliferation of primary human hepatocytes. (A) qRT‐PCR of ABTB1 in human hepatocytes transfected with miR‐125b‐5p mimic or inhibitor. (B) Western blot of ABTB1 and its quantification in primary human hepatocytes transfected with miR‐125b‐5p mimic, inhibitor, or scramble control. (C) Luciferase reporter assay of human 3′ UTR ABTB1 in the presence of either miR‐125b‐5p mimic or inhibitor. (D) WST‐1 assay of primary human hepatocytes transfected with miR‐125b‐5p mimic, inhibitor, or scramble control. (E) Ki67 immunofluorescence staining of primary human hepatocytes transfected with miR‐125b‐5p mimic, inhibitor, or scramble control and its quantification. (F) Western blot for human ABTB1 and its quantification on lysates of primary human hepatocytes transfected with either siRNA targeting human ABTB1 or miR‐125b‐5p inhibitor. (G) Ki67 immunofluorescence staining of primary human hepatocytes transfected with siRNA alone or siRNA and miR‐125b‐5p inhibitor and its quantification. Scale bars, 200 μm. (H,I) qRT‐PCR analysis of (H) FOXG1 and (I) CDKN1A expression in primary human hepatocytes transfected with miR‐125b‐5p mimic, inhibitor, or ABTB1 siRNA. *P < 0.05, **P < 0.01; data are presented as mean ± SEM (n = 4 per group). Abbreviation: CDKN1A, cyclin‐dependent kinase inhibitor 1A.
To address the question whether ABTB1 contributes to pro‐proliferative effects of miR‐125b‐5p, we transfected primary human hepatocytes with siRNA against human ABTB1 either alone or in combination with miR‐125b‐5p inhibitor. Western blot analysis confirmed reduced ABTB1 protein level following transfection with human ABTB1 siRNA alone and restored levels when cells were cotransfected with miR‐125b‐5p inhibitor and ABTB1 siRNA (Fig. 6F). Similar to the results observed in primary mouse hepatocytes, Ki67 staining showed that proliferation of human hepatocytes was either increased when transfected with ABTB1 siRNA or decreased when transfected with miR‐125b‐5p inhibitor. Importantly, the cotransfection of primary human hepatocytes with ABTB1 siRNA and miR‐125b‐5p inhibitor diminished the enhanced proliferation effect observed following ABTB1 siRNA alone (Fig. 6G). Likewise, the expression of FOXG1 and cyclin‐dependent kinase inhibitor 1A (CDKN1A) was also modulated following transfection of either miR‐125b‐5p or Abtb1 siRNA (Fig. 6H,I). Thus, ABTB1 contributes to the pro‐proliferative effect of miR‐125b‐5p through the FOXG1/p21 pathway in primary human hepatocytes.
Discussion
The robust termination at the end of the well‐orchestrated liver regeneration process ensures the re‐acquisition of the quiescence stage and restoration of liver mass while avoiding uncontrolled proliferation of hepatic cells. To identify potential posttranscriptional regulators of termination of liver regeneration, we performed miRNA profiling and identified miR‐125b‐5p as a regulator of the termination phase of liver regeneration. The expression of miR‐125b‐5p is decreased during the termination phase of normal liver regeneration. To investigate the functional relevance of this phenomenon, we overexpressed miR‐125b‐5p in hepatocytes using AAV. The overexpression of miR‐125b‐5p led to extension of the cell cycle during the termination phase and sustained restoration of liver mass at the end of liver regeneration.
Notably, our study is the first report that investigates the in vivo function of miR‐125b‐5p, a differentially regulated miRNA during the termination phase of liver regeneration. Two previous studies have reported up‐regulation of miR‐34a( 20 ) and down‐regulation of miR‐23b( 21 ) during this phase of liver regeneration; however, none of those studies investigated miRNA function in vivo. Our miRNA profiling analyses confirmed the deregulation of miR‐34a and miR‐23b during termination. The low basal level expression for miR‐34a in human liver( 13 ) and less than 2‐fold down‐regulation of miR‐23b led to the exclusion of these miRNAs from our functional analyses.
Mechanistically, we identified Abtb1 as a direct target of miR‐125b‐5p. Down‐regulation of Abtb1 by miR‐125b‐5p conferred the pro‐proliferation effects we observed in mice as well as in primary human hepatocytes. ABTB1, also called BPOZ, is a protein with an ankyrin repeat and BTB/POZ domain that is thought to be involved in protein–protein interactions.( 11 ) Using complementary DNA microarray analyses, Unoki and Nakamura( 11 ) identified ABTB1 as a mediator of phosphatase and tensin homolog–protein kinase B signaling. They also demonstrated exogenous expression of ABTB1 in colon cancer cell lines (SW480 and LoVo) suppressed tumor cell growth while inhibition of ABTB1 expression using antisense oligonucleotides resulted in growth enhancement in the SW480 cell line. In addition, it is reported that miR‐125b overexpression promotes myeloid cell proliferation by targeting ABTB1.( 18 ) Our results further identified the FOXG1/p21 pathway as a downstream mediator of ABTB1 in proliferating hepatocytes. FOXG1 has been shown to act as an oncoprotein that blocks TGFβ‐induced p21 expression in ovarian cancer cells.( 22 ) Additionally, FOXG1 was reported to inhibit the phosphatase activity of protein phosphatase 2A (PP2A) in neurons.( 23 ) Studies have shown that TGFβ signaling and PP2A‐regulated pathways control hepatocyte proliferation and its termination dynamically after PH.( 24 , 25 ) Of note, a previous study reported that p21−/− mice have similar amounts of proliferating cells in liver at 38 hours after PH compared to wild‐type mice and possess a higher liver to body weight ratio at 1 week after PH.( 26 ) Recently, hepatocyte nuclear factor 4 alpha, which activates p21 expression,( 27 ) was shown to resume function during the termination phase after PH.( 28 ) Similar to those findings, we provide evidence that miR‐125b‐5p regulates the ABTB1/FOXG11/p21 pathway and thus contributes to termination of the endogenous regenerative capacity at the end of liver regeneration.
It is noteworthy that hepatocyte proliferation returned to the quiescent state at the end of liver regeneration in the presence of miR‐125b‐5p overexpression, suggesting a comprehensive regulatory network in the termination phase. Thus, the regulatory function of miR‐125b‐5p during termination of liver regeneration most likely includes other targets along with Abtb1. For example, miR‐125b‐5p has been shown to regulate the expression of multiple Smads and receptor subunits by binding to their 3′ UTRs and thereby controlling TGFβ signaling,( 29 ) a key pathway that also regulates termination of liver regeneration.( 30 ) In concert with our findings, mice deficient in TGFβ in hepatocytes displayed accelerated liver regeneration.
Interestingly, according to current studies on hepatocellular carcinoma (HCC), miR‐125b‐5p is found to be down‐regulated in primary HCC and to exert an antiproliferative effect by directly targeting the oncogenes LIN28( 31 ) or sirtuin 7.( 32 ) These reports are contradictory to our finding that miR‐125b‐5p plays a role in promoting hepatocyte proliferation. The possible explanation for the dual role of miR‐125b‐5p could be its cellular context and microenvironment. More importantly, the multiple targets of a miRNA determine its functional diversity and sometimes can result in the observed opposing effects.
Taken together, we provide evidence that miR‐125b‐5p regulates hepatocyte proliferation during the termination phase of liver regeneration. Overexpression of miR‐125b‐5p in vivo increases the regenerative capacity of hepatocytes, suggesting its potential as a therapeutic strategy in liver diseases.
Supporting information
Fig S1
Fig S2
Fig S3
Supplementary Material
Acknowledgment
Open access funding enabled and organized by Projekt DEAL.
A.D.S. is supported by the Deutsche Forschungsgemeinschaft (DFG SH640/1‐2, SFB 738, and REBIRTH‐MWK), the Boehringer Ingelheim Foundation Plus 3 Program, and the China Scholarship Council (to T.Y.).
Potential conflict of interest: Nothing to report.
[Corrections added on September 23, 2020, after first online publication: the open access funding statement has been added.]
References
Author names in bold designate shared co‐first authorship.
- 1. Taki‐Eldin A, Zhou L, Xie HY, Zheng SS. Liver regeneration after liver transplantation. Eur Surg Res 2012;48:139‐153. [DOI] [PubMed] [Google Scholar]
- 2. Rychtrmoc D, Libra A, Buncek M, Garnol T, Cervinkova Z. Studying liver regeneration by means of molecular biology: how far we are in interpreting the findings? Acta Medica (Hradec Kralove) 2009;52:91‐99. [DOI] [PubMed] [Google Scholar]
- 3. Apte U, Gkretsi V, Bowen WC, Mars WM, Luo JH, Donthamsetty S, et al. Enhanced liver regeneration following changes induced by hepatocyte‐specific genetic ablation of integrin‐linked kinase. Hepatology 2009;50:844‐851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Carr BI, Hayashi I, Branum EL, Moses HL. Inhibition of DNA synthesis in rat hepatocytes by platelet‐derived type beta transforming growth factor. Cancer Res 1986;46:2330‐2334. [PubMed] [Google Scholar]
- 5. Jin J, Hong IH, Lewis K, Iakova P, Breaux M, Jiang Y, et al. Cooperation of C/EBP family proteins and chromatin remodeling proteins is essential for termination of liver regeneration. Hepatology 2015;61:315‐325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yuan Q, Loya K, Rani B, Mobus S, Balakrishnan A, Lamle J, et al. MicroRNA‐221 overexpression accelerates hepatocyte proliferation during liver regeneration. Hepatology 2013;57:299‐310. [DOI] [PubMed] [Google Scholar]
- 7. Song G, Sharma AD, Roll GR, Ng R, Lee AY, Blelloch RH, et al. MicroRNAs control hepatocyte proliferation during liver regeneration. Hepatology 2010;51:1735‐1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yang D, Yuan Q, Balakrishnan A, Bantel H, Klusmann JH, Manns MP, et al. MicroRNA‐125b‐5p mimic inhibits acute liver failure. Nat Commun 2016;7:11916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ng R, Song G, Roll GR, Frandsen NM, Willenbring H. A microRNA‐21 surge facilitates rapid cyclin D1 translation and cell cycle progression in mouse liver regeneration. J Clin Invest 2012;122:1097‐1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Szabo G, Bala S. MicroRNAs in liver disease. Nat Rev Gastroenterol Hepatol 2013;10:542‐552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Unoki M, Nakamura Y. Growth‐suppressive effects of BPOZ and EGR2, two genes involved in the PTEN signaling pathway. Oncogene 2001;20:4457‐4465. [DOI] [PubMed] [Google Scholar]
- 12. Kleine M, Riemer M, Krech T, DeTemple D, Jager MD, Lehner F, et al. Explanted diseased livers ‐ a possible source of metabolic competent primary human hepatocytes. PLoS One 2014;9:e101386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ward J, Kanchagar C, Veksler‐Lublinsky I, Lee RC, McGill MR, Jaeschke H, et al. Circulating microRNA profiles in human patients with acetaminophen hepatotoxicity or ischemic hepatitis. Proc Natl Acad Sci U S A 2014;111:12169‐12174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shaham L, Binder V, Gefen N, Borkhardt A, Izraeli S. MiR‐125 in normal and malignant hematopoiesis. Leukemia 2012;26:2011‐2018. [DOI] [PubMed] [Google Scholar]
- 15. Soldatow V, Peffer RC, Trask OJ, Cowie DE, Andersen ME, LeCluyse E, et al. Development of an in vitro high content imaging assay for quantitative assessment of CAR‐dependent mouse, rat, and human primary hepatocyte proliferation. Toxicol In Vitro 2016;36:224‐237. [DOI] [PubMed] [Google Scholar]
- 16. Wen Y, Feng D, Wu H, Liu W, Li H, Wang F, et al. Defective initiation of liver regeneration in osteopontin‐deficient mice after partial hepatectomy due to insufficient activation of IL‐6/Stat3 pathway. Int J Biol Sci 2015;11:1236‐1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 2010;466:835‐840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bousquet M, Nguyen D, Chen C, Shields L, Lodish HF. MicroRNA‐125b transforms myeloid cell lines by repressing multiple mRNA. Haematologica 2012;97:1713‐1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Seoane J, Le HV, Shen L, Anderson SA, Massague J. Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell 2004;117:211‐223. [DOI] [PubMed] [Google Scholar]
- 20. Chen H, Sun Y, Dong R, Yang S, Pan C, Xiang D, et al. Mir‐34a is upregulated during liver regeneration in rats and is associated with the suppression of hepatocyte proliferation. PLoS One 2011;6:e20238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yuan B, Dong R, Shi D, Zhou Y, Zhao Y, Miao M, et al. Down‐regulation of miR‐23b may contribute to activation of the TGF‐beta1/Smad3 signalling pathway during the termination stage of liver regeneration. FEBS Lett 2011;585:927‐934. [DOI] [PubMed] [Google Scholar]
- 22. Chan DW, Liu VW, To RM, Chiu PM, Lee WY, Yao KM, et al. Overexpression of FOXG1 contributes to TGF‐beta resistance through inhibition of p21WAF1/CIP1 expression in ovarian cancer. Br J Cancer 2009;101:1433‐1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chiola S, Do MD, Centrone L, Mallamaci A. Foxg1 overexpression in neocortical pyramids stimulates dendrite elongation via Hes1 and pCreb1 upregulation. Cereb Cortex 2019;29:1006‐1019. [DOI] [PubMed] [Google Scholar]
- 24. Hu J, Srivastava K, Wieland M, Runge A, Mogler C, Besemfelder E, et al. Endothelial cell‐derived angiopoietin‐2 controls liver regeneration as a spatiotemporal rheostat. Science 2014;343:416‐419. [DOI] [PubMed] [Google Scholar]
- 25. Lai SS, Zhao DD, Cao P, Lu K, Luo OY, Chen WB, et al. PP2Acalpha positively regulates the termination of liver regeneration in mice through the AKT/GSK3beta/Cyclin D1 pathway. J Hepatol 2016;64:352‐360. [DOI] [PubMed] [Google Scholar]
- 26. Buitrago‐Molina LE, Marhenke S, Longerich T, Sharma AD, Boukouris AE, Geffers R, et al. The degree of liver injury determines the role of p21 in liver regeneration and hepatocarcinogenesis in mice. Hepatology 2013;58:1143‐1152. [DOI] [PubMed] [Google Scholar]
- 27. Hwang‐Verslues WW, Sladek FM. Nuclear receptor hepatocyte nuclear factor 4alpha1 competes with oncoprotein c‐Myc for control of the p21/WAF1 promoter. Mol Endocrinol 2008;22:78‐90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huck I, Gunewardena S, Espanol‐Suner R, Willenbring H, Apte U. Hepatocyte nuclear factor 4 alpha activation is essential for termination of liver regeneration in mice. Hepatology 2019;70:666‐681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Emmrich S, Rasche M, Schoning J, Reimer C, Keihani S, Maroz A, et al. miR‐99a/100~125b tricistrons regulate hematopoietic stem and progenitor cell homeostasis by shifting the balance between TGFbeta and Wnt signaling. Genes Dev 2014;28:858‐874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bohm F, Kohler UA, Speicher T, Werner S. Regulation of liver regeneration by growth factors and cytokines. EMBO Mol Med 2010;2:294‐305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liang L, Wong CM, Ying Q, Fan DN, Huang S, Ding J, et al. MicroRNA‐125b suppressesed human liver cancer cell proliferation and metastasis by directly targeting oncogene LIN28B2. Hepatology 2010;52:1731‐1740. [DOI] [PubMed] [Google Scholar]
- 32. Kim JK, Noh JH, Jung KH, Eun JW, Bae HJ, Kim MG, et al. Sirtuin7 oncogenic potential in human hepatocellular carcinoma and its regulation by the tumor suppressors MiR‐125a‐5p and MiR‐125b. Hepatology 2013;57:1055‐1067. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Fig S2
Fig S3
Supplementary Material


