Abstract
Here, we present the first complete mitochondrial genome of Malayan Gaur (Bos gaurus hubbacki) inferred using next-generation sequencing. The mitogenome is 16,367 bp in length with the structural organization of a typical bovine mitochondrial arrangement comprising 13 protein-coding genes, 21 tRNAs, and 2 rRNAs. No internal stop codon was found in the protein-coding genes. Phylogenetic tree analysis revealed that Malayan gaur is more closely related to Burmese banteng instead of gaur.
Keywords: Malayan gaur, de novo assembly, mitogenome, next-generation sequencing
Introduction
Traditionally three subspecies have been recognized based on coloration and size (Duckworth et al. 2016): Bos gaurus gaurus; B. g. readei; and B. g. hubbacki. However, only two subspecies are recognized by the International Union for Conservation of Nature (IUCN) which were based on the measurements of skulls and horns (Groves and Grubb 2011; Duckworth et al. 2016). These two subspecies are B. g. gaurus which can be found in India, Nepal, and Bhutan; and B. g. laosiensis found in Myanmar, southern China, Laos, Vietnam, Cambodia, Thailand, and Peninsular Malaysia). Nonetheless, this dual classification is still inconclusive as the morphological measurements are based only on a few skull samples, and there is no supporting genetic analysis (Hassanin 2014).
In the past, wild gaur can be found in significant numbers in the states of Pahang, Perak, Kelantan, and Terengganu (Conry 1989). Nonetheless, due to poaching and habitat destruction, recently it is estimated that only around 270–300 individuals of wild gaur still exist in Peninsular Malaysia (DWNP 2012). Here, we present the first complete mitochondrial genome of B. gaurus isolate from Peninsular Malaysia with 16,367 bp in length. The full mitogenome reported here have the structural organization of a typical bovine mitochondrial arrangement comprising of 13 protein-coding genes, 21 tRNAs, and 2 rRNAs. No internal stop codon was found in the protein-coding genes. This annotated mitogenome has been deposited in GenBank under accession number MK770201.
Genomic DNA isolation and mitogenome sequencing
Tissue sample of a deceased male Malayan gaur with studbook number S0029 from Sungkai Wildlife Conservation Centre (Latitude: 4.034714 | Longitude: 101.369063) was used for extraction and analysis to generate the data presented here. The tissue samples are catalogued as WGRB-BGH40 and deposited in the Wildlife Genetic Resource Bank (WGRB) at National Wildlife Forensic Laboratory, Department of Wildlife and National Parks Peninsular Malaysia. Genomic DNA (gDNA) was used to generate sequences library of paired-end, 250-bp reads. It generated 2,541,912 of short-read sequences (SRS) totaling 635,478,000 bases of DNA.
Assembly and gene annotation
The raw SRS were screened using FastQC (Andrews 2010) and trimmed using BBduk (Bushnell 2015). Trimmed SRS were then assembled using repeated referenced mapping and de novo assembly. Repeated reference mapping was performed by employing end-to-end mapping and local mapping using BowTie2 plugin on Geneious Prime software (Biomatters, New Zealand) with high-sensitivity setting and re-iteration using a published mitochondrial genome of gaur as reference (JN632604). A total of 5782 contigs were assembled. NOVOPlasty (Dierckxsens et al. 2017) was used to perform de novo assembly of the trimmed SRS. Pair-wise alignment of sequences assembled from repeated referenced mapping and de novo were carried out in Geneious Prime and the consensus sequence from the alignment was selected as final sequence. Annotation generated by MITOS Webserver (Bernt et al. 2013) was used to identify protein-coding genes, ribosomal and transfer RNA gene and the result was compared with JN632604.
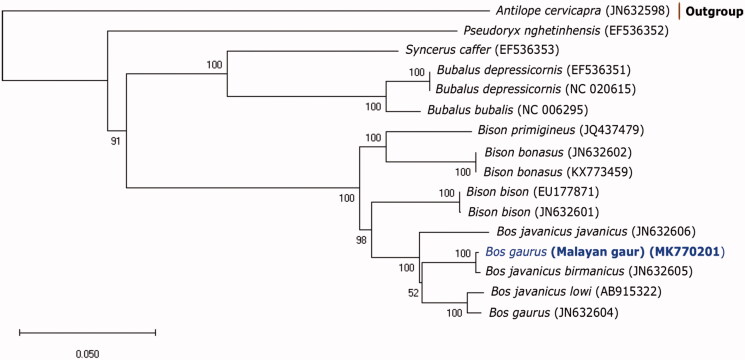
Phylogenetic tree (Figure 1) was constructed based on Maximum Likelihood by General Time Reversible model (Nei and Kumar 2000) with 1000 bootstrap value using MegaX software (Kumar et al. 2018). It showed that the Malayan Gaur was grouped together with the Burmese Banteng (JN632605) while the Burmese Gaur (JN632604) emerged as a sister group with the Borneon Banteng (Ishige et al. 2015). However, this grouping is only supported by 52% bootstrap value. More representatives from these two species may help to verify this grouping.
Figure 1.
Phylogenetic tree of complete mitogenome among members of Bovini tribe and a member of Antilopini as outgroup inferred using Maximum Likelihood method based on General Time Reversible model with 1000 bootstrap value. GenBank accession numbers for each mitogenomic sequences are shown in parentheses.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data; [accessed 2019 Jul 4]. http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenetics Evol. 69:313–319. [DOI] [PubMed] [Google Scholar]
- Bushnell B. 2015. BBMap v35.66; [accessed 2019 Jul 4]. https://sourceforge.net/projects/bbmap.
- Conry PJ. 1989. Gaur Bos gaurus and development in Malaysia. Biol Conserv. 49:47–65. [Google Scholar]
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data . Nucleic Acids Res. 45:e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckworth JW, Sankar K, Williams AC, Samba Kumar N, Timmins RJ. 2016. Bos gaurus. The IUCN Red List of Threatened Species 2016: e.T2891A46363646; [accessed 2019 Jul 4].
- DWNP 2012. Laporan Tahunan Jabatan PERHILITAN 2012. Kuala Lumpur: Department of Wildlife and National Parks, Peninsular Malaysia. [Google Scholar]
- Groves C, Grubb P. 2011. Ungulate taxonomy. Baltimore (MD): Johns Hopkins University Press. [Google Scholar]
- Hassanin A. 2014. Systematic and evolution of Bovini In: Melletti M, Burton J, editors Ecology, evolution and behaviour of wild cattle: implications for conservation. Cambridge: Cambridge University Press; p. 7–22. [Google Scholar]
- Ishige T, Gakuhari T, Hanzawa K, Kono T, Sunjoto I, Sukor JR, Ahmad AH, Matsubayashi H. 2015. Complete mitochondrial genomes of the tooth of a poached Borneon banteng (Bos javanicus lowi; Cetartiodactyla, Bovidea). Mitochondrial DNA. 27:2453–2454. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Kumar S. 2000. Molecular evolution and phylogenetics. New York: Oxford University Press. [Google Scholar]