Abstract
Asparagus setaceus is an important ornamental plant and is important to study the sex chromosome evolution in genus Asparagus. Here the complete chloroplast (cp) genome sequence of A. setaceus was determined. The cpDNA is 156,978 bp in length, contains a large single copy region of 85,311 bp and a small single copy region of 18,641 bp, which was separated by a pair of inverted repeat regions of 26,513 bp each. The cp genome contains 132 genes, including 88 protein-coding genes, 6 ribosomal RNA genes, and 38 transfer RNA genes. Phylogenetic analysis indicates that A. setaceus is closely related to Asparagus officinalis and Asparagus schoberioides.
Keywords: Asparagus setaceus, chloroplast genome, high-throughput sequencing, phylogenetic analysis
Asparagus setaceus is an important ornamental plant and belongs to genus Asparagus. The genus Asparagus consists of both dioecious and hermaphrodite plants, which made Asparagus an ideal genus for studying the sex chromosome origin and evolution. The transfer of chloroplast DNA into nuclear genomes form nuclear integrants of plastid DNAs (NUPTs), which likely play important roles in the sex chromosome evolution (VanBuren and Ming 2013; Steflova et al. 2014). As a hermaphrodite plant, A. setaceus is an important species for comparative studies of the sex chromosome evolution of the genus Asparagus. In this study, we assembled and characterized the complete chloroplast DNA of A. setaceus. The data presented here can provide a useful resource for further study of sex chromosome evolution of genus Asparagus and facilitate analysis of the evolution and genetics of this species and other species in the same genus.
Total genomic DNA was isolated from fresh leaves collected from an individual of A. setaceus grown in the greenhouse of Henan Normal Univeristy (Xinxiang, China; 113°91′E, 35°31'N). The voucher specimen (Li S.F., No. 2017110501) was deposited at the Biological Specimen Depository of Henan Normal University. Total DNA was used for the shotgun library construction and the subsequent high-throughput sequencing on the GridION X5 sequencing platform. A total of 123.6 Gb raw reads were obtained, quality-filtered, and assembled using Minimap2 (Li 2018) with the reference sequence of Asparagus officinalis (GenBank: LN896355.1). The chloroplast (cp) genome was annotated using DOGMA (Wyman et al. 2004) and corrected manually. The annotated cp genome sequence has been deposited into the GenBank with the accession number MK950153.
The complete cp genome of A. setaceus was 156,978 bp in length, consisting of a pair of inverted repeat regions of 26,513 bp each, a large single copy region of 85,311 bp, and a small single copy region of 18,641 bp. The overall GC content of the cp genome was 37.5%. A total of 132 genes were annotated, including 38 tRNA, 6 rRNA, and 88 protein-coding genes.
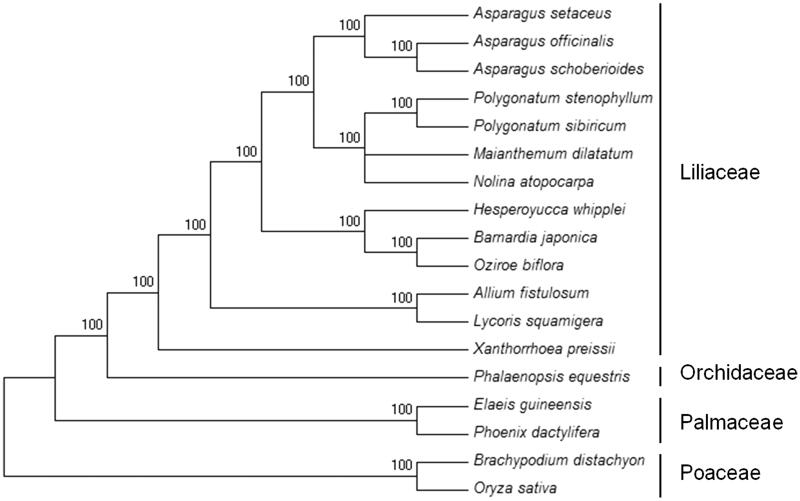
To confirm the phylogenetic position of A. setaceus, the complete chloroplast sequences of 13 species of Liliaceae (Monocotyledoneae) and 5 species of other Monocotyledons were aligned using MAFFT v.7 (Katoh and Standley 2013) and maximum-likelihood (ML) analysis was conducted using FastTree 2 (Price et al. 2010). The ML tree showed that A. setaceus is closely realted to A. officinalis and Asparagus schoberioides (Figure 1).
Figure 1.
Maximum-likelihood (ML) phylogenetic tree based on 18 complete chloroplast genomes. Accession numbers: Asparagus setaceus (MK950153); Asparagus officinalis (LN896355.1); Asparagus schoberioides (NC_035969.1); Polygonatum stenophyllum (KX822773.1); Polygonatum sibiricum (NC_029485.1); Maianthemum dilatatum (NC_039133.1); Nolina atopocarpa (KX931462.1); Hesperoyucca whipplei (KX931459.1); Barnardia japonica (MH287351.1); Oziroe biflora (KX931463.1); Allium fistulosum (NC_040222.1); Lycoris squamigera (NC_040164.1); Xanthorrhoea preissii (KX822774.1); Phalaenopsis equestris (NC_017609.1); Elaeis guineensis (NC_017602.1); Phoenix dactylifera (NC_013991.2); Brachypodium distachyon (NC_011032.1); Oryza sativa (NC_031333.1).
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 34:3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. 2010. FastTree 2-approximately maximum-likelihood trees for large alignments. PLoS One. 5:e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steflova P, Hobza R, Vyskot B, Kejnovsky E. 2014. Strong accumulation of chloroplast DNA in the Y chromosomes of Rumex acetosa and Silene latifolia. Cytogenet Genome Res. 142:59–65. [DOI] [PubMed] [Google Scholar]
- VanBuren R, Ming R. 2013. Organelle DNA accumulation in the recently evolved papaya sex chromosomes. Mol Genet Genom. 288:277–284. [DOI] [PubMed] [Google Scholar]
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255. [DOI] [PubMed] [Google Scholar]