Abstract
Alstonia scholaris is an evergreen tropical tree with significant medicinal values. To better understand its genetic and genomic profiles, we sequenced and assembled the completed plastome of A. scholaris. The plastome is 154,699 bp in length, consisting of a large (LSC, 85,364 bp) and a small single-copy region (SSC, 18,027 bp), which are separated by a pair of inverted repeat regions (IRs, 25,654 bp). It possesses 116 unique genes (82 protein-coding genes, 30 tRNAs, and 4 rRNAs). Phylogenetic analysis suggests that A. scholaris is sister to the clade including remaining Apocynaceae species.
Keywords: Alstonia scholaris, complete plastome, phylogenetic analysis
Alstonia scholaris (Linn.) R. Brown (Apocynaceae) is an evergreen tree distributed in South China, Indochina, New Guinea, and Austrilia (Li et al. 1995). The species is a medicinal plant traditionally used in China, India, Malaysia, and Thailand for the treatment of diarrhoea, dysentery, malaria, fever, and cardiac, as well as respiratory problems (Singh et al. 2017). Leaves of A. scholaris, bearing the Chinese name ‘Dengtaiye’, have been used as ethnomedicine to treat chronic respiratory diseases in the Yunnan province of China (Cai et al. 2010). However, little is known about the genetic and genomic profiles of the medicinally important plant, hindering the conservation and management of the germplasm resource. Here, we sequenced and assembled complete plastome of A. scholaris using high throughput Illumina sequencing technology.
Healthy and fresh leaves of A. scholaris were collected from Gengma, Yunnan, China (23°37′55.78″N, 99°19′18.40″E). Voucher specimen (Y. Ji 2017107) was deposited in the Herbarium of Kunming Institute of Botany, Chinese Academy of Sciences (KUN). Silica gel dried leaf tissues were used to extract total DNA by a modified CTAB method (Yang et al. 2014). Subsequently, we sheared the purified DNA by sonication to generate fragments of 500 bp length for constructing a paired-end library. Illumina libraries were prepared according to the manufacturer’s protocol (Illumina, San Diego, California, USA). Ensuing, paired-end sequencing was performed using the Illumina Hiseq 2000 (Illumina, San Diego, California, USA) sequencing platform at BGI (Wuhan, Hubei, China). The complete plastome sequences of Catharanthus roseus (GenBank Accession No. KC_563319) was downloaded as the reference for the plastome assembly following the method described by Jin et al (2018). The annotation of the plastome was performed in Geneious version 10.2.3 (Biomatters Ltd, Auckland, New Zealand) (Kearse et al. 2012). The validated plastome sequences of A. scholaris deposited in GenBank under Accession number MN176280.
The A. scholaris plastome is 154,699 bp in length and presents a typical quadripartite structure consisting of one large single-copy region (LSC, 85,364 bp), one small single-copy region (SSC, 18,027 bp), and a pair of inverted repeat regions (IRs, 25,654 bp). The overall G/C content in the A. scholaris plastome is 37.90%, and the corresponding value for LSC, SSC, and IR region were 36.00%, 31.90%, and 43.30%, respectively. The plastome encodes 133 genes, of those, 116 are unique genes (82 protein-coding genes, 30 tRNAs, and 4 rRNAs). Among these unique genes, 9 protein-coding genes (atpF, ndhA, ndhB, petB, petD, rpl16, rpl2, rpoC1, and rps12), and 6 tRNAs (trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA, and trnV-UAC) contain one intron, while three protein-coding genes (ycf3, clpP, and rps12) have two introns.
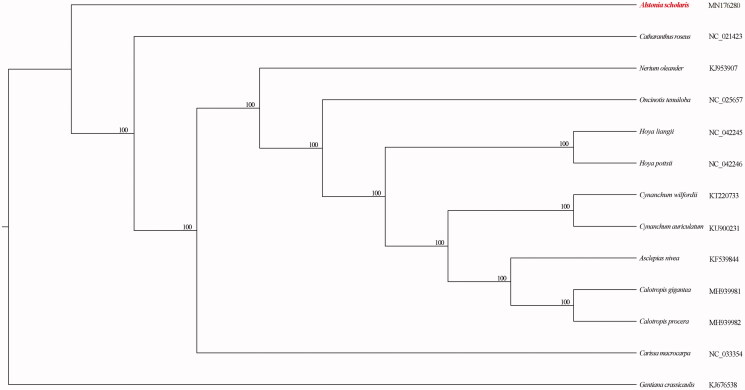
Eleven Apocynaceae complete plastomes were included in the phylogenetic analysis. We used RAxML (Stamatakis 2014) with 1000 bootstraps under the GTRCAT substitution model to reconstruct phylogenetic tree. Gentiana crassicaulis (Gentianaceae) was used to root the tree. The phylogeny robustly supported that A. scholaris is sister to the clade including remaining Apocynaceae species (Figure 1).
Figure 1.
Maximum-likelihood (ML) tree was reconstructed based on thirteen complete chloroplast plastomes. The numbers represent the bootstrap values.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Cai XH, Shang JH, Feng T, Luo XD. 2010. Novel alkaloids from alstonia scholaris. Z Naturforsch. 65:1164–1168. [Google Scholar]
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. bioRxiv. doi: 10.1101/256479. [DOI] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li PT, Antony JML, David JM. 1995. Apocynaceae In: Wu ZY, Raven PH, editors. Flora of China. Vol. 16 Beijing and St. Louis, China: Science Press and Missouri Botanical Garden Press; p. 154–156. [Google Scholar]
- Singh H, Arora R, Arora S, Singh B. 2017. Ameliorative potential of Alstonia scholaris (Linn.) R. Br. against chronic constriction injury-induced neuropathic pain in rats. BMC Complement Altern Med. 17:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. Raxml version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JB, Li DZ, Li HT. 2014. Highly effective sequencing whole chloroplast genomes of angiosperms by nine novel universal primer pairs. Mol Ecol Notes. 14:1024–1031. [DOI] [PubMed] [Google Scholar]