Abstract
Rosa chinensis known commonly as the Chinese rose, is a member of the genus Rosa native to Southwest China. In this study, the complete chloroplast genome of R. chinensis was sequenced and analyzed. Structural analysis of the complete chloroplast (cp) genome of R. chinensis that exhibits a typical quadripartite circular structure with 155,097 bp in size, which contains a large single-copy region (LSC) of 85,911 bp, a small single-copy region (SSC) of 17,270 bp and a pair of inverted repeat (IR) regions of 25,958 bp in each one. The cp genome of R. chinensis contains 130 genes, including 85 protein-coding genes, 37 tRNA genes and 8 rRNA genes. The phylogenetic Maximum-Likelihood (ML) analysis result shown that R. chinensis and Rosa odorata formed an independent clade with a 100% bootstrap support in phylogenetic relationship.
Keywords: Rosa chinensis, Chinese rose, Rosaceae, complete chloroplast genome, phylogenetic analysis
Chinese rose, Rosa chinensis is the most important ornamental plant with economic, cultural and symbolic value, which also is used as cut flowers as garden ornamental and for the perfume industry (Zhang and Zhu 2006). Rosa chinensis is the queen of flowers, and holds great symbolic and cultural value, which appeared as decoration on 5000 year-old Asian pottery (Wang 2007). Now, R. chinensis is greater economic importance than other flowers plants. It is widely cultivated around the world and sold as garden plants, in pots, or as cut flowers, the latter accounting for approximately 30% of the market (Nybom and Werlemark 2016). However, little information is available on the genome, genomics and molecular biology databases in Rosa. In this study, the complete chloroplast genome of R. chinensis is obtained and reported, which contributes to study the genetic diversity and the molecular breeding of the genus Rosa in the future.
The samples of R. chinensis were collected from flowers plantation in Putian district of Fujian province (Fujian, China, 119.07E; 25.49 N). The total cpDNA of R. chinensis form young leafs were extracted with the modified CTAB method and stored in Putian University (No. PTU004). The cpDNA was purified and fragmented using the NEB Next UltraTM II DNA Library Prep Kit (NEB, BJ, and CN) and was sequenced using the NGS method. Quality control was performed to remove low-quality reads and adapters using the FastQC (Andrews 2015). The chloroplast (cp) genome was assembled using the Plann (Huang and Cronk 2015) and annotated using the DOGMA (Wyman et al. 2004). The physical map of the chloroplast genome of R. chinensis was generated using OrganellarGenomeDRAW version 1.3.1 (Lohse et al. 2013). Here, the complete chloroplast genome sequence of R. chinensis was determined that the GenBank accession No. is MK8324431.
Structural analysis of the complete cp genome of R. chinensis that exhibits a typical quadripartite circular structure with 155,097 bp in size. It contains a large single-copy region (LSC) of 85,911 bp, a small single- copy region (SSC) of 17,270 bp and two inverted repeat regions (IRs) of 25,958 bp in each one. The overall nucleotide content of R. chinensis cp genome is: 31.0% of A, 31.8% of T, 18.9% of C, and 18.3% of G, with the total GC content is 37.2%. The cp genome of R. chinensis comprises 130 genes, which includes 85 protein-coding genes (PCGs), 37 transfer RNA (tRNA) genes and 8 ribosomal RNA (rRNA) genes. 18 genes were found duplicated in every one of the IR regions that includes 7 PCGs species (rpl2, rpl23, ycf2, ndhB, rps7, rps12 and ycf1), 7 tRNAs species (trnI-CAU, trnL-CAA, trnV-GAC, trnI-GAU, trnA-UGC, trnR-ACG and trnN-GUU) and 4 rRNAs species (rrn16, rrn23, rrn4.5 and rrn5).
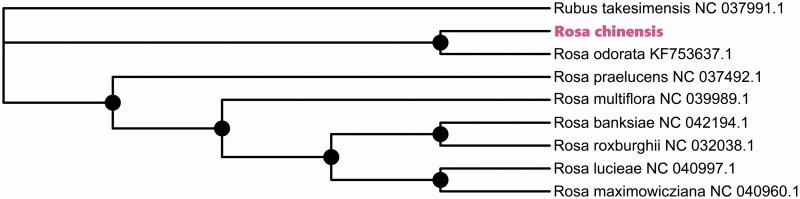
To identify the phylogenetic position of R. chinensis, phylogenetic relationship analysis was conducted. The phylogenetic tree was reconstructed using Maximum-Likelihood (ML) methods and performed using the RaxML (Stamatakis 2014), which the bootstrap value was calculated using 5000 replicates to assess node support and all the nodes were inferred with strong support. The evolutionary tree was constructed and edited using the MEGA X (Kumar et al. 2018). The results indicated that R. chinensis is located in the genus Rose and is closest related to R. odorata (KF753637.1) (Figure 1). This study can continue in-depth the genetic diversity and the molecular breeding of the genus Rosa in future.
Figure 1.
Phylogenetic relationship among 9 species based on the Maximum-Likelihood (ML) analysis of the complete chloroplast genome sequences from NCBI, using Rubus takesimensis as an outgroup. Bootstrap support values are indicated in each node.
Disclosure statement
No potential conflict of interest was reported by the authors.
Reference
- Andrews S. 2015. FastQC: a quality control tool for high throughput sequence data. http://www. bioinformatics.babraham.ac.uk/projects/fastqc/.
- Huang DI, Cronk QCB. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3:1500026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW–a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41:W575–W581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nybom H, Werlemark G. 2016. Realizing the potential of health-promoting rosehips from dogroses (Rosa sect. Caninae). CBC. 13:3–17. [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G. 2007. A study on the history of Chinese roses from ancient works and images. Acta Hortic. 751:347–356. [Google Scholar]
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255. [DOI] [PubMed] [Google Scholar]
- Zhang ZS, Zhu XZ. 2006. China rose. Beijing (China): China Forestry Publishing House; p. 170–172. [Google Scholar]