Abstract
Toona sinensis, also known as Xiangchun in Chinese, is a popular ‘tree vegetables’ and famous medicinal plant with good economic value. In our study, we sequenced the complete chloroplast (cp) genome of T. sinensis using the llumina sequencing platform. The cp genome of T. sinensis is a characteristic four-party structure with a length of 157,228 bp, which contains two 26,994 bp inverted repeats (IRs), an 85,971 bp large single copy (LSC), and a 17,269 bp small single copy (SSC). We identified a total of 126 genes, of which clouding 82 protein-coding genes, 36 tRNA genes, and eight rRNA genes. The phylogenetic analysis showed that T. sinensis was closely related to the congeneric T. ciliata.
Keywords: Toona sinensis, chloroplast genome, phylogenetic analysis
Toona sinensis (A. Juss.) M. Roem. (TS), commonly named Chinese Xiangchun, is a perennial, deciduous tree belongs to the family of Meliaceae. It is widely distributed in eastern and southeastern Asia, e.g., in central and southern China (Wu et al. 2012). Toona sinensis is compliment as ‘tree vegetables’ due to possesses a unique and pleasant flavor and is consumed as a popular seasonal vegetable in certain parts of eastern and southeastern Asia (Hu et al. 2016). Toona sinensis is also a famous medicinal plant in China. Its young leaves have various biological and pharmacological functions, including anti-cancer, anti-diabetic, anti-viral, and antioxidant properties (Yang et al. 2019). Its economic importance can exemplarily be seen in the fact that >1 million people are actually working on T. sinensis products in China. Its planting area is >1 billion square meters and cultivating >800 billion kg of T. sinensis sprouts per year (Zhai and Granvogl 2019). However, there is little genomic information has been reported. Herein, we first report the complete chloroplast genome of T. sinensis, which might provide comprehensive information relevant to the effective conservation and management of this species.
The tender leaves of T. sinensis were sampled from the school garden of Shanxi University (Shanxi, China; 112°34′12″ E, 37°43′48″ N) and were used for the total genomic DNA extraction with a DNeasy Plant Mini Kit (QIAGEN, Valencia, California, USA). The voucher specimen (liu2018015) and the DNA sample (TS-2) of T. sinensis were deposited in the Molecular and Physiological Ecology Laboratory, Institute of Loess Plateau, Shanxi University (Taiyuan, Shanxi, China). The whole-genome sequencing was conducted with 150 bp pair-end read on the Illumina Hiseq Platform (Illumina, San Diego, CA). In total, about 400 million high-quality clean reads are obtained and used for the cp genome de novo assembly by the program NOVOPlasty (Dierckxsens et al. 2017) and direct-viewing in Geneious R11 (Biomatters Ltd., Auckland, New Zealand). Annotation was performed with the program Plann (Huang and Cronk 2015) and Sequin (http://www.ncbi.nlm.nih.gov/). Together with gene annotations, the complete cp genome sequences were submitted to GenBank and deposited under the accession number MK954108. A neighbour-joining (NJ) tree with 1000 bootstrap replicates was inferred using MEGA version 6 (Tamura et al. 2013) from alignments created by the MAFFT (Katoh and Standley 2013) using plastid genomes of 21 species.
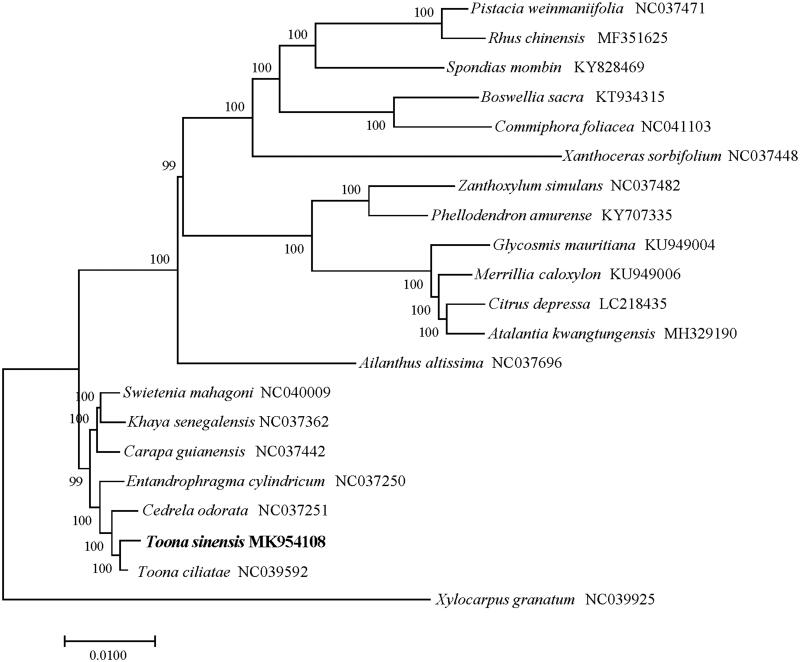
The chloroplast genome of T. sinensis is a typical quadripartite structure with a length of 157,228 bp, which contains two 26,994 bp inverted repeats (IRs), and the IRs were parted by a large single copy (LSC) (85,971bp) and a small single copy (SSC) (17,269 bp). We identified a total of 126 genes, of which clouding 82 protein-coding genes, 36 tRNA genes, and eight rRNA genes. Among the annotated genes, three of them contain three introns (trnA-UGC, trnI-GAU, rps12), one gene (clpP) contains two introns, and seven genes (trnI-CAU, trnL-CAA, trnV-GAC, trnN-GUU, trnR-ACG, ndhB, and ycf2) contain one intron. The overall GC content of the plastome is 37.9% while the corresponding values of the LSC, SSC, and IR regions are 36.1%, 32.1%, and 42.7%, respectively. The phylogenetic analysis suggested that T. sinensis was closely relevant to the species of T. ciliata (Figure 1).
Figure 1.
The neighbour-joining (NJ) tree based on the 21 chloroplast genomes. The bootstrap value based on 1000 replicates is shown on each node.
This absolute cp genome can be afterward used for population, phylogeny, and cp genetic project studies of T. sinensis and such cognition would be fundamental to the potential development value and provide some help for the large-scale breeding of T. sinensis.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Song Y, Mao X, Wang ZJ, Zhao QJ. 2016. Limonoids isolated from Toona sinensis and their radical scavenging, anti-inflammatory and cytotoxic activities. J Funct Foods. 20:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DI, Cronk Q. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3(8):1500026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30(12):2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YZ, Zhang XQ, Li SH, Chen JT. 2012. Biological characteristics and seedling raising techniques of Toona sinensis. Northern Horticult. 15:57–58. [Google Scholar]
- Yang WX, Cadwallader KR, Liu YP, Huang MQ, Sun BG. 2019. Characterization of typical potent odorants in raw and cooked Toona sinensis (A. Juss.) M. Roem. by instrumental-sensory analysis techniques. Food Chem. 282:153–163. [DOI] [PubMed] [Google Scholar]
- Zhai XT, Granvogl M. 2019. Characterization of the key aroma compounds in two differently dried Toona sinensis (A. Juss.) Roem. by means of the molecular sensory science concept. J Agric Food Chem. 67:9885–9894. [DOI] [PubMed] [Google Scholar]