Abstract
An abundance of protein structures has been solved in the last six decades that are paramount in defining the function of such proteins. For unsolved protein structures, however, predictions based on sequence and phylogenetic similarity can be useful for identifying key domains of interaction. Here, we describe expression and purification of a recombinant plant LRR-RLK ectodomain MIK1 using a modified baculovirus-mediated expression system with subsequent N-linked glycosylation analysis using LC-MS/MS and computational sequence-based analyses. Though highly ubiquitous, glycosylation site specificity and the degree of glycosylation influenced by genetic and exogenous factors are still largely unknown. Our experimental analysis of N-glycans on MIK1 identified clusters of glycosylation that may explicate the regions involved in MIK1 ectodomain binding. Whether these glycans are necessary for function is yet to be determined. Phylogenetic comparison using multiple sequence alignment between MIK1 and other LRR-RLKs, namely TDR in Arabidopsis thaliana, revealed conserved structural motifs that are known to play functional roles in ligand and receptor binding.
Keywords: LRR-RLK, post-translational modifications, N-glycosylation, LC-MS, phylogenetic analysis
Introduction
Post-translational modifications (PTMs) are complex processes that modulate protein activity and diversify protein function. PTMs can alter the biophysical characteristics of proteins by modifying their thermodynamic and structural properties, which play key roles in regulating cell signaling, enzyme activation, protein folding, protein trafficking, and protein stability[1].
A majority of the functional proteome is comprised of post-translationally modified proteins[2]. In both prokaryotes and eukaryotes, protein phosphorylation is a reversible post-translational modification that plays a regulatory role in signal transduction pathways[3]. The phosphorylation mechanism is generally catalyzed by a group of enzymes known as kinases. In plants, the kinome is much larger than that of animals; for example, the human genome contains approximately 500 kinases[4, 5], whereas the model plant Arabidopsis thaliana genome contains approximately 1000 serine-threonine kinases[6]. Plant kinases can be categorized into two groups: membrane-associated receptor-like kinases (RLKs) and receptor-like cytoplasmic kinases. These kinases are often associated with plant growth, development, and immune responses[7]. The leucine-rich repeats receptor-like kinase (LRR-RLK) family of membrane integral RKs, which contains more than 200 members in Arabidopsis, is considered to be the largest family of plant receptor kinases[8].
The design of LRR-RLKs includes an extracellular LRR domain, here referred to as ectodomain, a single-helix transmembrane domain (TM), and a cytoplasmic region kinase domain (KD) with serine-threonine specificity[8]. The ectodomain typically functions as the ligand-binding domain and is involved in a diverse range of exogenous interactions[9, 10]. The structural features of LRR domains are generally similar, usually containing 3–30 tandem LRRs, that form a super-helical structure. However, despite the structural similarity, LRR ectodomains can interact with various structurally diverse, ligands including steroid hormones[11], cysteine-rich peptides (CRPs)[12, 13], and post-translationally modified peptide ligands[10]. In Arabidopsis thaliana, based on the KD structures, LRR-RLKs have been classified into 23 subfamilies[14–16]. The biological functions of LRR-RLKs can be broadly subdivided into two major categories: 1) plant growth and development, and 2) plant defense and host-pathogen interactions.
In this paper, we focus on the expression, purification, and subsequent computational phylogenetic studies of MDIS1-INTERACTING RECEPTOR-LIKE KINASE1 (MIK1), a newly discovered A. thaliana LRR-RLK protein[17]. MIK1 regulates the plant reproduction process by facilitating ovule targeting and fertilization. Previous studies consider MIK1 to be an essential component of a signaling cascade in pollen tube formation that involves a chemoattractant CRP known as LURE[17]. LURE peptide is a chemoattractant that was first discovered in Torenia Fourneri[18] and has recently been found in A. thaliana[19]. In A. thaliana, the LURE family consists of six genes (AtLURE1.1-AtLURE1.6) encoding AtLURE1 peptides[20]. These peptides play a major role in proper pollen tube formation and development, pollen tube guidance, and the establishment of ovule-targeting signaling networks[17, 19]. In A. thaliana, genetic, biochemical, and mutational studies have identified MALE DISCOVERER1 (MDIS1), MDIS2, MIK1, and MIK2 as potential receptors for LURE[17]. Although MIK1 has major implications in pollen tube development and growth, our knowledge about its activation, how it interacts with peptide hormones, and how it translates the extracellular cues to downstream signaling cascade is limited. To understand the structural bases of MIK1 activation and how the LRR-RLK contributes to the overall process of plant reproduction, we recombinantly expressed and purified MIK1 ectodomain and set up crystallization trials. Unfortunately, the crystallization trials were unsuccessful, and we hypothesized that the presence of multiple N-linked glycans on the MIK1 ectodomain could be one of the reasons for not obtaining crystals.
Glycosylation is a common and essential PTM that occurs during or after protein synthesis[21, 22]. Glycosylation modifications covalently attach carbohydrate molecules to proteins in two major forms: N-linked glycosylation on the nitrogen of asparagine residues[23] and O-linked glycosylation on the hydroxyl oxygen of serine or threonine residues[24]. The consensus sequence where N-glycosylation linkages may be established is Asn-X-Ser/Thr (NXS/T motifs), where X can be any amino acid except proline[23–25].
Several NXS/T motifs have been found on the extracellular domains of LRR-RLKs in A. thaliana, including FLS2, EFR, TDR, PSKR1, BAK1, PRK3, PRK6, ERL1, and MIK1. In our study, the molecular weight (MW) of MIK1 ectodomain was approximately 75kDa, which is about 7kDa larger than its predicted MW of 68kDa. Similar discrepancies in MW between the expressed recombinant ectodomain of LRR proteins versus their predicted MW have been found in TDR, PRK3, PSKR1, and others[20]. This increase in MW suggests that these proteins were effectively glycosylated when expressed in insect cell-mediated expression system. Crystal structures of multiple LRR-RLK ectodomains (for example ERL1[12], TDR[10], PSKR1[26], and PRK3[27]) ectodomains have been resolved with N-linked glycans associated with it. Although glycosylation is inherently crucial for protein stability and folding, crystallization of glycoprotein is extremely difficult as the large chemical modification introduces heterogeneity, conformational flexibility, and increases the surface entropy.
Here, we used a modified baculovirus-mediated expression system to express and purify the ectodomain of recombinant MIK1. We also present a detailed identification of the N-glycosylation sites on the ectodomain of recombinant MIK1 coupled with computational analysis to address the questions regarding origin, phylogeny, and structural and functional similarities with other LRR-RLK ectodomains.
Materials and Methods
1.1. Modification of pFastBac1 vector
As previously reported, a pFastBac1 expression vector was modified with the addition of insect Hemolin secretion signal sequence[28]. The correct construct plasmid of the modified pFastBac1 vector was confirmed by DNA sequencing.
1.2. Cloning MIK1 into modified pFastBac1 vector
A. thaliana MIK1 gene fragments encoding residues 24–633 and modified with a C-terminal histidine tag were amplified by a conventional PCR method (95°C, 55°C, 72°C for 30 seconds each repeated for 30 cycles). Both the amplified MIK1 fragments and the modified pFastBac1 vector were digested with NotI and BamHI restriction endonucleases. The gene fragments were then ligated with the linearized vector and transformed into DH5α competent cells. The transformation mixture was plated onto corresponding antibiotic selection LB agar plates and incubated at 37°C overnight or until colonies formed. Colonies were confirmed for transformation efficacy by PCR and then by DNA sequencing.
1.3. Transfection and recombinant virus generation
The verified construct plasmid containing the MIK1 gene was transformed into DH10Bac competent cells following the Bac-to-Bac Baculovirus Expression System protocol (Invitrogen life technologies). The transformation plates were incubated at 37°C for 48 hours. On the transformed plates, two different colored colonies appeared: white colonies containing the MIK1 gene, and blue colonies that did contain the MIK1 gene. The bacmids/correct plasmids were isolated per the Bac-to-Bac protocol and verified by sequencing. The correct bacmids were used to transfect Sf9 monolayer cells through multiple passages as previously reported for amplification of the recombinant baculovirus.
1.4. Expression and purification strategy
High Five insect cells have been demonstrated to express a higher yield of recombinant protein than other cell lines, such as Sf9 cells, and they were used for the expression of MIK1 ectodomain. High Five cells were cultured and incubated with the freshly produced final passage of recombinant baculovirus. The cells in suspension were then transferred to a large culture for expression and secretion of recombinant MIK1. After 72 hours, the cultured media was centrifuged and the supernatant containing secreted MIK1 protein was collected for purification.
20ml Nickel-NTA resin was added to the supernatant to capture the his-tagged MIK1 protein. The recombinant MIK1 was captured and eluted following the standard NiNTA protocol. The purified protein was concentrated up to 20mg/ml and was subjected to gel-filtration chromatography to yield a homogeneous sample. 20mM Tris, pH 8, 200mM NaCl was used as our gel-filtration buffer.
1.5. Mass spectrometry analysis of N-glycosylation
For determining sites of N-linked glycosylation, samples digested with trypsin were subjected to LC-MS/MS analysis using an Orbitrap Elite mass spectrometry system coupled to an Easy nanoLC UPLC system for nanoscale separation (ThermoFisher). Proteolytic peptides were separated on a 75 μm i.d. x 15 cm capillary column packed in-house with Magic C18 stationary phase beads, 3 μm particle size (Michrom Bioresources), at a flow rate of 300 nL/min using a linear gradient from 2%A to 40%B (A=0.1% formic acid in water; B=0.1% formic acid in acetonitrile) over 60 min. The data were acquired using a Top-5 data-dependent acquisition (DDA) method with higher-collision energy dissociation (HCD) using a normalized collision energy of 27V. Following data collection, raw MS data files were processed with Proteome Discoverer (ThermoFisher), and processed files were searched against the TAIR10 database using Mascot (Matrix Science), allowing for identification of HexNAc modification on Asn residues for Endo F treated samples. Search tolerances of 10ppm for both precursor and product ions were used. For visualization of match assignments, search results were imported into Scaffold (Proteome Software).
1.6. Computational analysis
Multiple Sequence Alignment (MSA) was done using ClustalX2.1 and SeaView version 4 software. The resulting MSA was examined and adjusted manually by removing redundant sequences and sequences that did not align properly. To obtain a better alignment, columns and residues were also deleted as needed.
1.7. Amino acid conservation and protein structure analysis
Amino acid conservation between different clades was done by WebLogo 3[29]. WebLogo 3 was used for visualization and analysis of the amino acid residue conservation patterns in the MSA.
Results:
1.1. MIK1 expression and purification
Recombinant protein production is a convenient way to obtain large quantities of protein for biophysical and biochemical characterization. Our modified baculovirus-mediated insect cell protein expression system was used to obtain large quantities of recombinant MIK1 ectodomain protein. To induce high expression of secretory target protein, we introduced a Hemolin signal sequence preceding the target gene. We observed a significant increase in expression yield when using the Hemolin signal sequence when compared with the intrinsic signal peptide of the target gene.
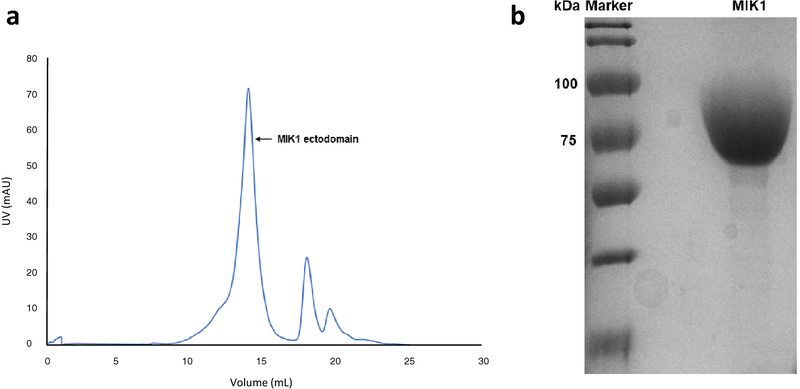
We expressed the extracellular domain of MIK1 from A. thaliana using our modified baculovirus-mediated insect cell expression. The modified pFastBac1 vector contained the MIK1 ectodomain gene, the Hemolin secretion signal sequence, and a C-terminal 6-histidine tag. The secreted protein was first purified by Ni-His affinity chromatography and then further purified by gel-filtration chromatography (Fig. 1a). The highly pure MIK1 protein was concentrated to 12 mg/ml using a 50kDa cut-off (Fig. 1b). The protein was subjected to crystallization trials but unfortunately did not yield any viable crystals. The recombinant MIK1 ectodomain has a predicted molecular weight of 68kDa. However, on SDS-PAGE, the apparent molecular weight of the purified recombinant protein is approximately 75kDa (Fig. 1b).
Fig. 1. Purification of MIK1 ectodomain.
(a) The recombinant ectodomain of MIK1 was expressed in baculovirus mediated insect cell expression system and purified using nickel-affinity chromatography followed by size exclusion chromatography. Fractions identified from the chromatogram to contain MIK1 were collected and concentrated to 12mg/ml. (b) Coomassie stained SDS-PAGE showing the concentrated MIK1 with high purity at 75 kDa
1.2. Analysis of N-glycosylation on recombinant MIK1
We had successfully crystallized and determined the structures of two plant glycoproteins, TDR and PRK3 ectodomains, but crystallizing MIK1 ectodomain has been very challenging. To promote crystallization, endoglycosidases such as Endo H and PNGase F were used to digest glycans from MIK1 ectodomain, but the trials did not yield any crystals. Therefore, we aim to identify the possible glycosylation sites by MS and then mutate the consensus N-glycosylation sites like NXS/T or mutate serine or threonine residues, which are responsible for O-glycosylation. We will screen a series of N-glycosylation mutants of the MIK1 ectodomain that will confer stability, proper folding, functionality, and encourage the formation of crystals.
Mass Spectrometry is a popular high-throughput technique to identify PTM sites and novel modifications due to its qualitative and quantitative capabilities. MS is widely being used as a high-throughput method to identify glycans and glycoprotein composition. In our study, MS was used to investigate the N-glycosylation sites on recombinant MIK1 ectodomain. Our study used trypsin and Endo H to digest known regions of the glycopeptide and the glycan tree itself, respectively, and found 8 N-glycosylated sites on the MIK1 ectodomain at N61, N82, N101, N271, N341, N381, N389, N417, and N535 (Table 1). We also used two different N-glycosylation prediction servers, NetNGlyc1.0 and GlyProt, and the UniProtKB database to compare our experimental results. All three databases predicted the same sixteen N-glycosylation sites based on the NXS/T motifs in the extracellular domain sequence, which align with the eight sites we experimentally identified (Table 1). Eight of the database-predicted sites did not show up in our experimental data and may be due to the poor fragmentation of peptide ions or co-elution of peptides having the same glycosylation moieties. It is also possible that these sites are not glycosylated due to exogenous factors during protein processing. The consensus motif NXS/T is necessary but not indicative of N-glycosylation. It is also possible that the N-glycan placement is due to the manner in which the MIK1 ectodomain binds to LURE and interacts with MDIS1 on the cell surface.
Table 1: Comparison of experimental versus database-derived identification of N-glycosylation sites on MIK1 ectodomain.
Consensus sequences for N-glycosylation sequence motifs were collected using three databases: UniProt, GlyProt, and NetNGlyc to compare with N-glycans identified experimentally using MS.
| Consensus Sequence | UniProt | GlyProt | NetNGlyc | Experimental Data | Position |
|---|---|---|---|---|---|
| NWT | 61 | 61 | 61 | 61 | LRR1 |
| NLT | 82 | 82 | 82 | 82 | LRR1 |
| NIS | 101 | 101 | 101 | LRR2 | |
| NES | 137 | 137 | 137 | LRR3 | |
| NAS | 146 | 146 | 146 | LRR4 | |
| NLS | 151 | 151 | 151 | LRR4 | |
| NLT | 155 | 155 | 155 | LRR4 | |
| NLT | 199 | 199 | 199 | LRR6 | |
| NFT | 271 | 271 | 271 | 271 | LRR9 |
| NNT | 341 | 341 | 341 | 341 | LRR12 |
| NLT | 381 | 381 | 381 | 381 | LRR14 |
| NNT | 389 | 389 | 389 | 389 | LRR14 |
| NGS | 417 | 417 | 417 | 417 | LRR15 |
| NLT | 535 | 535 | 535 | 535 | LRR20 |
| NNS | 557 | 557 | 557 | LRR21 | |
| NVS | 578 | 578 | 578 | LRR22 |
1.3. Phylogenetic analyses of MIK1 proteins:
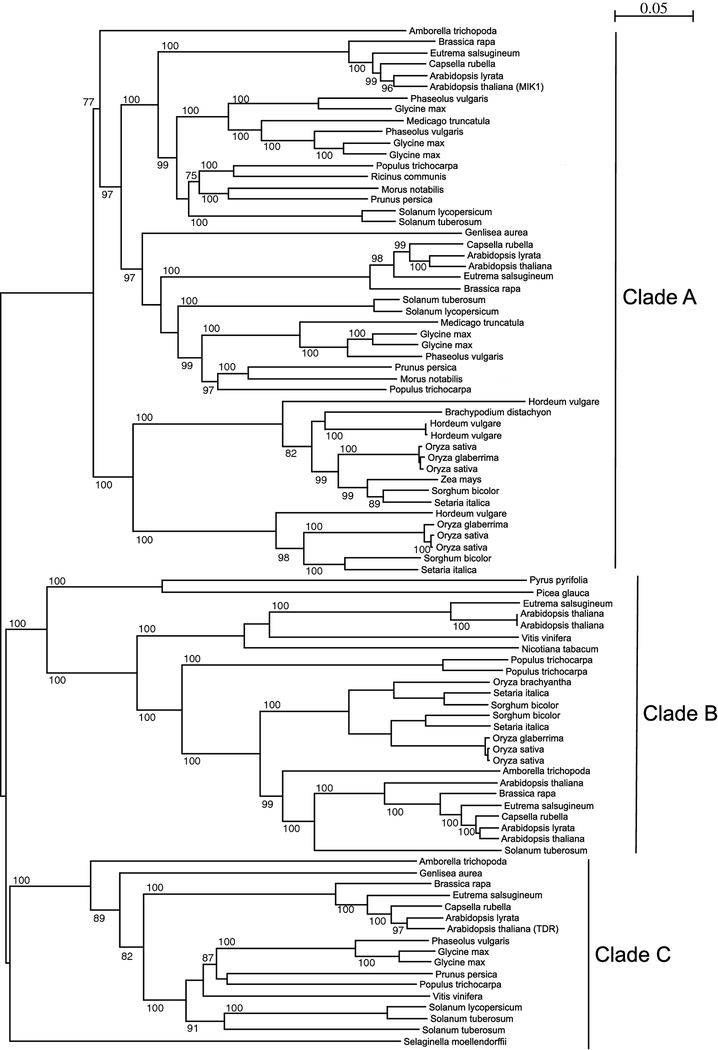
A multiple sequence alignment (MSA) on 51 protein sequences was performed to understand the phylogenetic relationship between MIK1 proteins from different plant species. These 51 sequences, originally from 31 different plants, were obtained from the database by performing BLAST. To comprehend the molecular evolution and functional phylogeny, these different plant species were also analyzed. These 31 plants can be grouped into three major clades: Poales, Asterids, and Rosids (Table 2). To decipher the phylogenetic relationship between MIK1 proteins obtained from these plants, we constructed a neighbor-joining (NJ) tree[30] (Fig. 3). The NJ tree, which is built on the basis of evolutionary distance data, branches out to generate three major clades: Clade A, Clade B, and Clade C. Also, based on the phylogenetic analysis, a clear evolutionary message comes across that differentiates MIK1 in two major groups: monocots and dicots (Table 2). As previously postulated by Shui and Bleecker (2001) and others, there is evidence in our phylogenetic tree of gene duplication and subsequent clade proliferation events that may have occurred before the separation of monocot and dicot.
Table 2: List of Clade A members.
Grouping Clade A members based on MSA of 51 plant protein sequences yielded three distinct clades. Groupings in Clade A distinguishes groups of Monocotyledons and Eudicots and clades of Poales, Rosids, and Asterids within each Clade A, Clade B, and Clade C.
| Species | Common Name | Group | Clade | UniProt |
|---|---|---|---|---|
| Setaria italica | Foxtail Millet | Monocotyledons | Poales | K3YZZ3 |
| Sorghum bicolor | Sorghum | Monocotyledons | Poales | C5XS73 |
| Oryza sativa (Japonica) | Rice | Monocotyledons | Poales | Q6Z8Y3 |
| Oryza sativa (Indica) | Rice | Monocotyledons | Poales | A2X010 |
| Oryza glaberrima | African Rice | Monocotyledons | Poales | I1NWH8 |
| Hordeum vulgare | House Barley | Monocotyledons | Poales | M0WT19 |
| Setaria italica | Foxtail Millet | Monocotyledons | Poales | K4A588 |
| Sorghum bicolor | Sorghum | Monocotyledons | Poales | C5WY01 |
| Zea mays | Maize | Monocotyledons | Poales | K7VHI1 |
| Oryza sativa (Japonica) | Rice | Monocotyledons | Poales | Q8H037 |
| Oryza glaberrima | African Rice | Monocotyledons | Poales | I1P7I1 |
| Oryza sativa (Indica) | Rice | Monocotyledons | Poales | A2XCF7 |
| Hordeum vulgare | House Barley | Monocotyledons | Poales | F2D364 |
| Hordeum vulgare | House Barley | Monocotyledons | Poales | M0UHW4 |
| Populus trichocarpa | Western Balsam Poplar | Eudicot | Rosids | B9I5Q8 |
| Prunus persica | Peach | Eudicot | Rosids | M5W7E1 |
| Phaseolus vulgaris | Kidney Beans | Eudicot | Rosids | V7BEC8 |
| Glycine max | Soybean | Eudicot | Rosids | I1NIS5 |
| Glycine max | Soybean | Eudicot | Rosids | I1LBP0 |
| Medicago truncatula | Barrel Medic | Eudicot | Rosids | G7I6U4 |
| Arabidopsis thaliana | Mouse-ear Cress | Eudicot | Rosids | PXL1/Q9FRS6 |
| Solanum tuberosum | Potato | Eudicot | Asterids | M0ZYB9 |
| Arabidopsis thaliana | Mouse-ear Cress | Eudicot | Rosids | MIK1/Q9M0G7 |
| Arabidopsis lyrata | Lyre-leaved Rock-cress | Eudicot | Rosids | D7MDA1 |
| Eutrema salsugineum | Saltwater Cress | Eudicot | Rosids | V4LUQ8 |
| Brassica rapa | Chinese Cabbage | Eudicot | Rosids | M4D3J2 |
| Prunus persica | Peach | Eudicot | Rosids | M5VVE7 |
| Ricinus communis | Castor Bean | Eudicot | Rosids | B9TAC6 |
| Populus trichocarpa | Western Balsam Poplar | Eudicot | Rosids | B9GRD1 |
| Phaseolus vulgaris | Kidney Bean | Eudicot | Rosids | V7D099 |
| Medicago truncatula | Barrel Medic | Eudicot | Rosids | G7KXH2 |
| Glycine max | Soybean | Eudicot | Rosids | I1L8H6 |
| Phaseolus vulgaris | Kidney Bean | Eudicot | Rosids | V7BEX0 |
Fig. 3. Phylogenetic tree depicting three clades based on ectodomain multiple sequence alignment (MSA).
A neighbor-joining (NJ) tree was generated in SeaView with MSA of 51 plant proteins ectodomains and displayed by species. Three distinct clades are differentiated, notably, with A. thaliana proteins MIK1 in Clade A and TDR in Clade C
In our BLAST search for Arabidopsis MIK1 sequence, we also obtained Arabidopsis TDR protein along with other closely related proteins. The two phylogenetic clades can be easily distinguished, as Clade A contains MIK1 protein, and the third clade, Clade C, contains TDR protein from A. thaliana (Fig. 3). It is evident from the tree that MIK1 and TDR may have a common ancestor, and they could have originated from the same gene and later acquired diverse functions. It can be inferred that MIK1 and TDR proteins are phylogenetically linked, and, interestingly, these two proteins share 43% sequence identity between each other.
1.4. Sequence-based structural analysis
By performing MSA on the various MIK1 sequences from different species, we were able to identify phylogenetic relationships and highly conserved sequences for our analysis. Overall, the MSA quality was high and later curated based on the ectodomain region of the receptor kinases. Thus, the resulting MSA does not contain the secretion signal sequence, transmembrane domain, or kinase domain. The amino acid residue cut-off for the MIK1 ectodomain was 24–633. Since TDR and MIK1 share significant amino acid sequence identity, the preliminary alignment analysis was conducted between the two ectodomain sequences. Contrary to MIK1, the crystal structure of TDR, in complex with its corresponding ligand TDIF and co-receptor ectodomain SERK2, has been solved. The interactions between TDR-SERK2 and TDR-TDIF have been described in detail. These studies acted as a base for our analysis of the MIK1 structure.
Based on phylogenetic analysis, the TDR and MIK1 ectodomains belong to two separate clades (Fig. 3). Two separate sequence WebLogos were generated for two clades (Clade Aectodomain and Clade Cectodomain), considering their ectodomain sequence alignment (Supplementary Fig. 1). We compared the WebLogos of Clade Aectodomain with Clade Cectodomain to determine how evolution has altered their overall sequence, structure, and function. A careful comparison between Clade Aectodomain and Clade Cectodomain has revealed multiple conserved regions containing the same or similar amino acids. Conserved sequence motifs found in both Clades included: RXR, DXS, where X can be any hydrophobic residue, GYN, LW/FN, and DL/FS regions. Notably, the RXR motif was found to be involved in ligand and co-receptor binding with TDR. Based on the TDR-TDIF-SERK2 complex structure, some of the residues present in these regions have significant structural implications for TDR ligand TDIF and co-receptor SERK2 binding. Moreover, both the clades contain two cysteine residues present in both the N and C terminal of the ectodomains and there are also similar arrangements in the TDR extracellular domain.
We further analyzed our MSA based on the TDR-TDIF complex structure. It is known that the TDR ectodomain concave surface residues interact with ligand TDIF and co-receptor SERK2. TDR residues R421, R423, W353, S305, F281, F279, Y234, S187, and Y188 are significant residues responsible for ligand binding. These residues are conserved within Clade Cectodomain. When we analyzed MIK1 in Clade Aectodomain, we found these residues are also relatively conserved or have been replaced by similar residues. In MIK1 ectodomain, equivalent residues such as R408, R410, W341, S292, Y221, S148, and F175 were presented (Supplementary Fig. 2). MIK1 residue W341 is equivalent to TDR residue W353, and it is conserved throughout Clade Aectodomain, but all species in Clade Cectodomain do not contain W in that position. Of note, soybean and kidney bean contain F in the equivalent position. S187 is fairly conserved throughout all species of Clade Cectodomain except for in potato, which is instead N187. Some species in Clade Aectodomain present G187 instead of S187, while others present N187 as well. In A. thaliana MIK1, S187 has been replaced by N187. PXL1, another LRR-RLK in A. thaliana and involved in vascular development, is also a member of Clade Aectodomain (Supplementary Fig. 3). PXL1 ectodomain presents R417, R419, W349, S301, Y230, S158, and Y184 residues, which are equivalent residues in TDR responsible for ligand binding.
TDR interaction with co-receptor SERK2, however, is mediated by R421, Y515, R516, S561, and K587 residues of the ectodomain. In Clade Cectodomain, R421 and Y515 are conserved; however, in some species, R516 has been substituted by a similar residue K516, S561 has been replaced by A561 in Western balsam poplar, and K587 is mostly variable, replaced by S587 or T587 throughout the clade. In Clade Aectodomain, R421 and Y515 are the only two residues that are conserved, whereas the rest of the residues are variable.
1.5. Conserved Surface Area
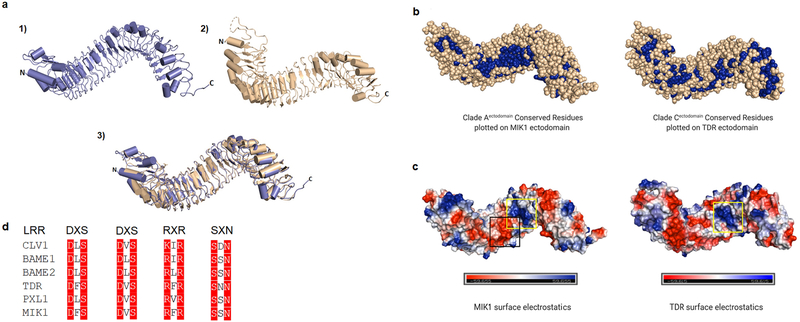
LRR ectodomain structures are overall conserved in a horseshoe shape, as seen in crystal structures of numerous proteins with LRR receptor domains such as FLS2, TDR, PSKR1, BRI1, and others. As discussed above, the conservation of residues, namely R421 and Y515, between Clade Aectodomain and Clade Cectodomain, and the role of these residues in protein-protein interaction, is a foundational phylogenetic connection between the two clades. The crystal structure of MIK1 ectodomain remains unavailable. We, therefore, used I-tasser, a protein modeling computational software, to predict the secondary structure (Fig. 4a). Without a solved structure, the use of computational tools to attain structural and functional insights from atomic models is highly informative in tandem with phylogenetic data. Given that the predicted MIK1 ectodomain structure is similar to that of TDR, we mapped conserved residues from Clade Aectodomain MSA analysis alongside the MIK1 predicted ectodomain structure and the conserved residues from Clade Cectodomain MSA analysis alongside the TDR ectodomain structure for comparison (Fig. 4b).
Fig. 4. Structural comparison between A. thaliana LRR-RLK MIK1 and TDR.
(a) LRR ectodomain structures for 1) MIK1 modeled using I-tasser and 2) TDR from the crystal structure illustrate a super-helical composition juxtaposed by 3) superposition of the two structures. MIK1 and TDR share approximately 43% sequence identity. (b) Conserved residues from Clade A members were plotted on the MIK1 ectodomain, and conserved residues from Clade C members were plotted on TDR ectodomain to analyze the conserved surface areas. Conserved residues are colored in blue spheres. (c) MIK1 and TDR ectodomains are shown with surface electrostatic potential. The positively charged interface is shown in blue, negative charge is shown in red, and neutral surface is shown in white. A positively charged surface patch is seen in both MIK1 and TDR concave region (yellow box). The positively charged surface area on TDR is responsible for TDIF peptide interaction. In MIK1, there is an additional region of negatively charged surface patch adjacent to the positively charged surface area which is absent in TDR. (black box). (d) Selected LRR-RLK sequences show widely conserved motifs, notably repeats of DXS, RXR and SXN where X can be any amino acid, that provide structural reference for MIK1 based on sequence similarity.
We found most of the conserved residues in MIK1 in the concave region of the ectodomain structure. A concentrated patch of conserved residues is present on the MIK1 concave surface, whereas the convex surface and C-terminal area are mostly variable (Fig. 4b). In the case of TDR, however, the C-terminal region is primarily conserved, and there were fewer parts of the concave region with conserved residues. We identified a conserved patch on the TDR surface that coincides with the TDIF binding region (Fig. 4b). To connect the binding domains of TDR to MIK1, we also analyzed the surface electrostatics of their ectodomain structures. The ligand-binding region of the TDR ectodomain varies from the neutral zone to the positively charged sections. In contrast, the MIK1 concave region has two distinct patches of a positive and negative charged area adjacent to each other (Fig. 4c).
Discussion
Here, we report a detailed protocol to recombinantly express and purify the ectodomain of a critical plant LRR-RLK, MIK1. We also identified some of the N-linked glycosylation sites on the MIK1 ectodomain. These verified glycosylation sites match the glycosylation sites predicted by various servers. There may be more glycosylation sites present on the MIK1 ectodomain, which remain undetected in our experimental setup. Moreover, our study was also not designed to detect any O-linked glycosylation sites. We suggest that these N-linked glycans are essential for the folding of the MIK1 ectodomain[31] and may act as a protective shield from protease actions[31–33]. It is also possible that these glycans play a crucial role in protein activity in the LRR-RLK protein family. Further studies on these glycosylation sites can shed light on their function and how they can impact the stability and folding of these proteins.
In addition, our computational analysis of the amino acid sequences showed that MIK1 and TDR share significant sequence similarity. Based on the ectodomain crystal structures and sequence conservation, we can say that the Clade Aectodomain and Clade Cectodomain are similar in structure and functions. We also identified conserved structural motifs on these two clades, RXR, DXS, SXN, to name a few. RXR motif is predominantly seen on the CLE binding receptors like TDR, BAM1, and others. Recent studies have identified that the RXR motif is highly conserved in the LRR XI subfamily[34]. The RXR motif is essential for the recognition of peptides such as CLEs[35] and RGFs[34] which feature His or Asn as the last residue. Interestingly, the LRR-RLK HAESA, TDR, and RK7 contain this RXR motif as well as a highly conserved DXS motif (Fig. 4d), which serves in recognition of the last residue of their respective ligands[35]. These LRR-RLKs commonly use SERKs as their co-receptors[36, 37]. Furthermore, the functional binding residues of TDR-TDIF interactions are either identical or mostly similar in the case of MIK1. Due to gene duplication and evolution, some residues on the interacting interface might have changed while the overall conservation of structural architecture and binding mode remains similar. MIK1 has been shown to be a receptor for AtLURE1.2[17], but, another in vitro study has shown no apparent interactions occur between MIK1 and AtLURE1.2[13]. Based on our sequence, structural, and computational analysis, it can be suggested that MIK1 may have functional redundancies that allow it to interact with both LURE1.2 and CLE peptides. Furthermore, these results also suggest that similar peptides may also function as ligands of other members of the LRR XI subfamily. It is also possible that MIK1 ectodomain and other members of Clade Aectodomain can also bind with the CLE family of small modified peptide hormones and use SERK family members as co-receptors for the propagation of signaling events.
The surface area analysis showed a patch of conserved residues colliding on the concave surface on the MIK1 modeled structure. Multiple studies on the LRR-RLK ectodomains have demonstrated that the plant peptide hormones tend to interact with the LRR-RLKs within the concave region[9]. Therefore, the presence of conserved residues on the MIK1 concave surface suggests a similar binding pattern and interaction mechanism. Furthermore, the electrostatic potential analysis showed a positive and a negative patch on the MIK1 concave surface, and, notably, the positively charged interface is similar to that of the TDR-TDIF interaction region, which leads us to conclude that peptide hormones with compatible structural properties could interact with MIK1 on the concave surface area.
Given the importance of LRR-RLKs in plant growth, development, and immunity, we propose that sequence-based and phylogenetic analyses can be highly revealing for preliminary studies of LRR-RLK proteins that are yet to be well-characterized. The recognition of conserved sequence motifs within families of proteins allows one to interpret surfaces of interaction based on related proteins, as was shown here for MIK1 and TDR. We hypothesize that MIK1 can also interact with CLE peptides and use the SERK family of proteins as a co-receptor for signal propagation. Moreover, our understanding of the role of glycosylation in the function, stability, and folding of LRR-RLKs have been limited. The LC-MS study of recombinant MIK1 LRR ectodomain has identified important N-glycosylation sites that match the predicted glycosylation sites on the native protein. The information gleaned from this study will likely prove useful in future studies to elucidate whether these glycosylation motifs play any functional role in ligand sensing and MIK1-mediated signal transduction, apart from protein folding.
Supplementary Material
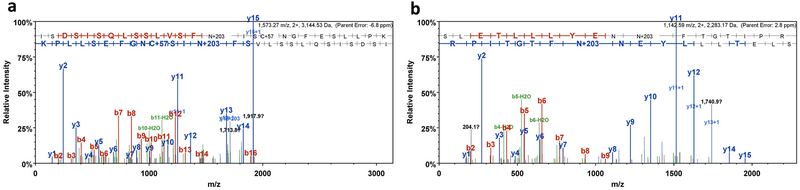
Fig. 2. Representative MS chromatogram identifying N-glycosylation sites on MIK1 ectodomain.
(a) MS spectrum showing the asparagine (N) residue of the ISDSISQLSSLVSFNISCNGFESLLPK peptide is glycosylated. (b) Identification of N-linked glycosylation on the asparagine residue present in the SLETLLLYENNFTGTIPR peptide. Glycosylated N residues are shown in bold letters
Acknowledgments:
We want to thank Dr. Haiyun Pan and Dr. Michael Goshe for their input in protein purification and mass spectrometry analysis.
Funding. This work was supported by the USDA National Institute of Food and Agriculture, Hatch project 1016752.
Footnotes
Competing Interests. The authors declare no competing interests.
Availability of data and material. The data that support the findings of this study that has not been reported here are available from the corresponding author upon request.
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
References
- 1.Walsh CT, Garneau-Tsodikova S, and Gatto GJ Jr., Protein posttranslational modifications: the chemistry of proteome diversifications. Angew Chem Int Ed Engl, 2005. 44(45): p. 7342–72. [DOI] [PubMed] [Google Scholar]
- 2.Khoury GA, Baliban RC, and Floudas CA, Proteome-wide post-translational modification statistics: frequency analysis and curation of the swiss-prot database. Sci Rep, 2011. 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ardito F, et al. , The crucial role of protein phosphorylation in cell signaling and its use as targeted therapy (Review). Int J Mol Med, 2017. 40(2): p. 271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manning G, et al. , The protein kinase complement of the human genome. Science, 2002. 298(5600): p. 1912–34. [DOI] [PubMed] [Google Scholar]
- 5.Kostich M, et al. , Human members of the eukaryotic protein kinase family. Genome Biol, 2002. 3(9): p. RESEARCH0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arabidopsis Genome I, Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature, 2000. 408(6814): p. 796–815. [DOI] [PubMed] [Google Scholar]
- 7.Park CJ, Caddell DF, and Ronald PC, Protein phosphorylation in plant immunity: insights into the regulation of pattern recognition receptor-mediated signaling . Front Plant Sci, 2012. 3: p. 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Torii KU, Leucine-rich repeat receptor kinases in plants: structure, function, and signal transduction pathways. Int Rev Cytol, 2004. 234: p. 1–46. [DOI] [PubMed] [Google Scholar]
- 9.Sun Y, et al. , Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex. Science, 2013. 342(6158): p. 624–8. [DOI] [PubMed] [Google Scholar]
- 10.Li ZJ, Chakraborty S, and Xu GZ, Differential CLE peptide perception by plant receptors implicated from structural and functional analyses of TDIF-TDR interactions. Plos One, 2017. 12(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.She J, et al. , Structural insight into brassinosteroid perception by BRI1. Nature, 2011. 474(7352): p. 472–U96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin G, et al. , A receptor-like protein acts as a specificity switch for the regulation of stomatal development. Genes Dev, 2017. 31(9): p. 927–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang X, et al. , Structural basis for receptor recognition of pollen tube attraction peptides. Nat Commun, 2017. 8(1): p. 1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shiu SH and Bleecker AB, Plant receptor-like kinase gene family: diversity, function, and signaling. Sci STKE, 2001. 2001(113): p. re22. [DOI] [PubMed] [Google Scholar]
- 15.Lehti-Shiu MD and Shiu SH, Diversity, classification and function of the plant protein kinase superfamily. Philos Trans R Soc Lond B Biol Sci, 2012. 367(1602): p. 2619–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J, et al. , Investigation of evolutionary and expressional relationships in the function of the leucine-rich repeat receptor-like protein kinase gene family (LRR-RLK) in the radish (Raphanus sativus L.). Sci Rep, 2019. 9(1): p. 6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang T, et al. , A receptor heteromer mediates the male perception of female attractants in plants. Nature, 2016. 531(7593): p. 241–4. [DOI] [PubMed] [Google Scholar]
- 18.Okuda S, et al. , Defensin-like polypeptide LUREs are pollen tube attractants secreted from synergid cells. Nature, 2009. 458(7236): p. 357–61. [DOI] [PubMed] [Google Scholar]
- 19.Takeuchi H and Higashiyama T, A species-specific cluster of defensin-like genes encodes diffusible pollen tube attractants in Arabidopsis. PLoS Biol, 2012. 10(12): p. e1001449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chakraborty S, et al. , Plant Leucine-Rich Repeat Receptor Kinase (LRR-RK): Structure, Ligand Perception, and Activation Mechanism. Molecules, 2019. 24(17). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Varki A, Biological roles of oligosaccharides: all of the theories are correct. Glycobiology, 1993. 3(2): p. 97–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lis H and Sharon N, Protein glycosylation. Structural and functional aspects. Eur J Biochem, 1993. 218(1): p. 1–27. [DOI] [PubMed] [Google Scholar]
- 23.Kornfeld R and Kornfeld S, Assembly of asparagine-linked oligosaccharides. Annu Rev Biochem, 1985. 54: p. 631–64. [DOI] [PubMed] [Google Scholar]
- 24.Hounsell EF, Davies MJ, and Renouf DV, O-linked protein glycosylation structure and function. Glycoconj J, 1996. 13(1): p. 19–26. [DOI] [PubMed] [Google Scholar]
- 25.Kukuruzinska MA and Robbins PW, Protein glycosylation in yeast: transcript heterogeneity of the ALG7 gene. Proc Natl Acad Sci U S A, 1987. 84(8): p. 2145–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J, et al. , Allosteric receptor activation by the plant peptide hormone phytosulfokine. Nature, 2015. 525(7568): p. 265–8. [DOI] [PubMed] [Google Scholar]
- 27.Chakraborty S, et al. , The Extracellular Domain of Pollen Receptor Kinase 3 is structurally similar to the SERK family of co-receptors. Sci Rep, 2018. 8(1): p. 2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chakraborty S, Trihemasava K, and Xu G, Modifying Baculovirus Expression Vectors to Produce Secreted Plant Proteins in Insect Cells . J Vis Exp, 2018(138). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crooks GE, et al. , WebLogo: a sequence logo generator. Genome Res, 2004. 14(6): p. 1188–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saitou N and Nei M, The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol, 1987. 4(4): p. 406–25. [DOI] [PubMed] [Google Scholar]
- 31.Ceriotti A, Duranti M, and Bollini R, Effects of N-glycosylation on the folding and structure of plant proteins. Journal of Experimental Botany, 1998. 49(324): p. 1091–1103. [Google Scholar]
- 32.Bosch D, et al. , N-glycosylation of plant-produced recombinant proteins. Curr Pharm Des, 2013. 19(31): p. 5503–12. [DOI] [PubMed] [Google Scholar]
- 33.Niu C, et al. , N-Glycosylation Improves the Pepsin Resistance of Histidine Acid Phosphatase Phytases by Enhancing Their Stability at Acidic pHs and Reducing Pepsin’s Accessibility to Its Cleavage Sites. Appl Environ Microbiol, 2016. 82(4): p. 1004–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song W, et al. , Signature motif-guided identification of receptors for peptide hormones essential for root meristem growth. Cell Res, 2016. 26(6): p. 674–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang H, et al. , Crystal structure of PXY-TDIF complex reveals a conserved recognition mechanism among CLE peptide-receptor pairs. Cell Res, 2016. 26(5): p. 543–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Han Z, Sun Y, and Chai J, Structural insight into the activation of plant receptor kinases. Curr Opin Plant Biol, 2014. 20: p. 55–63. [DOI] [PubMed] [Google Scholar]
- 37.Meng X, et al. , Ligand-Induced Receptor-like Kinase Complex Regulates Floral Organ Abscission in Arabidopsis. Cell Rep, 2016. 14(6): p. 1330–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.