Abstract
The complete mitochondrial genome of the widespread and common Indo-Pacific sea star Archaster typicus has been sequenced in this study. The mitogenome is 16,230 base pairs (bp) in length, with 13 protein coding genes (PCGs), 22 tRNAs and 2 rRNAs. Gene order of its PCGs and rRNAs matches those of nine other asteroid taxa included for comparison in this study, and it has a similar nucleotide composition of 33.08% A, 26.38% T, 25.53% C and 15.01% G nucleotides. Phylogenetic analyses place A. typicus as the sister group to Acanthaster spp., consistent with previous inferences.
Keywords: Echinodermata, intertidal, phylogeny, sand star, Valvatida
Distributed across the Indo-Pacific are three sea star species from the family Archasteridae (Sukarno and Jangoux 1977), of which Archaster typicus Müller & Troschel, 1840 is the most frequently encountered (Chan et al. 2018). Given its ubiquity, A. typicus has become one of the most well-studied sea star species, with numerous studies ranging from the examination of its reproductive biology (Run et al. 1988) to characterization of the metabolites produced (Yang et al. 2011). However, genomic data for A. typicus remain limited. Therefore, we here sequenced its mitochondrial genome and performed a phylogenetic analysis along with 12 other echinoderm mitogenomes.
Tube feet were subsampled from one A. typicus specimen on 7 December 2017 from the intertidal zone of Cyrene Reef, Singapore (1°15′22.9″N, 103°44′49.0″E). Total genomic DNA was extracted using E.Z.N.A Mollusc DNA Kit (Omega Bio-tek), and subsequently purified using DNA Clean and Concentrator (Zymo Research). Tissue samples and genomic DNA have been deposited in the cryogenic collection of Lee Kong Chian Natural History Museum (catalogue no.: HS0082/333081/333082). Genomic DNA was sheared using BioRuptor Pico (Diagenode) and libraries were prepared using NEBNext Ultra II Library Prep Kit (New England BioLabs). Sequencing was performed in 21.26% of an Illumina MiSeq run (250 × 250 bp).
A total of 3,768,666 raw reads were trimmed using Trimmomatic v0.38 (Bolger et al. 2014) under default settings and assembled using SPAdes v3.12.0 (Bankevich et al. 2012). Assembled mitochondrial contigs were identified using BLASTn (e-value 10−6) against two Acanthaster (Acanthasteridae) mitogenomes (Yasuda et al. 2006). Matched contigs were assembled into one contiguous sequence using CAP3 (Huang and Madan 1999). Putative circularity of the assembly was verified using circules.py v0.5 (Hahn et al. 2013).
The complete mitogenome of A. typicus is 16,230 bp in length, comprising 33.08% A, 26.38% T, 25.53% C and 15.01% G nucleotides (GenBank Accession No. MN052674). MITOS2 (Bernt et al. 2013) (RefSeq 81 Metazoa; Genetic Code 9) annotated 13 protein-coding genes (PCGs), 22 tRNA genes, and 2 rRNA genes. Besides NAD1 which has an initiation codon of GTG (valine), all PCGs have an ATG (methionine) initiation codon. Termination codon for all PCGs is TAA, with cytochrome b being TA(A).
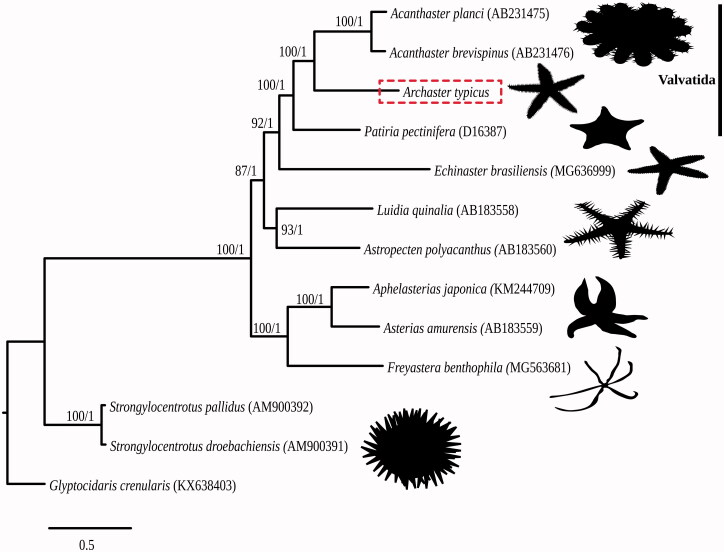
For phylogenetic reconstruction, 12 additional mitogenomes – including nine from Asteroidea and three from Echinoidea outgroups – were reannotated using MITOS2 (Bernt et al. 2013). Annotated PCGs and rRNA sequences were extracted, aligned using MAFFT-L-INS-I v7.271 (Katoh and Standley 2013), and concatenated into a single matrix (14,712 bp). Maximum likelihood phylogeny was inferred using RAxML v8.2.11 (Stamatakis 2014) with 100 random starting trees (GTRGAMA model) and 1000 bootstrap pseudoreplicates. Bayesian analysis was conducted using MrBayes v3.2.6 (Ronquist et al. 2012) (Figure 1). Consistent with the close relationship between Archasteridae and Acanthasteridae recovered by previous analyses (Knott and Wray 2000; Mah and Blake 2012; see also Matsubara et al. 2004), A. typicus is sister to Acanthaster spp. with maximum node support, and is within the order Valvatida.
Figure 1.
Maximum likelihood phylogeny of Asteroidea with Echinodea as outgroup. Bootstrap support and posterior probability values are shown adjacent to each node.
Acknowledgements
We would like to thank National Parks Board, Singapore for the permit (number NP/RP15-088) under which the sample was collected. The computational work for this article was partially performed on resources of the National Supercomputing Centre, Singapore (https://nscc.sg).
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Bankevich A, Nurk S, Antipov D, Gurevich A, Dvorkin M, Kulikov AS, Lesin V, Nikolenko S, Pham S, Prjibelski A, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YKS, Toh TC, Huang D. 2018. Distinct size and distribution patterns of the sand-sifting sea star, Archaster typicus, in an urbanised marine environment. Zool Stud. 57:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Madan A. 1999. CAP3: a DNA sequence assembly program. Genome Res. 9:868–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott KE, Wray GA. 2000. Controversy and consensus in asteroid systematics: new insights to ordinal and familial relationships. Am Zool. 40:382–392. [Google Scholar]
- Mah CL, Blake DB. 2012. Global diversity and phylogeny of the Asteroidea (Echinodermata). PLoS One. 7:e35644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara M, Komatsu M, Wada H. 2004. Close relationship between Asterina and Solasteridae (Asteroidea) supported by both nuclear and mitochondrial gene molecular phylogenies. Zool Sci. 21:785–793. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Run J-Q, Chen C-P, Chang K-H, Chia F-S. 1988. Mating behaviour and reproductive cycle of Archaster typicus (Echinodermata: Asteroidea). Mar Biol. 99:247–253. [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukarno P, Jangoux M. 1977. Révision du genre Archaster Müller et Troschel. Rev Zool Africaine. 91:817–844. [Google Scholar]
- Yang X-W, Chen X-Q, Dong G, Zhou X-F, Chai X-Y, Li Y-Q, Yang B, Zhang W-D, Liu Y. 2011. Isolation and structural characterisation of five new and 14 known metabolites from the commercial starfish Archaster typicus. Food Chem. 124:1634–1638. [Google Scholar]
- Yasuda N, Hamaguchi M, Sasaki M, Nagai S, Saba M, Nadaoka K. 2006. Complete mitochondrial genome sequences for crown-of-thorns starfish Acanthaster planci and Acanthaster brevispinus. BMC Genomics. 7:17. [DOI] [PMC free article] [PubMed] [Google Scholar]