Abstract
Although we are just beginning to understand the mechanisms that regulate the epigenome, aberrant epigenetic programming has already emerged as a hallmark of hematologic malignancies including acute myeloid leukemia (AML) and B-cell lymphomas. Although these diseases arise from the hematopoietic system, the epigenetic mechanisms that drive these malignancies are quite different. Yet, in all of these tumors, somatic mutations in transcription factors and epigenetic modifiers are the most commonly mutated set of genes and result in multilayered disruption of the epigenome. Myeloid and lymphoid neoplasms generally manifest epigenetic allele diversity, which contributes to tumor cell population fitness regardless of the underlying genetics. Epigenetic therapies are emerging as one of the most promising new approaches for these patients. However, effective targeting of the epigenome must consider the need to restore the various layers of epigenetic marks, appropriate biological end points, and specificity of therapeutic agents to truly realize the potential of this modality.
Epigenetic instructions are the equivalent of software programs that instruct cellular phenotypes, including those of tumor cells. Along these lines, it has been shown that aberrant epigenetic programming occurs universally in hematologic malignancies, and somatic mutations in genes encoding epigenetic modifiers and transcription factors are the most abundant class of genetic lesions in these tumors. Therefore, hematological malignancies provide an excellent viewpoint for exploring how the many layers of epigenetic mechanisms interact to mediate transformed phenotypes. Herein, we focus on, and provide the basis to compare and contrast, epigenetic mechanisms underlying pathogenesis of acute myeloid leukemia (AML) and the common lymphomas derived from germinal center (GC) B cells.
EPIGENETIC MECHANISMS IN AML
Aberrant Cytosine Methylation Profiles Are a Hallmark of AML
A significant subset of recurrent mutations in AML affect epigenetic modifications of DNA and/or histones (Fig. 1) (Abbas et al. 2010; Marcucci et al. 2010, 2012; Paschka et al. 2010; Hollink et al. 2011; Shen et al. 2011; Gaidzik et al. 2012; Patel et al. 2012; Weissmann et al. 2012; Cancer Genome Atlas Research 2013; Gao et al. 2013; Metzeler et al. 2016; Papaemmanuil et al. 2016; Terada et al. 2018). Cytosine methylation (5mC) is critical for gene silencing, imprinting, X-chromosome inactivation, genome stability, and cell fate determination (Bird 2002). Tissue-specific 5mC patterns are established primarily by de novo DNA methyltransferases DNMT3A and DNMT3B (Okano et al. 1999) and are subsequently maintained by DNMT1 in a replication-dependent manner (Fig. 2A) (Robert et al. 2003). CpG islands (CGIs), comprising <10% of all CpGs, are found in ∼70% of promoters and are predominantly unmethylated, whereas a majority of remaining CpGs (60%–80%) are methylated in human cells (Saxonov et al. 2006; Smith and Meissner 2013). Disruption of cytosine methylation patterning at gene promoters is a hallmark of AML and is clearly linked to aberrant gene silencing (Figueroa et al. 2010b; Cancer Genome Atlas Research 2013). However, more recent studies indicate that critical 5mC changes in AML are also present at gene enhancers, in which their effects on gene expression are more nuanced (Glass et al. 2017). Importantly, 5mC profiles allow AMLs to be classified into biologically defined subtypes with distinct clinical outcomes (Figueroa et al. 2010a,b). Some of these profiles are linked to specific somatic mutations, whereas others are independent of mutations and are dependent on underlying disease-driving mechanisms such as overexpression of EVI1 (Lugthart et al. 2011) or silencing of myeloid lineage transcription factors such as CEBPα (Figueroa et al. 2009). Importantly, a subset of genes appears to be almost universally aberrantly methylated and silenced in AML patients regardless of somatic mutations (Figueroa et al. 2010b). It is possible that these epigenetic alleles are “epi-drivers” that are required for normal hematopoietic cells to manifest a leukemic phenotype in cooperation with somatic mutations and may explain why AMLs manifest relatively few genetic lesions. 5mC redistribution plays a critical role in myeloid differentiation (Bröske et al. 2009; Ji et al. 2010; Bock et al. 2012) and might be prone to disruption by such epi-drivers.
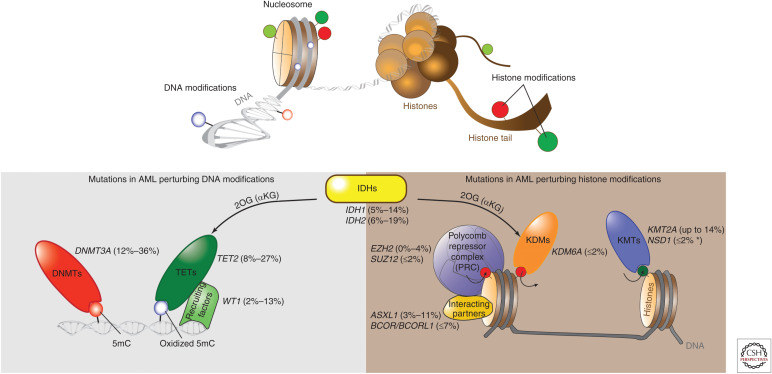
Figure 1.
Frequent mutations of epigenetic regulators in acute myeloid leukemia (AML). Upper panel illustrates epigenetic modifications on the DNA and histone layer of the epigenome. Lower panel shows gene mutations that impact DNA and/or histone modifications in at least 1% of AML cases. Ranges of reported mutation frequency are indicated in parentheses (Abbas et al. 2010; Marcucci et al. 2010, 2012; Paschka et al. 2010; Hollink et al. 2011; Shen et al. 2011; Gaidzik et al. 2012; Patel et al. 2012; Weissmann et al. 2012; Cancer Genome Atlas Research 2013; Gao et al. 2013; Metzeler et al. 2016; Papaemmanuil et al. 2016; Terada et al. 2018). The asterisk indicates frequency in adult AML. KMT, lysine methyltransferase; KDM, lysine demethylase; 2OG, 2-oxoglutarate, also called α-ketoglutarate (αKG).
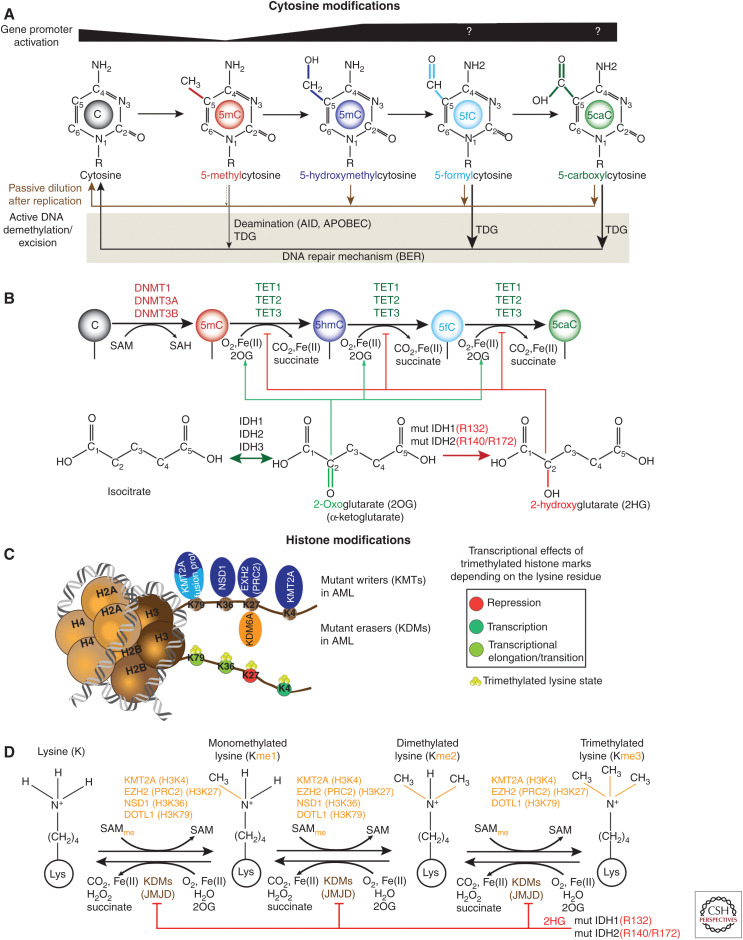
Figure 2.
Epigenetic features and perturbations in acute myeloid leukemia (AML). (A) Diagram showing the structure of methylated cytosine (5mC) and oxidized mCs (5hmC, 5fC, 5caC) as well as their transcriptional association at promoters. Active DNA demethylation is indicated by black arrows below modified cytosines. 5fC and 5caC can be excised directly by the thymine DNA glycosylase (TDG), resulting in an abasic site that is eventually replaced with unmethylated cytosine by the base excision repair (BER) machinery. Activation-induced cytidine deaminase (AID) or APOBEC can deaminated 5mC to thymine that is subsequently removed by TDG and repaired by BER. On the other hand, passive demethylation (brown arrows) is an alternative process that can occur through dilution following cell replication. DNMT1 can directly replicate the 5mC pattern onto the newly synthesized daughter strand unlike the pattern of oxidized mCs. (For review, see Rasmussen and Helin 2016.) (B) Enzymatic processes involved in cytosine methylation and oxidation of 5mC. DNMTs catalyze the addition of a methyl group to cytosine using S-adenosyl methionine (SAM) as methyl donor. Ten-eleven translocation (TET) proteins oxidize methylated cytosines to 5hmC, 5fC, and 5caC in an iterative manner using 2OG (αKG) as co-substrate. Mutant IDH1/2 generates 2HG that inhibits competitively the enzymatic activity of TET proteins. (C) Nucleosome model shows lysine substrate residues on histone H3 for writers (KMTs) and erasers (KDMs) that are found mutated in AML. Chromosomal translocations involving KMT2A (KMT2A-re) often result in fusion proteins that associate with the H3K79 histone methyltransferase DOT1L. (D) Enzymatic processes of lysine (de)methylation for selected KMTs. Besides TET proteins, 2HG can also impair the enzymatic activity Jumonji domain (JMJD)-containing KDMs.
AMLs Feature Highly Recurrent Mutations of Proteins That Modify Cytosine Residues
DNMT3A Mutations
Most mutant DNMT3A AML show either a heterozygous missense (R882H/C) mutation that affects the catalytic domain or truncating mutations (Cancer Genome Atlas Research 2013; Yang et al. 2015). R882 hotspot mutations impair DNMT3A methyltransferase activity and are linked to hypomethylation at specific CpGs (Russler-Germain et al. 2014). Dnmt3a knockout mice manifest severe differentiation block and enhanced self-renewal potential of hematopoietic stem cells (HSCs) (Challen et al. 2011). This is associated with DNA hypomethylation at borders of so-called “canyons” at regulatory regions of self-renewal genes (Jeong et al. 2014) consisting of large hypomethylated regions that contain histone activating marks (H3K4me3), repressive marks (H3K27me3), or both (Xie et al. 2013; Jeong et al. 2014). Noncompetitive transplantation experiments reveal that loss of DNMT3A predisposes murine HSCs to malignant transformation (Mayle et al. 2015), consistent with findings in humans, in which DNMT3A mutations establish a reservoir of preleukemic stem cells that can evolve to AML (Shlush et al. 2014). In addition, mutant DNMT3AR882 impairs chromatin remodeling and nucleosome eviction during chemotherapy, conferring resistance against anthracyclines that could explain the poor prognosis of DNMT3Amut AML (Ley et al. 2010; Shen et al. 2011; Guryanova et al. 2016).
Somatic Mutations of TET2 in AML
Removal of 5mC is initiated by members of the ten-eleven translocation (TET) family of dioxygenases through successive oxidization of 5mC to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC) (Fig. 2A) (Tahiliani et al. 2009; He et al. 2011; Ito et al. 2011). Apart from DNA demethylation by passive dilution following cell replication, 5fC and 5caC can be actively excised by thymine-DNA glycosylase (TDG) and repaired by the base excision repair (BER) pathway to regenerate unmodified cytosines (Maiti and Drohat 2011; Rasmussen and Helin 2016). In addition, activation-induced cytidine deaminase (AID) and APOBEC can deaminate 5mC but not 5hmC, contributing to 5mC erasure (Fig. 2A) (Nabel et al. 2012). Recent findings suggest that genomic 5hmC might also play an active role as an epigenetic mark that regulates transcription through modulation of chromatin accessibility and recruitment/hinderance of specific factors (e.g., MBD proteins binds to 5hmC DNA with weaker affinity than to 5mC DNA) (Hashimoto et al. 2012; Shen et al. 2013; Bachman et al. 2014; Ngo et al. 2016). In murine embryonic stem cells (ESCs), 5hmC function is involved in maintaining the active state of gene enhancers (Hon et al. 2014). Importantly, TET2 is primarily localized at CpG-sparse distal regulatory elements, suggesting a main function in enhancer regulation (Rasmussen et al. 2019). In agreement with this, deletion of TET2 in hematopoietic cells reduces 5hmC primarily at enhancers leading to down-regulation of tumor-suppressor genes (Rasmussen et al. 2015; Duy et al. 2019). TET2 is frequently mutated in AML, largely by frameshift or nonsense mutations (Chou et al. 2011; Weissmann et al. 2012). Loss of TET2 impairs differentiation and increases self-renewal potential of HSCs but requires cooperating mutations such as FLT3-ITD for full leukemic transformation (Delhommeau et al. 2009; Moran-Crusio et al. 2011; Quivoron et al. 2011; Shih et al. 2015). Notably, loss of TET2 alone has little effect on 5mC, but TET2 can synergistically induce DNA hypermethylation in the presence of a cooperating oncogene such as FLT3-ITD (Shih et al. 2015). Transformed cells derived from Tet2-deleted Flt3-ITD mice or human AML cells show DNA hypermethylation and suppression of GATA2, a hematopoietic master regulator, whose reconstitution impaires leukemia growth (Shih et al. 2015; Duy et al. 2019). Recruitment of TET2 to chromatin depends on transcription factors such as WT1 (Rampal et al. 2014; Wang et al. 2015). TET2 functions as a tumor suppressor in AML and requires the interaction with WT1 to suppress leukemia growth (Rampal et al. 2014; Wang et al. 2015). Overexpression of WT1 increased global levels of 5hmC, whereas reduced 5hmC levels were observed when WT1 was silenced. Recurrent missense mutations of TET2 can compromise its binding to WT1 and thereby fail to inhibit leukemia proliferation (Wang et al. 2015). Finally, loss of TET2 inactivates enhancers not only by reducing 5hmC but also through reduced enhancer histone marks such H3K4me1 mediated through LSD1 (KDM1A) (Duy et al. 2019). Hence, loss of TET2 impairs multiple layers of the epigenome.
Somatic Mutations of IDH1 and IDH2
AML somatic mutations in the IDH1 and IDH2 genes are virtually always heterozygous and affect IDH1 on R132 or IDH2 on the R140, R172 residues (Mardis et al. 2009; Abbas et al. 2010; Paschka et al. 2010). IDH1 (cytosolic protein) and IDH2 (mitochondrial protein) are metabolic enzymes that catalyze the interconversion of isocitrate to α-ketoglutarate (αKG) (Fig. 2B). αKG is necessary for oxidation of 5mC by TET enzymes (Fig. 2B). IDH1/2 hotspot mutations yield an enzymatic gain of function that instead favors the synthesis and accumulation of (R)-2-hydroxyglutarate (2HG) (Dang et al. 2009; Gross et al. 2010; Ward et al. 2011). 2HG competitively inhibits αKG-dependent dioxygenases such as TET enzymes and the Jumonji domain histone lysine demethylases, resulting in loss of 5hmC, gain of 5mC, and gain of histone methylation (Fig. 2B,C) (Figueroa et al. 2010a; Chowdhury et al. 2011; Xu et al. 2011; Lu et al. 2012; Rampal et al. 2014). Mutant IDH1/2 impairs HSC differentiation and it is notable that IDH1/2 mutations are almost entirely mutually exclusive with TET2 mutations in AML, suggesting that their effects on 5hmC and/or 5mC are dominant transforming effects in myeloid cells (Figueroa et al. 2010a). However, unlike Tet2 knockout mice, mutant IDH1R132 knock-in mice show reduced long-term (LT) HSCs and an altered DNA damage response caused by down-regulation of the ATM kinase independent of TET2 (Inoue et al. 2016). It was proposed that ATM down-regulation was mediated by 2HG inhibition of KDM4, resulting in a repressive chromatin environment suppressing ATM (Inoue et al. 2016). Moreover, 2HG can impair all TET enzymes, thereby causing a more pleiotropic effect than just mutant TET2 alone. Indeed, IDHmut AML shows a greater DNA hypermethylation phenotype than TET2mut AML (Rampal et al. 2014). Altogether, this suggests that the leukemogenic mechanisms are not equivalent and may contribute to different therapeutic responses of IDHmut AML compared to TET2mut (Patel et al. 2012; Duy et al. 2019). Moreover, IDHmut and DNMT3Amut AMLs feature opposing cytosine methylation profiles, and double-mutant IDH1/DNMT3A AML show almost complete loss of these profiles, thus questioning the relevance of DNA methylation (Glass et al. 2017). How these mutations cooperate is still unknown, but double-mutant IDH1/DNMT3A AML features up-regulation of RAS signatures and unique sensitivity to MEK inhibition ex vivo relative to AMLs with either single mutation (Glass et al. 2017).
Somatic Mutation of Chromatin Modifiers in AML
Loss-of-function and missense mutations have been reported in core components of the Polycomb repressive complex 2 (PRC2) SUZ12 and EZH2, which trimethylates histone H3 lysine 27 (H3K27me3) to induce transcriptional repression (Figs. 1 and 2C,D) (Ernst et al. 2010, 2012; Cancer Genome Atlas Research 2013). PRC2 induces stable silencing of early development genes, as well as transiently repressed bivalent chromatin promoters (H3K4me3, H3K27me3) (Margueron and Reinberg 2011). This complexity is exemplified by the finding that Ezh2 deletion preceding transduction of oncogenes like MLL-AF9 or AML1-ETO9a accelerates leukemia progression, whereas deletion of Ezh2 in already established AMLs with these oncogenes had the opposite effect and, in this case, attenuated disease progression (Basheer et al. 2019). Thus, EZH2 has a tumor-suppressive function at initiation of AML but a tumor-supportive one in maintenance of AML. Epigenetic changes induced by impaired function of PRC2 allows AML cells to tolerate increased stress including chemotherapy treatment (Göllner et al. 2017; Duy and Melnick 2018; Maganti et al. 2018). PRC2 functions can also be disrupted in the presence of inactivating somatic mutations in ASXL1, which directs PRC2 to target loci (Fig. 1) (Abdel-Wahab et al. 2012). Disrupting mutations in BCOR and BCORL1 have been reported and likely impact Polycomb-like complexes (Grossmann et al. 2011; Metzeler et al. 2016; Chittock et al. 2017). Mutations in KDM6A (UTX), which demethylates H3K27me3, have been reported in a small cohort of AML (Figs. 1 and 2D) (Cancer Genome Atlas Research 2013; Metzeler et al. 2016). Knockout of UTX induces spontaneous AML and results primarily in bidirectional changes of H3K27Ac as well as chromatin remodeling that is associated with inactivation of tumor-suppressive GATA programs and activation of oncogenic ETS programs (Gozdecka et al. 2018).
The histone methyltransferase KMT2A (MLL) is frequently mutated by partial tandem duplications (PTDs) or chromosomal rearrangements, the latter resulting in loss of the catalytic SET domain (Caligiuri et al. 1998; Grimwade et al. 2001; Krivtsov and Armstrong 2007). KMT2A methylates H3K4, which plays a role in the formation and activity of enhancers and promoters (Fig. 2C,D) (Ernst et al. 2011). The duplicated region in MLL-PTD includes the CXXC domain, which preferentially binds to nonmethylated CpGs (Schichman et al. 1994; Birke et al. 2002) that can potentially perturb coordination with DNA methylation. Mice with MLL-PTD show dysregulation of homeobox (Hox) gene expression but require additional mutations such as the Flt3-ITD for full leukemogenesis (Dorrance et al. 2008; Zorko et al. 2012). MLL rearrangements (KMT2A-re) result frequently in fusion with genes encoding super elongation complex proteins like AF9, AF10, and ENL (Krivtsov and Armstrong 2007). This complex associates with the histone methyltransferase DOT1L that targets H3K79 (Fig. 2C,D). MLL-transformed AML samples show aberrant H3K79 methylation, leading to ongoing expression of Hox genes. The NSD1 H3K36 histone methyltransferase is sometimes fused with NUP98, which could affect several gene regulatory functions (Fig. 2C,D) (Wagner and Carpenter 2012).
An additional layer of epigenetic regulation in AML is conferred by the 3D conformation of chromatin that brings distant genes into spatial proximity with each other or that forms loops between enhancers and promoters (Kagey et al. 2010). Many of these interactions are mediated by the cohesin complex, a ring-like structure composed of SMC1A, SMC3, RAD21, and STAG1/STAG2. The cohesin complex is responsible for connecting sister chromatids, regulation of transcription, and DNA repair (Haarhuis et al. 2014; Kim et al. 2016). Mutations in the cohesin complex are found between 6% and 18% in AML (Ding et al. 2012; Cancer Genome Atlas Research 2013; Kon et al. 2013; Thol et al. 2014; Thota et al. 2014; Tsai et al. 2017) and induce expansion of hematopoietic stem/progenitor cells and impair myeloid differentiation (Mullenders et al. 2015; Viny et al. 2015; Tothova et al. 2017). ASXL1 also interacts with the cohesin complex for proper gene regulation and might indicate a potential role in chromatin conformation (Li et al. 2017).
Epigenetic Therapy of AML
Targeting Specific Epigenetic Mechanisms
Aberrant DNA hypermethylation can be removed by DNMT inhibitors (DNMTi) (Fig. 3A) (Jones and Baylin 2007), although these drugs have pleiotropic effects and impact methylation on a genome-wide level. Specific inhibitors were recently developed against mutant IDH1/2 (AG120/AG221) and show a promising overall response rate of ∼40% in patients with relapsed/refractory IDH-mutant AML (Stein et al. 2017; DiNardo et al. 2018); however, their impact on epigenetic reprogramming and remodeling in patient samples remains to be delineated. Despite the initial response to IDH inhibitors, multiple drug resistance pathways have been reported including isoform switching between mutant IDH1 and mutant IDH2, second-site target mutations, co-occurring NRAS mutations, and selection of ancestral or terminal clones (Amatangelo et al. 2017; Harding et al. 2018; Intlekofer et al. 2018; Quek et al. 2018). Although protein loss (e.g., truncating TET2 mutations) cannot be targeted directly, approaches to compensate its function present an option. This has been shown with vitamin C, which enhances the catalytic activity of the residual wild-type TET proteins and suppresses leukemia growth (Fig. 3A) (Cimmino et al. 2017). A related approach was proposed for mutant DNMT3A by increasing intracellular levels of SAM (Fig. 3A) (Adema et al. 2017).
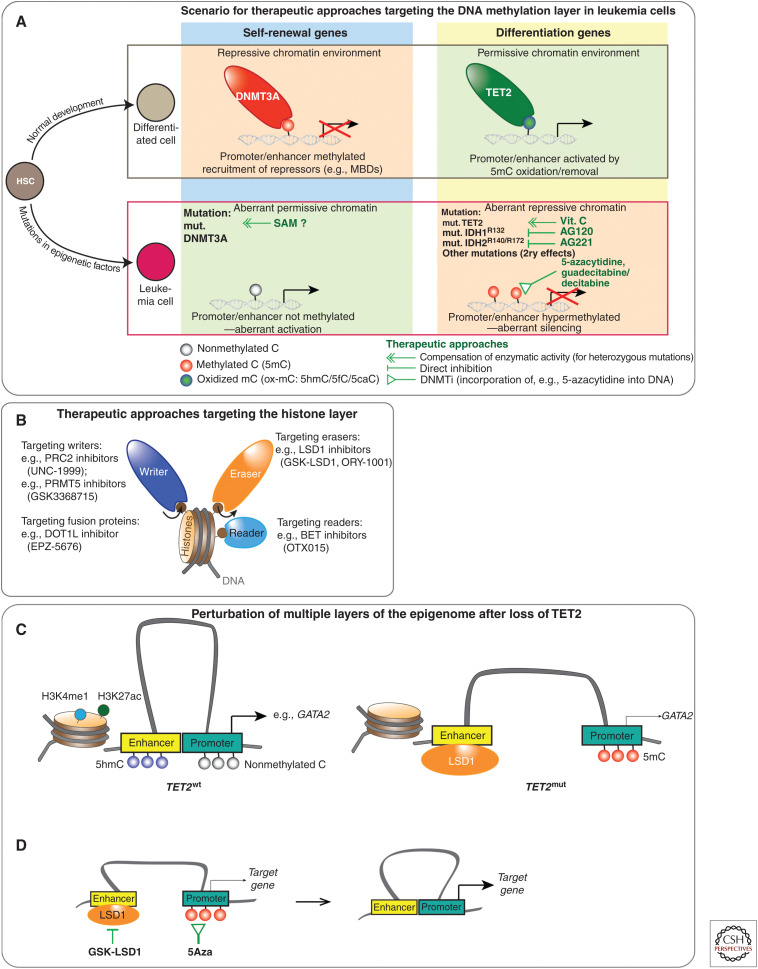
Figure 3.
Epigenetic therapy and combination therapy targeting cooperating layers of the epigenome in acute myeloid leukemia (AML). (A) Scenario illustrating the effect of distinct epigenetic mutations (indicated in bold letters) in leukemia cells versus normal differentiated cells derived from hematopoietic stem cells (HSCs). Epigenetic therapy and strategies targeting aberrant DNA methylation are shown by green lines. Loss of DNA methylation at self-renewal genes in DNMT3Amut AML may be potentially reversible by increasing intracellular levels of SAM (methyl donor). Loss of TET2 function can be compensated with vitamin C treatment. Specific inhibitors (AG120, AG221) against mutant IDH1/2 can block production of the TET inhibitor 2HG. Direct DNA hypermethylation (mutant TET2 or IDH1/2) or indirect hypermethylation due to secondary effects from other disease-driving mutations can be treated with DNMT inhibitors like 5-azacytidine or guadecitabine/decitabine (Issa et al. 2015; Gardin and Dombret 2017). (B) Drugs to target histone-modifying enzymes (writers+erasers) as well as readers (e.g., BET inhibitors) that are critical for AML maintenance and present vulnerabilities of the disease. Specific LSD1 inhibitors include GSK-LSD1 and ORY-1001 (Mohammad et al. 2015; Maes et al. 2018). Although PRC2 is deleted in a subset of AML, other AML subtypes such as KMT2A-re (MLL-rearranged) leukemias depend on functional PRC2. UNC1999, a dual inhibitor of EZH1/2 EZH1/2, impaired the growth of KMT2A-re in preclinical studies (Xu et al. 2015). DOT1L inhibitors (e.g., EPZ-5676) were also developed to target KMT2A(MLL)-re AML (Chen et al. 2016). The BRD 2/3/4 inhibitor OTX015 induces apoptosis in a variety of AMLs (Coudé et al. 2015). Inhibition of PRMT1 and PRMT5 show an antileukemia effect in distinct AMLs (Shia et al. 2012; Tarighat et al. 2016; Fedoriw et al. 2019). (C) Scheme illustrating the disruption of multiple layers of the epigenome in TET2mut cells. Loss of TET2 facilitates recruitment of the H3K4me1/2 histone demethylase LSD1 that inactivates enhancers at target genes. In addition, loss of TET2 results in promoter methylation at target genes such as GATA2. (D) Concept of targeting cooperating layers of the epigenome at enhancers and promoters in TET2mut AML to reconstitute enhancer–promoter interactions. Removal of 5mC promoter methylation by 5Aza treatment combined with LSD1 inhibition (GSK-LSD1) facilitates interactions of the LSD1-occupied enhancer and its target promoter, resulting in up-regulation of target genes like GATA2.
Targeting Epigenetic Mechanisms on Histones
LSD1 has emerged as a promising therapeutic target in AML (Fig. 3B) (Harris et al. 2012; Schenk et al. 2012; McGrath et al. 2016; Maes et al. 2018). LSD1 inhibitors (LSD1i) present also another option in TET2mut AML, in which it reverses aberrant repression of LSD1-inactivating enhancers (Duy et al. 2019). Moreover, inhibition of LSD1 can overcome nongenetic acquired drug resistance against BET inhibitors by modulating enhancer dependencies of key survival genes, showing further the critical role of enhancers in drug tolerance in AML (Bell et al. 2019). BET proteins can bind to acetylated lysines on histones (Fig. 3B) and facilitate transcriptional activation and promoter–enhancer interactions (Florence and Faller 2001; Wu and Chiang 2007). BRD4, a BET family member, has been implicated in AML, perhaps linked to maintaining MYC expression (Zuber et al. 2011). The role of PRC2 (EZH2) in AML is complex and not clear (Duy and Melnick 2018; Basheer et al. 2019). The use of UNC1999, a dual inhibitor of EZH1/2, impaired the growth of MLL(KMT2A)-rearranged leukemia cell lines suggesting that inhibition of PRC2 is detrimental in these leukemias (Fig. 3B) (Xu et al. 2015). Inhibition of DOT1L (EPZ-5676) also targets MLL-rearranged AML (Fig. 3B) (Chen et al. 2016). In addition, inhibition of protein arginine methyltransferase 1 (PRMT1) and 5 (PRMT5) displays an antileukemic effect in certain AMLs (Fig. 3B) (Shia et al. 2012; Tarighat et al. 2016; Fedoriw et al. 2019). Overall, it is important to note that all these histone-regulating proteins are not frequently mutated or overexpressed in AML and also play distinct roles in normal cells. Thus, targeting these factors requires a careful approach in a therapeutic window that will preferentially interfere with key nononcogene dependencies underlying the malignant program of AML.
Epigenetic Combination Therapy
Because the epigenome consists of many functionally interdependent mechanisms, it is inherently challenging to fully correct aberrant epigenetic programming by hitting only a single epigenetic target. This consideration may explain in part the relatively modest activity of epigenetic therapies to date (Fennell et al. 2019). Combining epigenetic therapies is challenging, however, because they may antagonize in vivo, such as occurred with the combination of DNMT inhibitors (DNMTi) + histone deacetylase inhibitors (HDACi) in AML patients (Prebet et al. 2014), and certain compounds such as HDACi have profoundly pleiotropic effects and hence significant toxicity in humans. Moreover, because little is known yet about how epigenetic mechanisms play into AML, it is difficult to predict which patients might respond to a given therapy, with rare exceptions such as HDACi. On the other hand, it is difficult to assess the efficacy of epigenetic therapies in the laboratory because AML cell lines do not reflect the genetic and epigenetic spectrum of AML cases. However, recent studies deploying organoid type cultures of primary AML cells are better suited to test epigenetic agents, many of which mediate their effects through slow and gradual effects. The unexpected enhanced activity of LSD1i+DNMTi against TET2 leukemia was discovered through such an approach, in which a large cohort of genetically characterized primary AMLs were screened using an organoid system (Duy et al. 2019). In TET2mut AML, full activation of aberrantly silenced tumor suppressors such as GATA2 could only be restored by simultaneously reversing gene promoter hypermethylation using DNMTi and restoring active enhancer marks for the same genes using LSD1i (because loss of TET2/5hmC results in LSD1-mediated enhancer repression; Fig. 3C,D).
Combination of Epigenetic with Nonepigenetic Drugs
AML mutations in nonepigenetic factors (e.g., FLT3) provide a rationale to combine epigenetic with specific nonepigenetic drugs. Supporting this, combination of DNMTi and FLT3 inhibitors improved therapeutic response in FLT3-ITD-mutant AML (Ravandi et al. 2013; Muppidi et al. 2015; Strati et al. 2015; Chang et al. 2016; Shih et al. 2017). Also, combination of DNMTi with drugs targeting nonmutant proteins like BCL2 (venetoclax) improved therapeutic efficacy particularly in elderly patients with AML (Tsao et al. 2012; Bogenberger et al. 2015; Shih et al. 2017; Aldoss et al. 2018; DiNardo et al. 2019). However, it is unclear whether this combination therapy is driven by epigenetic effects or rather by partial DNA damage caused by DNMTi treatment that results in apoptosis in leukemia cells after inhibition of BCL2. Because DNMTis also show immunomodulatory effects like induction of PD-1 and IFN-γ signaling, clinical trials exploring the combination of DNMTi with immune checkpoint inhibitor (ICI) drugs (e.g., nivolumab) are underway (Daver et al. 2019). Although CTLA-4 and PD-1 immune checkpoint drugs have revolutionized the management of certain cancer types like melanoma (Postow et al. 2015; Robert et al. 2015), other cancer types (e.g., pancreatic cancer) remain unaffected by ICI drugs (Brahmer et al. 2012). Given that the mutational burden of AML is one of the lowest compared with other cancers (Lawrence et al. 2013), the recognition of AML cells by the immune system is presumably less efficient compared with cancers with high mutational burden such as melanoma.
EPIGENETIC MECHANISMS IN B-CELL MALIGNANCIES
Most Lymphomas Arise from GC B Cells, Which Are Epigenetically Programmed to Resemble Tumor Cells
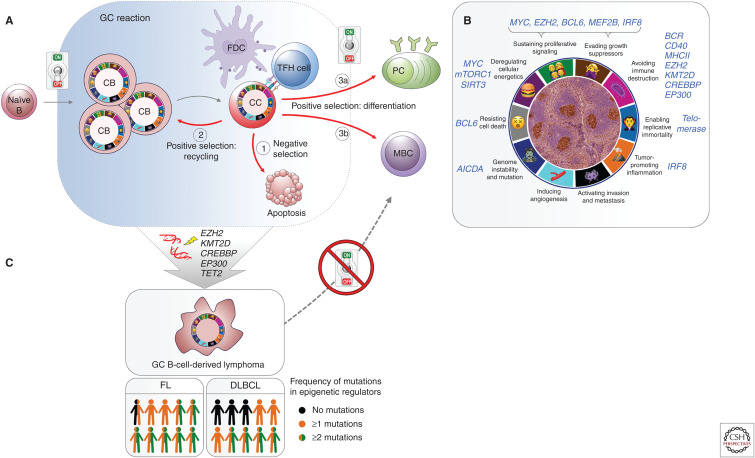
B-cell lymphomas arise from a wide variety of functionally distinct B-cell subsets to yield a bewildering number of lymphoma subtypes. Some of the more common B-cell neoplasms, their mutations, and their cell of origin are depicted in Figure 4. However, a majority of B-cell lymphomas (diffuse large B-cell lymphomas [DLBCLs] and follicular lymphomas [FLs]) arise from B cells transiting the GC reaction, which will be the focus of this section. GCs are transient structures that form in response to T-cell-dependent antigen (Hatzi and Melnick 2014). Naive B cells are epigenetically “primed”—that is, feature active chromatin marks—to induce expression of genes required for plasma cell differentiation, on receiving activation signals. However, after T-cell-directed activation, a subset of naive B cells is able to transiently silence these terminal differentiation genes and instead migrate within lymphoid follicles to form nests of highly proliferative GC B cells called “centroblasts,” which undergo somatic hypermutation of their immunoglobulin genes because of the actions of AICDA. After several rounds of division these GC B cells stop proliferating and compete to interact with a limiting number of T follicular helper (TFH) cells to survive. These nonproliferating GC B cells are called “centrocytes,” and interact with TFH cells through a number of receptor–ligand processes collectively called the “immune synapse” (Papa and Vinuesa 2018). Only the few GC B cells that generate high-affinity B-cell receptors interact strongly with TFH cells and can then restore their original epigenetic programming to differentiate into plasma or memory B cells or undergo further somatic hypermutation. However, the vast majority of GC B cells undergo apoptosis (Fig. 5A) (Mesin et al. 2016).
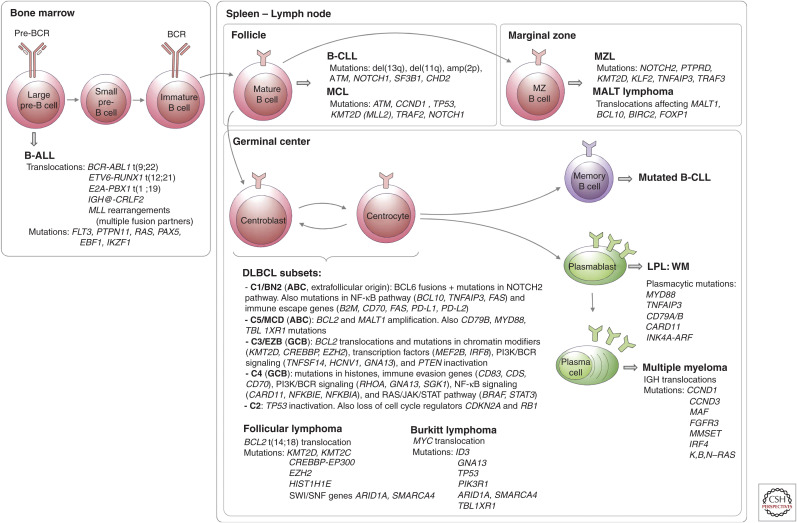
Figure 4.
Frequent mutations during B-cell development. In the bone marrow, rearrangements of the immunoglobulin genes of B-cell precursors to form a B-cell receptor (BCR) generate DNA breaks that are occasionally resolved aberrantly, leading to chromosomal translocations (Fugmann et al. 2000). These are the most common genetic alterations in B-precursor acute lymphoblastic leukemia (B-ALL) (Mullighan 2012). The most frequent mature B-cell neoplasms that have their origin outside the germinal center (GC) are B-cell chronic lymphocytic leukemia (B-CLL), mantle cell lymphoma (MCL), marginal zone lymphoma (MZL), and mucosa-associated lymphoid tissue (MALT) lymphoma. B-CLL and MCL differ in their molecular pathways, genomic alterations, and clinical behavior, being more aggressive in naive-like- than memory-like-derived tumors. The pathogenesis of the two malignancies involves the BCR signaling, tumor cell microenvironment interactions, genomic alterations, and epigenome modifications (Zhang et al. 2014; Landau et al. 2015). MALT lymphoma is the commonest MZL type and presents recurrent chromosomal translocations, which usually lead to activation of the NF-κB pathway. Nodal and splenic MZLs share recurrent mutations affecting the Notch pathway and the transcription factor KLF2, but differ for the inactivation of two tumor-suppressor genes, detected exclusively (PTPRD) or much more commonly (KMT2D/MLL2) in the nodal type (Rossi et al. 2012; Spina et al. 2016). The presence of immunoglobulin mutations is evidence that the cell of origin of the tumor passed through the GC microenvironment. Follicular lymphomas, Burkitt lymphomas, and DLBCLs express GC B cell signature genes. In the GC, two molecular processes remodel DNA: immunoglobulin class switch recombination (CSR) and somatic hypermutation (SHM), mechanisms that predispose to chromosomal translocations and mutations (Muramatsu et al. 2000). DLBCL is a clinically and genetically heterogeneous disease and accounts for 35% of non-Hodgkin lymphomas. Based on transcriptional profiles, DLBCL is further classified into activated B-cell (ABC) and germinal center B-cell (GCB) subtypes (Alizadeh et al. 2000; Rosenwald et al. 2002). ABC-DLBCLs derive from B cells that are committed to plasmablastic differentiation (Victora et al. 2012). These tumors have increased NF-κB activity, genetic alterations in NF-κB modifiers and components of the BCR pathway, and perturbed terminal B-cell differentiation (Lenz et al. 2008; Ngo et al. 2011). GCB-DLBCLs originate from light-zone GC B cells (Alizadeh et al. 2000; Victora et al. 2012). These tumors have frequent alterations in chromatin-modifying enzymes, PI3 K signaling, and genetic alterations of BCL2 (Pfeifer et al. 2013; Basso and Dalla-Favera 2015). Modifications in these pathways could favor epigenetic reprogramming and escape from cellular immunity. Recent genomic profiles have identified sub-ABC and GCB-DLBCL clusters: C1-C5 in one study (of which two are GCB-, two are ABC-subtypes, and the fifth is mostly characterized by genomic instability and TP53 mutations) (Chapuy et al. 2018), BN2, MCD, N1 (mostly ABC), and EZB (mostly GCB) in a different study (Schmitz et al. 2018). Follicular lymphoma (FL) is characterized by a unique histology in which tumor B cells form follicle-like structures with large numbers of nonmalignant immune cells infiltrating within the follicular and interfollicular regions (Kridel et al. 2012). The most frequent genetic event is the t(14;18) translocation that places BCL2 under control of the immunoglobulin heavy-chain enhancer, which occurs in 90% of FL patients. Mutations in epigenetic modifiers (KMT2D, CREBBP, and EZH2) are also a hallmark of FL (Green 2018). These mutations result in altering normal B-cell differentiation programs and impeding GC exit (Green et al. 2015). Burkitt lymphoma is characterized by deregulation of the MYC gene through its translocation to one of the immunoglobulin loci (Love et al. 2012). LPL, lymphoplasmacytic lymphoma; WM, Waldenstrom macroglobulinemia.
Figure 5.
Vulnerable points of germinal center (GC) B cells that give advantage to lymphomagenesis. (A) Chromatin-based epigenetic switches transiently poise the active plasma/memory B-cell program to enable the GC phenotype to emerge in a reversible manner. Cell fate decisions during a normal GC reaction: (1) negatively selected centrocytes (CC) undergo apoptosis; positively selected centrocytes may (2) recycle to centroblast (CB) and reenter the dark zone, or (3) differentiate to (3a) plasma cells (PC) or (3b) memory B cells (MBC). (B) GC B cells feature the typical hallmarks of transformed cells through epigenetic mechanisms, without requiring somatic mutations. This “pseudo-malignant” state can be reversed to normal state also through epigenetic switching mechanisms. (Sketch adapted from Hanahan and Weinberg 2011.) An immunohistochemistry (IHC) picture of GCs identified with peanut agglutinin (PNA) stain in a murine splenic section is shown in the middle. (C) Mutations in epigenetic modifiers maintain B cells in the GC phenotype, allowing the development of GC-derived B-cell lymphomas. Although mutations of one or more chromatin modifier genes occur within 96% of follicular lymphoma (FL) and ∼70% diffuse B-cell lymphoma (DLBCL) patients, 76% FL, and ∼40% DLBCL cases feature at least two mutations in epigenetic regulators (Green et al. 2015; Ortega-Molina et al. 2015; Reddy et al. 2017; Chapuy et al. 2018; Schmitz et al. 2018).
Importantly, the phenotype of GC B cells features many hallmarks that are considered pathognomonic for cancer (Hanahan and Weinberg 2011). These include (1) sustained proliferation and self-renewal (through repression of cell cycle checkpoint genes) (Phan and Dalla-Favera 2004; Phan et al. 2005; Cato et al. 2011; Béguelin et al. 2013, 2017); (2) potential immortalization (induction of telomerase) (Hu et al. 1997; Herrera et al. 2000; Norrback et al. 2001); (3) silencing of DNA damage checkpoint genes (repression of ATR, CHEK1, and TP53) (Phan and Dalla-Favera 2004; Ranuncolo et al. 2007, 2008); (4) genome instability and mutagenesis (caused by AICDA) (Muramatsu et al. 2000; McHeyzer-Williams et al. 2015; Teater et al. 2018); (5) resistance to cell death (induction of stress response pathways) (Cerchietti et al. 2009; Fernando et al. 2019); (6) deregulated energetics (efficient anaplerosis to support massive growth needs) (Doughty et al. 2006; Jellusova et al. 2017; Waters et al. 2018); (7) blockade of terminal differentiation (Béguelin et al. 2013, 2016); and (8) evasion from immune surveillance (down-regulation of antigen presentation and immune synapse genes) (Fig. 5B) (Basso et al. 2004; Good-Jacobson et al. 2010).
However, unlike cancer cells in which the transformed phenotype is primarily irreversible and caused by somatic mutations, in GC B cells this phenotype is solely induced through epigenetic reprogramming, which is reversible once B cells exit from the GC reaction (Bunting et al. 2016; Rivas and Melnick 2019). One of the more prominent epigenetic features of the GC reaction is the transient silencing of promoters and enhancers for genes involved in proliferation checkpoints, BCR signaling, CD40 signaling, interferon response, antigen presentation, and plasma cell differentiation (Béguelin et al. 2013, 2016; Hatzi et al. 2013; Ortega-Molina et al. 2015; Jiang et al. 2017). Most of this effect is coordinated by the transcriptional repressor BCL6, which is induced in the GC reaction and silenced on GC exit. Many of the mutations occurring in DLBCL and FL result in strengthening this effect (Fig. 5C).
Somatic Mutations in Epigenetic Modifiers Are a Hallmark of GC-Derived B-Cell Lymphomas
More than 70% of the genes mutated in DLBCL and FL are epigenetic modifiers and components of transcription factor complexes (Green et al. 2015; Ortega-Molina et al. 2015; Reddy et al. 2017; Chapuy et al. 2018; Schmitz et al. 2018). These include the H3K4 methyltransferase KMT2D, the H3 acetyltransferases CREBBP and EP300, the H3K27 methyltransferase EZH2, and TET2. Approximately 95% of FL patients manifest at least one of these mutations (Green et al. 2015), as do a majority of patients with GCB-DLBCL (Reddy et al. 2017; Chapuy et al. 2018; Schmitz et al. 2018). Each one of these genes is required to either establish the GC phenotype or to facilitate exit from the GC reaction. All are acquired early during pathogenesis and are considered founder mutations (Green et al. 2015; Pasqualucci and Dalla-Favera 2015). Mutations in TET2 are the earliest and only DLBCL mutations that originate in HSCs (Quivoron et al. 2011; Dominguez et al. 2018).
Wild-Type and Mutant EZH2 in the GC Reaction
EZH2 is induced and required for the formation of GC B cells (Velichutina et al. 2010; Béguelin et al. 2013; Caganova et al. 2013), where it deposits the H3K27me3 mark at active promoters (marked with H3K4me3) to form bivalent chromatin (Béguelin et al. 2013). The H3K27me3 mark then attracts the repressive Polycomb-like BCOR complex (Béguelin et al. 2016). However, a second tether is needed for BCOR recruitment and is provided by BCL6 (Huynh et al. 2000; Ghetu et al. 2008; Béguelin et al. 2016). This EZH2-BCOR-BCL6 combinatorial tethering process explains how EZH2 represses GC context-specific target genes, thanks to DNA sequence-specific binding of BCL6. Heterozygous somatic mutation of EZH2 occurs in up to 30% of FL and GCB-DLBCL patients and primarily affects the EZH2 SET (histone methyltransferase) domain at residue Y641 (Morin et al. 2010; Bödör et al. 2013; Okosun et al. 2014; Reddy et al. 2017). Unlike AML, in which EZH2 mutations result in loss of function, these mutations confer a gain of function that makes EZH2 more efficient at H3K27 trimethylation, but less efficient at H3K27 monomethylation (Sneeringer et al. 2010; Yap et al. 2011; McCabe et al. 2012). Mice engineered to express heterozygous mutant Ezh2 in GC B cells develop hyperplasia and B-cell lymphoma (Béguelin et al. 2013, 2016). This may be a consequence of more profound and less reversible silencing of EZH2 target genes involved in cell cycle checkpoints and GC exit. In contrast homozygous expression of mutant Ezh2 phenocopies the EZH2 knockout phenotype, showing that the WT EZH2 allele is required to cooperate with the mutant allele (Béguelin et al. 2016).
Somatic Mutations of CREBBP and EP300
These often manifest as missense mutations that inactivate their histone acetyltransferase domain, or truncations leading to loss of the allele (Cerchietti et al. 2010; Morin et al. 2011; Pasqualucci et al. 2011; Green et al. 2015; Jiang et al. 2017). CREBBP and EP300 maintain H3K27 acetylation mark at enhancers for genes involved in immune synapse and antigen presentation functions in B cells (Jiang et al. 2017). However, during the GC reaction BCL6 toggles these enhancers to a poised configuration by recruiting the SMRT/HDAC3 complex (Hatzi et al. 2013) (different than the function of BCL6 at promoters with BCOR and EZH2). CREBBP and p300 reactivate these enhancers when B cells are induced to exit the GC reaction. However, their loss-of-function mutation in lymphoma impairs this function and results in unopposed repression of these genes by HDAC3 and biological dependence on this protein (Jiang et al. 2017; Mondello et al. 2019). Loss of CREBBP or EP300 in GC B cells accelerates lymphomagenesis in mice (Garcia-Ramírez et al. 2017; Hashwah et al. 2017; Jiang et al. 2017; Zhang et al. 2017). The MHC class II genes are critical CREBBP targets that are aberrantly and persistently silenced by HDAC3 in CREBBP-mutant lymphoma cells (Hashwah et al. 2017; Jiang et al. 2017; Zhang et al. 2017).
Somatic Mutations of KMT2D
KMT2D has functions analogous to CREBBP in maintaining H3K4 monomethylation of gene enhancers in B cells. During the GC reaction, BCL6 represses KMT2D regulated enhancers in part through direct recruitment of LSD1, which in turn recruits the CoREST complex (Hatzi et al. 2019). In B cells, exiting the GC reaction KMT2D is required to enable the functionality of enhancers that normally respond to CD40 and BCR signaling. Lack of KMT2D leads to aberrant repression of these genes, failure of B cells to differentiate and exit the GC reaction, and eventually lymphomagenesis (Ortega-Molina et al. 2015; Zhang et al. 2015).
TET2 Mutations in the GC Context
TET2 mutations in DLBCL are similar in nature to those occurring in myeloid neoplasms (Quivoron et al. 2011). Loss of TET2 in GC B cells results in loss of enhancer 5hmC including at the PRDM1 locus, which is a master regulator of plasma cell differentiation. In addition, there is repression of many of the same genes that are normally induced by CREBBP on GC exit. Indeed, TET2 and CREBBP mutations are generally mutually exclusive, suggesting they represent a similar pathogenic hit. Along these lines, TET2 loss of function results in failure of CREBBP to mediate H3K27 acetylation at gene enhancers and makes lymphoma cells biologically dependent on HDAC3 (Dominguez et al. 2018).
The Immune Synapse as a Focal Point for Epigenetic Deregulation in Lymphoma
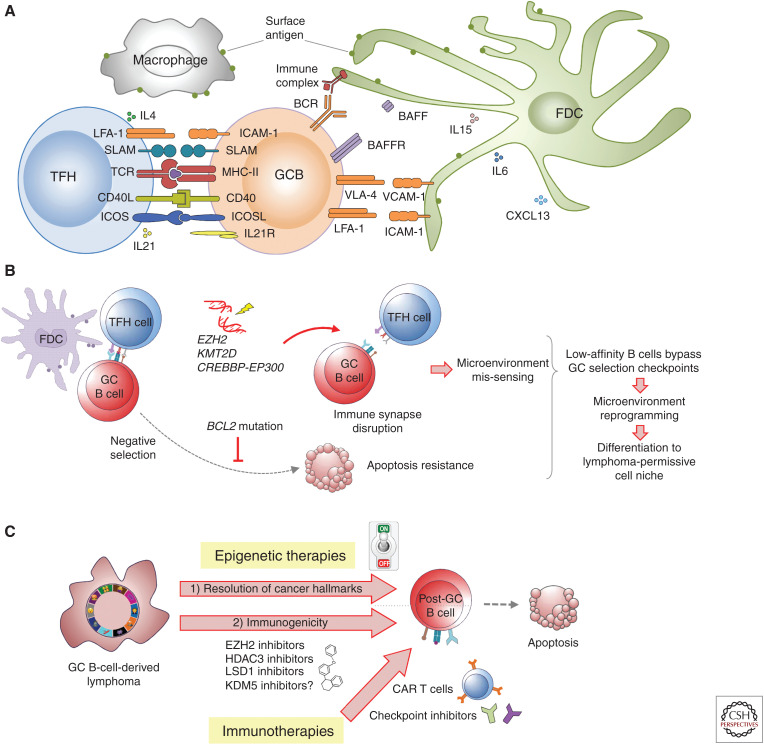
Reversion of the GC B-cell phenotype is essential to immune homeostasis. This effect is largely dependent on the immune synapse and interaction with TFH and follicular dendritic cells, involving CD40, BCR, MHC class II, IL21, and other signaling pathways (Fig. 6A) (Papa and Vinuesa 2018). Through unknown signals, the immune synapse reverses the poised state of gene promoters and enhancers driven by BCL6 with BCOR, EZH2, HDAC3, and LSD1 during the GC reaction. This is impaired by somatic mutations of EZH2, CREBBP, EP300, KMT2D, and TET2. Mutant EZH2 mediates aberrant silencing of cell cycle checkpoint inhibitors, MHC I and MHC II, and other GC exit genes (Béguelin et al. 2013; Ennishi et al. 2019). CREBBP, KMT2D, EP300, and TET2 loss of function disables enhancers that respond to CD40, BCR, and MHC class II genes (Ortega-Molina et al. 2015; Zhang et al. 2015, 2017; Hashwah et al. 2017; Jiang et al. 2017; Dominguez et al. 2018). Thus, the primary effect of epigenetic founder mutations in DLBCL and FL is mostly about “maintaining” GC B cells in an inherently oncogenic state by suppressing their ability to interface with the immune microenvironment. This also likely explains how mutant GC B cells evade immune surveillance to give rise to DLBCLs and FLs (Nicholas et al. 2016). Moreover, it is possible that aberrant immune synapse interactions with mutant GC B cells could epigenetically reprogram components of the immune microenvironment to form a lymphoma-permissive cell niche (Fig. 6B).
Figure 6.
The immune synapse and epigenetic therapy. Like germinal center (GC) B cells, the microenvironment in B-cell malignancies is crucial for the provision of survival and proliferation signals. (A) In both cases, normal GC B cells and malignant B cells interact with T cells, dendritic cells, macrophages, and lymphoid stromal cells (follicular dendritic cells) (Papa and Vinuesa 2018). (B) Mutations in epigenetic regulators and oncogenes such as BCL2 allow low-affinity B cells to survive, leading to the initiation of a prosurvival, immunosuppressive microenvironment in the lymphoid tissue. (C) Although epigenetic therapy can overcome the effect of founder mutations, the microenvironment makes critical contributions to both disease progression and drug resistance/disease relapse. The combination of epigenetic therapy with checkpoint inhibitor therapy can lead to potent antilymphoma effects.
Cytosine Methylation Patterning in Lymphomagenesis
Unlike the case of AML, 5mC patterning is not established as a driver of disease pathogenesis in GC-derived lymphomas. To date, DNA methylation profiling studies in DLBCL and FL have failed to identify robust and specific patterns of cytosine methylation indicative of underlying pathogenesis or with links to particular somatic mutations, with the possible exception of TET2. As compared with naive B cells, human and murine GC B cells manifest a characteristic hypomethylation signature (Dominguez et al. 2015, 2018; Teater et al. 2018). This appears to most likely represent a genomic “scar” effect of AICDA-mediated cytosine deamination of methylated residues that are replaced with unmethylated cytosines during replication, because AICDA knockout GC B cells fail to manifest this phenotype (Dominguez et al. 2015). Maintenance of DNA methylation is clearly important because, as expected, reduction of DNMT1 levels in mice severely impair GC formation (Shaknovich et al. 2011). DNA methylation profiling studies in cohorts of DLBCL patients have shown that progressive shifting of 5mC distribution from the normal GC pattern is linked to inferior clinical outcomes (Chambwe et al. 2014). Moreover, aberrant methylation and silencing of genes such as SMAD1 have been causally linked to chemotherapy resistance in DLBCL and can be overcome by administering DNA methyltransferase inhibitors (Clozel et al. 2013).
The Role of Epigenetic Therapy in GC-Derived Lymphomas
Nonspecific Epigenetic Therapies
The most experience to date with “epigenetic” therapy in lymphoma involves first-generation, pan-HDAC inhibitors. By and large these studies have been disappointing (Sermer et al. 2019). This may not be entirely surprising because these drugs are pleiotropic in their actions, have toxic side effects and there are no definitive, causal data indicating that they affect lymphoma cells through a particular epigenetic effect. The one bright spot has been in the setting of T-cell lymphomas, albeit for unknown biological reasons. BET inhibitors, which block binding of BRD4 to acetylated lysines, have shown toxicity against lymphoma cells in laboratory models but modest effects in patients (Chapuy et al. 2013; Amorim et al. 2016). On the other hand, DNMTis are well-tolerated in lymphoma patients in combination with chemo-immunotherapy and yielded favorable outcomes in phase I/II studies (Clozel et al. 2013). DNMTis were shown to reverse chemotherapy resistance in patients through the ex vivo study of sequential lymphoma biopsies obtained on patients enrolled in DNMTi trials (Clozel et al. 2013), warranting the expansion of these studies into the phase III setting.
Precision Epigenetic Therapy
Given the specific role of aberrant epigenetic modifiers in DLBCL and FL, there is great interest to deploy therapeutic agents that could reverse these effects. The only such approach to reach the clinic are EZH2 inhibitors, based on the strong rationale that EZH2 is essential to GC B cells and its mutant forms further enhance dependency on EZH2. Early clinical trials (albeit in patients with aggressive disease) show that EZH2i are well-tolerated and confer a significant clinical benefit (Gulati et al. 2018). Although there are a number of PRC2 inhibitors moving into clinical trials, most data available to date come from studies of the selective EZH2 inhibitor tazemetostat. A phase I, dose-escalation study in DLBCL patients previously treated with R-CHOP (NCT02889523) reported favorable safety results. However, in April 2018, the Food and Drug Administration (FDA) placed a temporary hold on the enrollment of new U.S. patients into this clinical trial because of a pediatric patient who developed a T-cell lymphoma while taking the drug. Another phase II clinical trial enrolls subjects with DLBCL and FL for the determination of efficacy and safety of tazemetostat monotherapy in combination with prednisolone (NCT01897571). For FL, it was reported that tazemetostat achieved objective response rates of 71% in patients with EZH2 mutations and 33% in patients with wild-type EZH2. These data are encouraging because the patients enrolled in this study had already undergone at least two previous therapies.
Despite achieving this milestone, there are currently no biomarkers that can predict whether a given patient will be responsive nor is the optimal timing and duration of dosing established. The fact that DLBCLs with EZH2 mutations manifest silencing of MHC I, MHC II, and reduced infiltrating CD4 and CD8 cells has pointed toward a potential immunomodulatory effect of EZH2i (Ennishi et al. 2019). However, it will be necessary to assess whether EZH2i might suppress antitumor immunity, given that PRC2 also plays critical roles in T-cell activation.
Because CREBBP- and TET2-mutant DLBCLs are addicted to HDAC3, there is a rationale for deploying selective HDAC3i to the clinic. HDAC3 has been biochemically purified as a component of the SMRT complex, >90% of which is bound with BCL6 in lymphoma B cells, pointing to the likely highly selective impact of HDAC3 inhibitors (Hatzi et al. 2013). Indeed, treatment of DLBCL cell lines or primary patient specimens with HDAC3i resulted in strong induction of MHC class II and triggered T-cell-mediated killing of lymphoma cells (Mondello et al. 2019). In contrast to pan-HDACis, HDAC3is lack toxicity against hematopoietic and T cells (Mondello et al. 2019) and hence represent a promising approach to restore T-cell-mediated immune surveillance against CREBBP-mutant DLBCL or FL cells. In the case of KMT2D-mutant lymphomas, there may be a rationale for targeting LSD1, although it would be required to use compounds that degrade LSD1 because purely enzymatic inhibition of this histone demethylase is insufficient to counteract its full affect in suppressing KMT2D target genes (Hatzi et al. 2019). It is also proposed that compounds that target the KDM5 family of H3K4 demethylases might also offer selective activity in the setting of KMT2D-mutant patients (Mondello et al. 2018).
Although these more precise epigenetic therapies represent a logical and promising opportunity, their most effective use will be in combination regimens, especially with immunotherapies. The success of immunotherapies such as checkpoint inhibitors and even CAR T cells is generally mixed in the setting of FL and DLBCL, perhaps linked to their epigenetic down-regulation of genes needed for B cells to effectively interface with T cells. Epigenetic therapies that reverse these effects might potently synergize with these immunotherapy modalities (Fig. 6C).
EPIGENETIC ALLELES, CLONAL EVOLUTION, AND THERAPY RESISTANCE IN HEME MALIGNANCIES
AMLs and lymphoma are composed of genetically distinct clones, and clonal complexity is linked to inferior clinical outcome, presumably because this endows tumors with greater population fitness. Because 5mC patterning is maintained with 1000-fold less fidelity than genome sequencing during replication, there is far more opportunity to develop clonal variance in epigenetic marks than in genetic composition. Indeed, foci of CpGs with variable methylation with populations of cells were identified by two initial studies (Hansen et al. 2011; Shaknovich et al. 2011), one in solid tumor cells and one in normal GC B cells. Epigenetic heterogeneity in GC B cells was considered as a possible precursor to malignant transformation: Perhaps sampling of epigenetic states during the GC reaction could yield selection advantages and the formation of premalignant clonal precursor cells (Shaknovich et al. 2011). Indeed, the first study of epigenetic heterogeneity was performed in GC-derived lymphomas (FL and DLBCL) and showed more severe 5mC heterogeneity in these tumors as compared with GC B cells (De et al. 2013). The extent of 5mC heterogeneity was associated with shorter survival, suggesting that it contributes to lymphoma population fitness (De et al. 2013).
Linking together the concepts of epigenetic heterogeneity and tumor clonality, several groups developed the concept of “epigenetic alleles,” defined as the DNA methylation status of groups of four consecutive CpGs present on the same DNA strand (Li et al. 2016). Analysis of DLBCL patients indicated that epigenetic allele diversity is linked to risk for relapse (Pan et al. 2015). Relapsed DLBCLs showed selection epigenetic allele states, which suggests that particular 5mC patterns were best suited to tolerate exposure to chemotherapy (Pan et al. 2015). Epigenetic allele studies in CLL also showed a link between 5mC diversity and outcome (Landau et al. 2014). Loci that manifested epigenetic allele diversity in CLL patients were also associated with deregulated transcription, providing further evidence that epigenetic heterogeneity has functional implications (Landau et al. 2014). Notably, 5mC heterogeneity in the context of normal GC B cells and GC-derived lymphomas was shown to be caused at least in part by AICDA (Dominguez et al. 2015; Teater et al. 2018). This is likely due to stochastic deamination of 5mCs in B cells, which eventually leads to AICDA dose-dependent 5mC heterogeneity (Dominguez et al. 2015; Teater et al. 2018).
A large-scale integrative analysis of exomes, whole-genome sequencing, methylomes, and transcriptomes in matched paired AML specimens showed that epigenetic allele diversity was an independent risk factor for time to relapse and, most remarkably, was completely independent of genetic clonality (Li et al. 2016). Genes linked to promoters with epigenetic allele diversity manifested diversity in their transcriptional output, and after relapse there was also evidence of selection for distinct epigenetic clonal states (Li et al. 2016). Taken together, epigenetic allele diversity or 5mC heterogeneity have emerged as critically important biological features of lymphoid and myeloid neoplasms that cannot be predicted based on tumor genetics. It is intriguing to speculate that DNMT inhibitors might actually suppress epigenetic clonal diversity, perhaps explaining in part how these drugs enhance the effect of chemotherapy drugs in AMLs and DLBCLs.
CONCLUDING REMARKS
It is evident that epigenetic mechanisms play a fundamental role in the pathogenesis of hematologic malignancies. However, as of yet, we only have a very superficial view of the depth and breadth of epigenetic mechanisms and very few therapeutic approaches that are amenable for clinical translation. We need to dig deeper into how epigenetic marks connect with each other and the role of novel layers of the epigenome. It is also necessary to consider the potential for epigenetic heterogeneity among these layers and within given patients. There is a need to understand bidirectional epigenetic programming between tumor and microenvironment and the host immune system, and how epigenetic drugs could have beneficial or deleterious effects on these various cellular compartments. This is an important and richly rewarding avenue of research that will require concerted effort to truly harness from the therapeutic standpoint.
ACKNOWLEDGMENTS
A.M. is supported by National Cancer Institute (NCI) R35 CA220499, NCI 1UG CA233332, NCI R01 CA198089, the Leukemia & Lymphoma Society (LLS) TRP 6572-19, LLS SCOR 7013-17, the Follicular Lymphoma Consortium, and the Chemotherapy Foundation. W.B. is supported through LLS SCOR 7012-16, LLS TRP 6572-19, and NCI R35 CA220499, C.D. is a recipient of the LLS Fellow Award LLS 5486 and is supported by NCI R01 CA198089.
Footnotes
Editors: Michael G. Kharas, Ross L. Levine and Ari M. Melnick
Additional Perspectives on Leukemia and Lymphoma: Molecular and Therapeutic Insights available at www.perspectivesinmedicine.org
REFERENCES
- Abbas S, Lugthart S, Kavelaars FG, Schelen A, Koenders J, Zeilemaker A, van Putten WJL, Rijneveld A, Löwenberg B, Valk PJM. 2010. Acquired mutations in the genes encoding IDH1 and IDH2 both are recurrent aberrations in acute myeloid leukemia (AML): prevalence and prognostic value. Blood 116: 2122–2126. 10.1182/blood-2009-11-250878 [DOI] [PubMed] [Google Scholar]
- Abdel-Wahab O, Adli M, LaFave LM, Gao J, Hricik T, Shih AH, Pandey S, Patel JP, Chung YR, Koche R, et al. 2012. ASXL1 mutations promote myeloid transformation through loss of PRC2-mediated gene repression. Cancer Cell 22: 180–193. 10.1016/j.ccr.2012.06.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adema V, Balasubramanian SK, Hirsch CM, Przychodzen BP, Phillips JG, Lindner D, Radivoyevitch T, Mukherjee S, Nazha A, Carraway HE, et al. 2017. Novel therapeutic targets for DNMT3A mutant myeloid neoplasms. Blood 130: 106 10.1182/blood-2017-05-786277 [DOI] [PubMed] [Google Scholar]
- Aldoss I, Yang D, Aribi A, Ali H, Sandhu K, Al Malki MM, Mei M, Salhotra A, Khaled S, Nakamura R, et al. 2018. Efficacy of the combination of venetoclax and hypomethylating agents in relapsed/refractory acute myeloid leukemia. Haematologica 103: e404–e407. 10.3324/haematol.2018.188094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, et al. 2000. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 403: 503–511. 10.1038/35000501 [DOI] [PubMed] [Google Scholar]
- Amatangelo MD, Quek L, Shih A, Stein EM, Roshal M, David MD, Marteyn B, Farnoud NR, de Botton S, Bernard OA, et al. 2017. Enasidenib induces acute myeloid leukemia cell differentiation to promote clinical response. Blood 130: 732–741. 10.1182/blood-2017-04-779447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amorim S, Stathis A, Gleeson M, Iyengar S, Magarotto V, Leleu X, Morschhauser F, Karlin L, Broussais F, Rezai K, et al. 2016. Bromodomain inhibitor OTX015 in patients with lymphoma or multiple myeloma: a dose-escalation, open-label, pharmacokinetic, phase 1 study. Lancet Haematol 3: e196–e204. 10.1016/S2352-3026(16)00021-1 [DOI] [PubMed] [Google Scholar]
- Bachman M, Uribe-Lewis S, Yang X, Williams M, Murrell A, Balasubramanian S. 2014. 5-Hydroxymethylcytosine is a predominantly stable DNA modification. Nat Chem 6: 1049–1055. 10.1038/nchem.2064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basheer F, Giotopoulos G, Meduri E, Yun H, Mazan M, Sasca D, Gallipoli P, Marando L, Gozdecka M, Asby R, et al. 2019. Contrasting requirements during disease evolution identify EZH2 as a therapeutic target in AML. J Exp Med 216: 966–981. 10.1084/jem.20181276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basso K, Dalla-Favera R. 2015. Germinal centres and B cell lymphomagenesis. Nat Rev Immunol 15: 172–184. 10.1038/nri3814 [DOI] [PubMed] [Google Scholar]
- Basso K, Klein U, Niu H, Stolovitzky GA, Tu Y, Califano A, Cattoretti G, Dalla-Favera R. 2004. Tracking CD40 signaling during germinal center development. Blood 104: 4088–4096. 10.1182/blood-2003-12-4291 [DOI] [PubMed] [Google Scholar]
- Béguelin W, Popovic R, Teater M, Jiang Y, Bunting KL, Rosen M, Shen H, Yang SN, Wang L, Ezponda T, et al. 2013. EZH2 is required for germinal center formation and somatic EZH2 mutations promote lymphoid transformation. Cancer Cell 23: 677–692. 10.1016/j.ccr.2013.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Béguelin W, Teater M, Gearhart MD, Calvo Fernández MT, Goldstein RL, Cárdenas MG, Hatzi K, Rosen M, Shen H, Corcoran CM, et al. 2016. EZH2 and BCL6 cooperate to assemble CBX8-BCOR complex to repress bivalent promoters, mediate germinal center formation and lymphomagenesis. Cancer Cell 30: 197–213. 10.1016/j.ccell.2016.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Béguelin W, Rivas MA, Calvo Fernandez MT, Teater M, Purwada A, Redmond D, Shen H, Challman MF, Elemento O, Singh A, et al. 2017. EZH2 enables germinal centre formation through epigenetic silencing of CDKN1A and an Rb-E2F1 feedback loop. Nat Commun 8: 877 10.1038/s41467-017-01029-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell CC, Fennell KA, Chan YC, Rambow F, Yeung MM, Vassiliadis D, Lara L, Yeh P, Martelotto LG, Rogiers A, et al. 2019. Targeting enhancer switching overcomes non-genetic drug resistance in acute myeloid leukaemia. Nat commun 10: 2723 10.1038/s41467-019-10652-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. 2002. DNA methylation patterns and epigenetic memory. Genes Dev 16: 6–21. 10.1101/gad.947102 [DOI] [PubMed] [Google Scholar]
- Birke M, Schreiner S, Garcia-Cuéllar MP, Mahr K, Titgemeyer F, Slany RK. 2002. The MT domain of the proto-oncoprotein MLL binds to CpG-containing DNA and discriminates against methylation. Nucleic Acids Res 30: 958–965. 10.1093/nar/30.4.958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock C, Beerman I, Lien WH, Smith ZD, Gu H, Boyle P, Gnirke A, Fuchs E, Rossi DJ, Meissner A. 2012. DNA methylation dynamics during in vivo differentiation of blood and skin stem cells. Mol Cell 47: 633–647. 10.1016/j.molcel.2012.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bödör C, Grossmann V, Popov N, Okosun J, O'Riain C, Tan K, Marzec J, Araf S, Wang J, Lee AM, et al. 2013. EZH2 mutations are frequent and represent an early event in follicular lymphoma. Blood 122: 3165–3168. 10.1182/blood-2013-04-496893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogenberger JM, Delman D, Hansen N, Valdez R, Fauble V, Mesa RA, Tibes R. 2015. Ex vivo activity of BCL-2 family inhibitors ABT-199 and ABT-737 combined with 5-azacytidine in myeloid malignancies. Leuk Lymphoma 56: 226–229. 10.3109/10428194.2014.910657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahmer JR, Tykodi SS, Chow LQ, Hwu WJ, Topalian SL, Hwu P, Drake CG, Camacho LH, Kauh J, Odunsi K, et al. 2012. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med 366: 2455–2465. 10.1056/NEJMoa1200694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bröske AM, Vockentanz L, Kharazi S, Huska MR, Mancini E, Scheller M, Kuhl C, Enns A, Prinz M, Jaenisch R, et al. 2009. DNA methylation protects hematopoietic stem cell multipotency from myeloerythroid restriction. Nat Genet 41: 1207–1215. 10.1038/ng.463 [DOI] [PubMed] [Google Scholar]
- Bunting KL, Soong TD, Singh R, Jiang Y, Béguelin W, Poloway DW, Swed BL, Hatzi K, Reisacher W, Teater M, et al. 2016. Multi-tiered reorganization of the genome during B cell affinity maturation anchored by a germinal center-specific locus control region. Immunity 45: 497–512. 10.1016/j.immuni.2016.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caganova M, Carrisi C, Varano G, Mainoldi F, Zanardi F, Germain PL, George L, Alberghini F, Ferrarini L, Talukder AK, et al. 2013. Germinal center dysregulation by histone methyltransferase EZH2 promotes lymphomagenesis. J Clin Invest 123: 5009–5022. 10.1172/JCI70626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caligiuri MA, Strout MP, Lawrence D, Arthur DC, Baer MR, Yu F, Knuutila S, Mrozek K, Oberkircher AR, Marcucci G, et al. 1998. Rearrangement of ALL1 (MLL) in acute myeloid leukemia with normal cytogenetics. Cancer Res 58: 55–59. [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. 2013. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 368: 2059–2074. 10.1056/NEJMoa1301689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cato MH, Chintalapati SK, Yau IW, Omori SA, Rickert RC. 2011. Cyclin D3 is selectively required for proliferative expansion of germinal center B cells. Mol Cell Biol 31: 127–137. 10.1128/MCB.00650-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerchietti LC, Lopes EC, Yang SN, Hatzi K, Bunting KL, Tsikitas LA, Mallik A, Robles AI, Walling J, Varticovski L, et al. 2009. A purine scaffold Hsp90 inhibitor destabilizes BCL-6 and has specific antitumor activity in BCL-6–dependent B cell lymphomas. Nat Med 15: 1369–1376. 10.1038/nm.2059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerchietti LC, Hatzi K, Caldas-Lopes E, Yang SN, Figueroa ME, Morin RD, Hirst M, Mendez L, Shaknovich R, Cole PA, et al. 2010. BCL6 repression of EP300 in human diffuse large B cell lymphoma cells provides a basis for rational combinatorial therapy. J Clin Invest 120: 4569–4582. 10.1172/JCI42869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challen GA, Sun D, Jeong M, Luo M, Jelinek J, Berg JS, Bock C, Vasanthakumar A, Gu H, Xi Y, et al. 2011. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat Genet 44: 23–31. 10.1038/ng.1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambwe N, Kormaksson M, Geng H, De S, Michor F, Johnson NA, Morin RD, Scott DW, Godley LA, Gascoyne RD, et al. 2014. Variability in DNA methylation defines novel epigenetic subgroups of DLBCL associated with different clinical outcomes. Blood 123: 1699–1708. 10.1182/blood-2013-07-509885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang E, Ganguly S, Rajkhowa T, Gocke CD, Levis M, Konig H. 2016. The combination of FLT3 and DNA methyltransferase inhibition is synergistically cytotoxic to FLT3/ITD acute myeloid leukemia cells. Leukemia 30: 1025–1032. 10.1038/leu.2015.346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapuy B, McKeown MR, Lin CY, Monti S, Roemer MG, Qi J, Rahl PB, Sun HH, Yeda KT, Doench JG, et al. 2013. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer Cell 24: 777–790. 10.1016/j.ccr.2013.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapuy B, Stewart C, Dunford AJ, Kim J, Kamburov A, Redd RA, Lawrence MS, Roemer MGM, Li AJ, Ziepert M, et al. 2018. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat Med 24: 679–690. 10.1038/s41591-018-0016-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Zhu H, Stauffer F, Caravatti G, Vollmer S, Machauer R, Holzer P, Möbitz H, Scheufler C, Klumpp M, et al. 2016. Discovery of novel Dot1L inhibitors through a structure-based fragmentation approach. ACS Med Chem Lett 7: 735–740. 10.1021/acsmedchemlett.6b00167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chittock EC, Latwiel S, Miller TC, Müller CW. 2017. Molecular architecture of Polycomb repressive complexes. Biochem Soc Trans 45: 193–205. 10.1042/BST20160173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou WC, Chou SC, Liu CY, Chen CY, Hou HA, Kuo YY, Lee MC, Ko BS, Tang JL, Yao M, et al. 2011. TET2 mutation is an unfavorable prognostic factor in acute myeloid leukemia patients with intermediate-risk cytogenetics. Blood 118: 3803–3810. 10.1182/blood-2011-02-339747 [DOI] [PubMed] [Google Scholar]
- Chowdhury R, Yeoh KK, Tian YM, Hillringhaus L, Bagg EA, Rose NR, Leung IK, Li XS, Woon EC, Yang M, et al. 2011. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep 12: 463–469. 10.1038/embor.2011.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimmino L, Dolgalev I, Wang Y, Yoshimi A, Martin GH, Wang J, Ng V, Xia B, Witkowski MT, Mitchell-Flack M, et al. 2017. Restoration of TET2 function blocks aberrant self-renewal and leukemia progression. Cell 170: 1079–1095.e20. 10.1016/j.cell.2017.07.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clozel T, Yang S, Elstrom RL, Tam W, Martin P, Kormaksson M, Banerjee S, Vasanthakumar A, Culjkovic B, Scott DW, et al. 2013. Mechanism-based epigenetic chemosensitization therapy of diffuse large B-cell lymphoma. Cancer Discov 3: 1002–1019. 10.1158/2159-8290.CD-13-0117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coudé M-M, Braun T, Berrou J, Dupont M, Bertrand S, Masse A, Raffoux E, Itzykson R, Delord M, Riveiro ME, et al. 2015. BET inhibitor OTX015 targets BRD2 and BRD4 and decreases c-MYC in acute leukemia cells. Oncotarget 6: 17698–17712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, et al. 2009. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462: 739–744. 10.1038/nature08617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daver N, Garcia-Manero G, Basu S, Boddu PC, Alfayez M, Cortes JE, Konopleva M, Ravandi-Kashani F, Jabbour E, Kadia T, et al. 2019. Efficacy, safety, and biomarkers of response to azacitidine and nivolumab in relapsed/refractory acute myeloid leukemia: a nonrandomized, open-label, phase II study. Cancer Discov 9: 370–383. 10.1158/2159-8290.CD-18-0774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De S, Shaknovich R, Riester M, Elemento O, Geng H, Kormaksson M, Jiang Y, Woolcock B, Johnson N, Polo JM, et al. 2013. Aberration in DNA methylation in B-cell lymphomas has a complex origin and increases with disease severity. PLoS Genet 9: e1003137 10.1371/journal.pgen.1003137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhommeau F, Dupont S, Valle VD, James C, Trannoy S, Massé A, Kosmider O, Le Couedic JP, Robert F, Alberdi A, et al. 2009. Mutation in TET2 in myeloid cancers. N Engl J Med 360: 2289–2301. 10.1056/NEJMoa0810069 [DOI] [PubMed] [Google Scholar]
- DiNardo CD, Stein EM, de Botton S, Roboz GJ, Altman JK, Mims AS, Swords R, Collins RH, Mannis GN, Pollyea DA, et al. 2018. Durable remissions with ivosidenib in IDH1-mutated relapsed or refractory AML. N Engl J Med 378: 2386–2398. 10.1056/NEJMoa1716984 [DOI] [PubMed] [Google Scholar]
- DiNardo CD, Pratz K, Pullarkat V, Jonas BA, Arellano M, Becker PS, Frankfurt O, Konopleva M, Wei AH, Kantarjian HM, et al. 2019. Venetoclax combined with decitabine or azacitidine in treatment-naive, elderly patients with acute myeloid leukemia. Blood 133: 7–17. 10.1182/blood-2018-08-868752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, Ritchey JK, Young MA, Lamprecht T, McLellan M, et al. 2012. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 481: 506–510. 10.1038/nature10738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez PM, Teater M, Chambwe N, Kormaksson M, Redmond D, Ishii J, Vuong B, Chaudhuri J, Melnick A, Vasanthakumar A, et al. 2015. DNA methylation dynamics of germinal center B cells are mediated by AID. Cell Rep 12: 2086–2098. 10.1016/j.celrep.2015.08.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez PM, Ghamlouch H, Rosikiewicz W, Kumar P, Beguelin W, Fontan L, Rivas MA, Pawlikowska P, Armand M, Mouly E, et al. 2018. TET2 deficiency causes germinal center hyperplasia, impairs plasma cell differentiation, and promotes B-cell lymphomagenesis. Cancer Discov 8: 1632–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorrance AM, Liu S, Chong A, Pulley B, Nemer D, Guimond M, Yuan W, Chang D, Whitman SP, Marcucci G, et al. 2008. The Mll partial tandem duplication: differential, tissue-specific activity in the presence or absence of the wild-type allele. Blood 112: 2508–2511. 10.1182/blood-2008-01-134338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doughty CA, Bleiman BF, Wagner DJ, Dufort FJ, Mataraza JM, Roberts MF, Chiles TC. 2006. Antigen receptor-mediated changes in glucose metabolism in B lymphocytes: role of phosphatidylinositol 3-kinase signaling in the glycolytic control of growth. Blood 107: 4458–4465. 10.1182/blood-2005-12-4788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duy C, Melnick A. 2018. Untangling the role of Polycomb complexes in chemotherapy resistance. Cancer Discov 8: 1348–1351. 10.1158/2159-8290.CD-18-1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duy C, Teater M, Garrett-Bakelman FE, Lee TC, Meydan C, Glass JL, Li M, Hellmuth JC, Mohammad HP, Smitheman KN, et al. 2019. Rational targeting of cooperating layers of the epigenome yields enhanced therapeutic efficacy against AML. Cancer Discov 9: 872–889. 10.1158/2159-8290.CD-19-0106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ennishi D, Takata K, Béguelin W, Duns G, Mottok A, Farinha P, Bashashati A, Saberi S, Boyle M, Meissner B, et al. 2019. Molecular and genetic characterization of MHC deficiency identifies EZH2 as therapeutic target for enhancing immune recognition. Cancer Discov 9: 546–563. 10.1158/2159-8290.CD-18-1090 [DOI] [PubMed] [Google Scholar]
- Ernst T, Chase AJ, Score J, Hidalgo-Curtis CE, Bryant C, Jones AV, Waghorn K, Zoi K, Ross FM, Reiter A, et al. 2010. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet 42: 722–726. 10.1038/ng.621 [DOI] [PubMed] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, et al. 2011. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473: 43–49. 10.1038/nature09906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst T, Pflug A, Rinke J, Ernst J, Bierbach U, Beck JF, Hochhaus A, Gruhn B. 2012. A somatic EZH2 mutation in childhood acute myeloid leukemia. Leukemia 26: 1701–1703. 10.1038/leu.2012.16 [DOI] [PubMed] [Google Scholar]
- Fedoriw A, Rajapurkar SR, O'Brien S, Gerhart SV, Mitchell LH, Adams ND, Rioux N, Lingaraj T, Ribich SA, Pappalardi MB, et al. 2019. Anti-tumor activity of the type I PRMT inhibitor, GSK3368715, synergizes with PRMT5 inhibition through MTAP loss. Cancer Cell 36: 100–114.e25. 10.1016/j.ccell.2019.05.014 [DOI] [PubMed] [Google Scholar]
- Fennell KA, Bell CC, Dawson MA. 2019. Epigenetic therapies in acute myeloid leukemia: where to from here? Blood 134: 1891–1901. 10.1182/blood.2019003262 [DOI] [PubMed] [Google Scholar]
- Fernando TM, Marullo R, Pera Gresely B, Phillip JM, Yang SN, Lundell-Smith G, Torregroza I, Ahn H, Evans T, Győrffy B, et al. 2019. BCL6 evolved to enable stress tolerance in vertebrates and is broadly required by cancer cells to adapt to stress. Cancer Discov 9: 662–679. 10.1158/2159-8290.CD-17-1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa ME, Wouters BJ, Skrabanek L, Glass J, Li Y, Erpelinck-Verschueren CA, Langerak AW, Löwenberg B, Fazzari M, Greally JM, et al. 2009. Genome-wide epigenetic analysis delineates a biologically distinct immature acute leukemia with myeloid/T-lymphoid features. Blood 113: 2795–2804. 10.1182/blood-2008-08-172387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, et al. 2010a. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 18: 553–567. 10.1016/j.ccr.2010.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa ME, Lugthart S, Li Y, Erpelinck-Verschueren C, Deng X, Christos PJ, Schifano E, Booth J, van Putten W, Skrabanek L, et al. 2010b. DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell 17: 13–27. 10.1016/j.ccr.2009.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florence B, Faller DV. 2001. You bet-cha: a novel family of transcriptional regulators. Front Biosci 6: D1008–D1018. [DOI] [PubMed] [Google Scholar]
- Fugmann SD, Lee AI, Shockett PE, Villey IJ, Schatz DG. 2000. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu Rev Immunol 18: 495–527. 10.1146/annurev.immunol.18.1.495 [DOI] [PubMed] [Google Scholar]
- Gaidzik VI, Paschka P, Späth D, Habdank M, Köhne CH, Germing U, von Lilienfeld-Toal M, Held G, Horst HA, Haase D, et al. 2012. TET2 mutations in acute myeloid leukemia (AML): results from a comprehensive genetic and clinical analysis of the AML study group. J Clin Oncol 30: 1350–1357. 10.1200/JCO.2011.39.2886 [DOI] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. 2013. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6: l1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Ramírez I, Tadros S, González-Herrero I, Martin-Lorenzo A, Rodriguez-Hernández G, Moore D, Ruiz-Roca L, Blanco O, Alonso-López D, Rivas JL, et al. 2017. Crebbp loss cooperates with Bcl2 overexpression to promote lymphoma in mice. Blood 129: 2645–2656. 10.1182/blood-2016-08-733469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardin C, Dombret H. 2017. Hypomethylating agents as a therapy for AML. Curr Hematol Malig Rep 12: 1–10. 10.1007/s11899-017-0363-4 [DOI] [PubMed] [Google Scholar]
- Ghetu AF, Corcoran CM, Cerchietti L, Bardwell VJ, Melnick A, Privé GG. 2008. Structure of a BCOR corepressor peptide in complex with the BCL6 BTB domain dimer. Mol Cell 29: 384–391. 10.1016/j.molcel.2007.12.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass JL, Hassane D, Wouters BJ, Kunimoto H, Avellino R, Garrett-Bakelman FE, Guryanova OA, Bowman R, Redlich S, Intlekofer AM, et al. 2017. Epigenetic identity in AML depends on disruption of nonpromoter regulatory elements and is affected by antagonistic effects of mutations in epigenetic modifiers. Cancer Discov 7: 868–883. 10.1158/2159-8290.CD-16-1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göllner S, Oellerich T, Agrawal-Singh S, Schenk T, Klein HU, Rohde C, Pabst C, Sauer T, Lerdrup M, Tavor S, et al. 2017. Loss of the histone methyltransferase EZH2 induces resistance to multiple drugs in acute myeloid leukemia. Nat Med 23: 69–78. 10.1038/nm.4247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good-Jacobson KL, Szumilas CG, Chen L, Sharpe AH, Tomayko MM, Shlomchik MJ. 2010. PD-1 regulates germinal center B cell survival and the formation and affinity of long-lived plasma cells. Nat Immunol 11: 535–542. 10.1038/ni.1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozdecka M, Meduri E, Mazan M, Tzelepis K, Dudek M, Knights AJ, Pardo M, Yu L, Choudhary JS, Metzakopian E, et al. 2018. UTX-mediated enhancer and chromatin remodeling suppresses myeloid leukemogenesis through noncatalytic inverse regulation of ETS and GATA programs. Nat Genet 50: 883–894. 10.1038/s41588-018-0114-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green MR. 2018. Chromatin modifying gene mutations in follicular lymphoma. Blood 131: 595–604. 10.1182/blood-2017-08-737361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green MR, Kihira S, Liu CL, Nair RV, Salari R, Gentles AJ, Irish J, Stehr H, Vicente-Dueñas C, Romero-Camarero I, et al. 2015. Mutations in early follicular lymphoma progenitors are associated with suppressed antigen presentation. Proc Natl Acad Sci 112: E1116–E1125. 10.1073/pnas.1501199112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimwade D, Walker H, Harrison G, Oliver F, Chatters S, Harrison CJ, Wheatley K, Burnett AK, Goldstone AH. 2001. The predictive value of hierarchical cytogenetic classification in older adults with acute myeloid leukemia (AML): analysis of 1065 patients entered into the United Kingdom Medical Research Council AML11 trial. Blood 98: 1312–1320. 10.1182/blood.V98.5.1312 [DOI] [PubMed] [Google Scholar]
- Gross S, Cairns RA, Minden MD, Driggers EM, Bittinger MA, Jang HG, Sasaki M, Jin S, Schenkein DP, Su SM, et al. 2010. Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations. J Exp Med 207: 339–344. 10.1084/jem.20092506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossmann V, Tiacci E, Holmes AB, Kohlmann A, Martelli MP, Kern W, Spanhol-Rosseto A, Klein HU, Dugas M, Schindela S, et al. 2011. Whole-exome sequencing identifies somatic mutations of BCOR in acute myeloid leukemia with normal karyotype. Blood 118: 6153–6163. 10.1182/blood-2011-07-365320 [DOI] [PubMed] [Google Scholar]
- Gulati N, Béguelin W, Giulino-Roth L. 2018. Enhancer of zeste homolog 2 (EZH2) inhibitors. Leuk Lymphoma 59: 1574–1585. 10.1080/10428194.2018.1430795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guryanova OA, Shank K, Spitzer B, Luciani L, Koche RP, Garrett-Bakelman FE, Ganzel C, Durham BH, Mohanty A, Hoermann G, et al. 2016. DNMT3A mutations promote anthracycline resistance in acute myeloid leukemia via impaired nucleosome remodeling. Nat Med 22: 1488–1495. 10.1038/nm.4210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haarhuis JH, Elbatsh AM, Rowland BD. 2014. Cohesin and its regulation: on the logic of X-shaped chromosomes. Dev Cell 31: 7–18. 10.1016/j.devcel.2014.09.010 [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. 2011. Hallmarks of cancer: the next generation. Cell 144: 646–674. 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- Hansen KD, Timp W, Bravo HC, Sabunciyan S, Langmead B, McDonald OG, Wen B, Wu H, Liu Y, Diep D, et al. 2011. Increased methylation variation in epigenetic domains across cancer types. Nat Genet 43: 768–775. 10.1038/ng.865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding JJ, Lowery MA, Shih AH, Schvartzman JM, Hou S, Famulare C, Patel M, Roshal M, Do RK, Zehir A, et al. 2018. Isoform switching as a mechanism of acquired resistance to mutant isocitrate dehydrogenase inhibition. Cancer Discov 8: 1540–1547. 10.1158/2159-8290.CD-18-0877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris WJ, Huang X, Lynch JT, Spencer GJ, Hitchin JR, Li Y, Ciceri F, Blaser JG, Greystoke BF, Jordan AM, et al. 2012. The histone demethylase KDM1A sustains the oncogenic potential of MLL-AF9 leukemia stem cells. Cancer Cell 21: 473–487. 10.1016/j.ccr.2012.03.014 [DOI] [PubMed] [Google Scholar]
- Hashimoto H, Liu Y, Upadhyay AK, Chang Y, Howerton SB, Vertino PM, Zhang X, Cheng X. 2012. Recognition and potential mechanisms for replication and erasure of cytosine hydroxymethylation. Nucleic Acids Res 40: 4841–4849. 10.1093/nar/gks155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashwah H, Schmid CA, Kasser S, Bertram K, Stelling A, Manz MG, Müller A. 2017. Inactivation of CREBBP expands the germinal center B cell compartment, down-regulates MHCII expression and promotes DLBCL growth. Proc Natl Acad Sci 114: 9701–9706. 10.1073/pnas.1619555114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzi K, Melnick A. 2014. Breaking bad in the germinal center: how deregulation of BCL6 contributes to lymphomagenesis. Trends Mol Med 20: 343–352. 10.1016/j.molmed.2014.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzi K, Jiang Y, Huang C, Garrett-Bakelman F, Gearhart MD, Giannopoulou EG, Zumbo P, Kirouac K, Bhaskara S, Polo JM, et al. 2013. A hybrid mechanism of action for BCL6 in B cells defined by formation of functionally distinct complexes at enhancers and promoters. Cell Rep 4: 578–588. 10.1016/j.celrep.2013.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzi K, Geng H, Doane AS, Meydan C, LaRiviere R, Cardenas M, Duy C, Shen H, Vidal MNC, Baslan T, et al. 2019. Histone demethylase LSD1 is required for germinal center formation and BCL6-driven lymphomagenesis. Nat Immunol 20: 86–96. 10.1038/s41590-018-0273-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, et al. 2011. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333: 1303–1307. 10.1126/science.1210944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera E, Martinez-A C, Blasco MA. 2000. Impaired germinal center reaction in mice with short telomeres. EMBO J 19: 472–481. 10.1093/emboj/19.3.472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollink IH, van den Heuvel-Eibrink MM, Arentsen-Peters ST, Pratcorona M, Abbas S, Kuipers JE, van Galen JF, Beverloo HB, Sonneveld E, Kaspers GJ, et al. 2011. NUP98/NSD1 characterizes a novel poor prognostic group in acute myeloid leukemia with a distinct HOX gene expression pattern. Blood 118: 3645–3656. 10.1182/blood-2011-04-346643 [DOI] [PubMed] [Google Scholar]
- Hon GC, Song CX, Du T, Jin F, Selvaraj S, Lee AY, Yen CA, Ye Z, Mao SQ, Wang BA, et al. 2014. 5mC oxidation by Tet2 modulates enhancer activity and timing of transcriptome reprogramming during differentiation. Mol Cell 56: 286–297. 10.1016/j.molcel.2014.08.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu BT, Lee SC, Marin E, Ryan DH, Insel RA. 1997. Telomerase is up-regulated in human germinal center B cells in vivo and can be re-expressed in memory B cells activated in vitro. J Immunol 159: 1068–1071. [PubMed] [Google Scholar]
- Huynh KD, Fischle W, Verdin E, Bardwell VJ. 2000. BCoR, a novel corepressor involved in BCL-6 repression. Genes Dev 14: 1810–1823. [PMC free article] [PubMed] [Google Scholar]
- Inoue S, Li WY, Tseng A, Beerman I, Elia AJ, Bendall SC, Lemonnier F, Kron KJ, Cescon DW, Hao Z, et al. 2016. Mutant IDH1 downregulates ATM and alters DNA repair and sensitivity to DNA damage independent of TET2. Cancer Cell 30: 337–348. 10.1016/j.ccell.2016.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intlekofer AM, Shih AH, Wang B, Nazir A, Rustenburg AS, Albanese SK, Patel M, Famulare C, Correa FM, Takemoto N, et al. 2018. Acquired resistance to IDH inhibition through trans or cis dimer-interface mutations. Nature 559: 125–129. 10.1038/s41586-018-0251-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issa JJ, Roboz G, Rizzieri D, Jabbour E, Stock W, O'Connell C, Yee K, Tibes R, Griffiths EA, Walsh K, et al. 2015. Safety and tolerability of guadecitabine (SGI-110) in patients with myelodysplastic syndrome and acute myeloid leukaemia: a multicentre, randomised, dose-escalation phase 1 study. Lancet Oncol 16: 1099–1110. 10.1016/S1470-2045(15)00038-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. 2011. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333: 1300–1303. 10.1126/science.1210597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jellusova J, Cato MH, Apgar JR, Ramezani-Rad P, Leung CR, Chen C, Richardson AD, Conner EM, Benschop RJ, Woodgett JR, et al. 2017. GSK3 is a metabolic checkpoint regulator in B cells. Nat Immunol 18: 303–312. 10.1038/ni.3664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong M, Sun D, Luo M, Huang Y, Challen GA, Rodriguez B, Zhang X, Chavez L, Wang H, Hannah R, et al. 2014. Large conserved domains of low DNA methylation maintained by Dnmt3a. Nat Genet 46: 17–23. 10.1038/ng.2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Ehrlich LIR, Seita J, Murakami P, Doi A, Lindau P, Lee H, Aryee MJ, Irizarry RA, Kim K, et al. 2010. Comprehensive methylome map of lineage commitment from haematopoietic progenitors. Nature 467: 338–342. 10.1038/nature09367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Ortega-Molina A, Geng H, Ying HY, Hatzi K, Parsa S, McNally D, Wang L, Doane AS, Agirre X, et al. 2017. CREBBP inactivation promotes the development of HDAC3-dependent lymphomas. Cancer Discov 7: 38–53. 10.1158/2159-8290.CD-16-0975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. 2007. The epigenomics of cancer. Cell 128: 683–692. 10.1016/j.cell.2007.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. 2010. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467: 430–435. 10.1038/nature09380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JS, He X, Orr B, Wutz G, Hill V, Peters JM, Compton DA, Waldman T. 2016. Intact cohesion, anaphase, and chromosome segregation in human cells harboring tumor-derived mutations in STAG2. PLoS Genet 12: e1005865 10.1371/journal.pgen.1005865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kon A, Shih LY, Minamino M, Sanada M, Shiraishi Y, Nagata Y, Yoshida K, Okuno Y, Bando M, Nakato R, et al. 2013. Recurrent mutations in multiple components of the cohesin complex in myeloid neoplasms. Nat Genet 45: 1232–1237. 10.1038/ng.2731 [DOI] [PubMed] [Google Scholar]
- Kridel R, Sehn LH, Gascoyne RD. 2012. Pathogenesis of follicular lymphoma. J Clin Invest 122: 3424–3431. 10.1172/JCI63186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivtsov AV, Armstrong SA. 2007. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer 7: 823–833. 10.1038/nrc2253 [DOI] [PubMed] [Google Scholar]
- Landau DA, Clement K, Ziller MJ, Boyle P, Fan J, Gu H, Stevenson K, Sougnez C, Wang L, Li S, et al. 2014. Locally disordered methylation forms the basis of intratumor methylome variation in chronic lymphocytic leukemia. Cancer Cell 26: 813–825. 10.1016/j.ccell.2014.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau DA, Tausch E, Taylor-Weiner AN, Stewart C, Reiter JG, Bahlo J, Kluth S, Bozic I, Lawrence M, Böttcher S, et al. 2015. Mutations driving CLL and their evolution in progression and relapse. Nature 526: 525–530. 10.1038/nature15395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, Carter SL, Stewart C, Mermel CH, Roberts SA, et al. 2013. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499: 214–218. 10.1038/nature12213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz G, Davis RE, Ngo VN, Lam L, George TC, Wright GW, Dave SS, Zhao H, Xu W, Rosenwald A, et al. 2008. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science 319: 1676–1679. 10.1126/science.1153629 [DOI] [PubMed] [Google Scholar]
- Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, Kandoth C, Payton JE, Baty J, Welch J, et al. 2010. DNMT3A mutations in acute myeloid leukemia. N Engl J Med 363: 2424–2433. 10.1056/NEJMoa1005143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Garrett-Bakelman FE, Chung SS, Sanders MA, Hricik T, Rapaport F, Patel J, Dillon R, Vijay P, Brown AL, et al. 2016. Distinct evolution and dynamics of epigenetic and genetic heterogeneity in acute myeloid leukemia. Nat Med 22: 792–799. 10.1038/nm.4125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Zhang P, Yan A, Guo Z, Ban Y, Li J, Chen S, Yang H, He Y, Li J, et al. 2017. ASXL1 interacts with the cohesin complex to maintain chromatid separation and gene expression for normal hematopoiesis. Sci Adv 3: e1601602 10.1126/sciadv.1601602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love C, Sun Z, Jima D, Li G, Zhang J, Miles R, Richards KL, Dunphy CH, Choi WW, Srivastava G, et al. 2012. The genetic landscape of mutations in Burkitt lymphoma. Nat Genet 44: 1321–1325. 10.1038/ng.2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, Edwards CR, Khanin R, Figueroa ME, Melnick A, et al. 2012. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature 483: 474–478. 10.1038/nature10860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugthart S, Figueroa ME, Bindels E, Skrabanek L, Valk PJ, Li Y, Meyer S, Erpelinck-Verschueren C, Greally J, Lowenberg B, et al. 2011. Aberrant DNA hypermethylation signature in acute myeloid leukemia directed by EVI1. Blood 117: 234–241. 10.1182/blood-2010-04-281337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes T, Mascaró C, Tirapu I, Estiarte A, Ciceri F, Lunardi S, Guibourt N, Perdones A, Lufino MMP, Somervaille TCP, et al. 2018. ORY-1001, a potent and selective covalent KDM1A inhibitor, for the treatment of acute leukemia. Cancer Cell 33: 495–511.e12. 10.1016/j.ccell.2018.02.002 [DOI] [PubMed] [Google Scholar]
- Maganti HB, Jrade H, Cafariello C, Manias Rothberg JL, Porter CJ, Yockell-Lelièvre J, Battaion HL, Khan ST, Howard JP, Li Y, et al. 2018. Targeting the MTF2–MDM2 axis sensitizes refractory acute myeloid leukemia to chemotherapy. Cancer Discov 8: 1376–1389. 10.1158/2159-8290.CD-17-0841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti A, Drohat AC. 2011. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites. J Biol Chem 286: 35334–35338. 10.1074/jbc.C111.284620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcucci G, Maharry K, Wu YZ, Radmacher MD, Mrózek K, Margeson D, Holland KB, Whitman SP, Becker H, Schwind S, et al. 2010. IDH1 and IDH2 gene mutations identify novel molecular subsets within de novo cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. J Clin Oncol 28: 2348–2355. 10.1200/JCO.2009.27.3730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcucci G, Metzeler KH, Schwind S, Becker H, Maharry K, Mrozek K, Radmacher MD, Kohlschmidt J, Nicolet D, Whitman SP, et al. 2012. Age-related prognostic impact of different types of DNMT3A mutations in adults with primary cytogenetically normal acute myeloid leukemia. J Clin Oncol 30: 742–750. 10.1200/JCO.2011.39.2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, Koboldt DC, Fulton RS, Delehaunty KD, McGrath SD, et al. 2009. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med 361: 1058–1066. 10.1056/NEJMoa0903840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. 2011. The Polycomb complex PRC2 and its mark in life. Nature 469: 343–349. 10.1038/nature09784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayle A, Yang L, Rodriguez B, Zhou T, Chang E, Curry CV, Challen GA, Li W, Wheeler D, Rebel VI, et al. 2015. Dnmt3a loss predisposes murine hematopoietic stem cells to malignant transformation. Blood 125: 629–638. 10.1182/blood-2014-08-594648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe MT, Graves AP, Ganji G, Diaz E, Halsey WS, Jiang Y, Smitheman KN, Ott HM, Pappalardi MB, Allen KE, et al. 2012. Mutation of A677 in histone methyltransferase EZH2 in human B-cell lymphoma promotes hypertrimethylation of histone H3 on lysine 27 (H3K27). Proc Natl Acad Sci 109: 2989–2994. 10.1073/pnas.1116418109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath JP, Williamson KE, Balasubramanian S, Odate S, Arora S, Hatton C, Edwards TM, O'Brien T, Magnuson S, Stokoe D, et al. 2016. Pharmacological inhibition of the histone lysine demethylase KDM1A suppresses the growth of multiple acute myeloid leukemia subtypes. Cancer Res 76: 1975–1988. 10.1158/0008-5472.CAN-15-2333 [DOI] [PubMed] [Google Scholar]
- McHeyzer-Williams LJ, Milpied PJ, Okitsu SL, McHeyzer-Williams MG. 2015. Class-switched memory B cells remodel BCRs within secondary germinal centers. Nat Immunol 16: 296–305. 10.1038/ni.3095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesin L, Ersching J, Victora GD. 2016. Germinal center B cell dynamics. Immunity 45: 471–482. 10.1016/j.immuni.2016.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzeler KH, Herold T, Rothenberg-Thurley M, Amler S, Sauerland MC, Görlich D, Schneider S, Konstandin NP, Dufour A, Bräundl K, et al. 2016. Spectrum and prognostic relevance of driver gene mutations in acute myeloid leukemia. Blood 128: 686–698. 10.1182/blood-2016-01-693879 [DOI] [PubMed] [Google Scholar]
- Mohammad HP, Smitheman KN, Kamat CD, Soong D, Federowicz KE, Van Aller GS, Schneck JL, Carson JD, Liu Y, Butticello M, et al. 2015. A DNA hypomethylation signature predicts antitumor activity of LSD1 inhibitors in SCLC. Cancer Cell 28: 57–69. 10.1016/j.ccell.2015.06.002 [DOI] [PubMed] [Google Scholar]
- Mondello P, Toska E, Teater M, Fontan L, Durant M, Casalena G, De Stanchina E, Inghirami G, Baselga J, Weill Melnick A. 2018. Targeting KDM5 demethylases counteracts KMT2D loss of function in diffuse large B-cell lymphoma. In AACR advances in malignant lymphoma, Boston AACR, Philadelphia. [Google Scholar]
- Mondello P, Tadros S, Teater M, Fontan L, Chang A, Jain N, Singh S, Ma MCJ, Yang H, Toska E, et al. 2019. Selective inhibition of HDAC3 targets synthetic vulnerabilities and activates immune surveillance in lymphoma. Cancer Discov. 10.1158/2159-8290.CD-19-0116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C, Figueroa ME, Vasanthakumar A, Patel J, Zhao X, et al. 2011. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell 20: 11–24. 10.1016/j.ccr.2011.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin RD, Johnson NA, Severson TM, Mungall AJ, An J, Goya R, Paul JE, Boyle M, Woolcock BW, Kuchenbauer F, et al. 2010. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet 42: 181–185. 10.1038/ng.518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin RD, Mendez-Lago M, Mungall AJ, Goya R, Mungall KL, Corbett RD, Johnson NA, Severson TM, Chiu R, Field M, et al. 2011. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature 476: 298–303. 10.1038/nature10351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullenders J, Aranda-Orgilles B, Lhoumaud P, Keller M, Pae J, Wang K, Kayembe C, Rocha PP, Raviram R, Gong Y, et al. 2015. Cohesin loss alters adult hematopoietic stem cell homeostasis, leading to myeloproliferative neoplasms. J Exp Med 212: 1833–1850. 10.1084/jem.20151323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullighan CG. 2012. Molecular genetics of B-precursor acute lymphoblastic leukemia. J Clin Invest 122: 3407–3415. 10.1172/JCI61203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muppidi MR, Portwood S, Griffiths EA, Thompson JE, Ford LA, Freyer CW, Wetzler M, Wang ES. 2015. Decitabine and sorafenib therapy in FLT-3 ITD-mutant acute myeloid leukemia. Clin Lymphoma Myeloma Leuk 15: S73–S79. 10.1016/j.clml.2015.02.033 [DOI] [PubMed] [Google Scholar]
- Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. 2000. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell 102: 553–563. 10.1016/S0092-8674(00)00078-7 [DOI] [PubMed] [Google Scholar]
- Nabel CS, Jia H, Ye Y, Shen L, Goldschmidt HL, Stivers JT, Zhang Y, Kohli RM. 2012. AID/APOBEC deaminases disfavor modified cytosines implicated in DNA demethylation. Nat Chem Biol 8: 751–758. 10.1038/nchembio.1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo VN, Young RM, Schmitz R, Jhavar S, Xiao W, Lim KH, Kohlhammer H, Xu W, Yang Y, Zhao H, et al. 2011. Oncogenically active MYD88 mutations in human lymphoma. Nature 470: 115–119. 10.1038/nature09671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo TT, Yoo J, Dai Q, Zhang Q, He C, Aksimentiev A, Ha T. 2016. Effects of cytosine modifications on DNA flexibility and nucleosome mechanical stability. Nat Commun 7: 10813 10.1038/ncomms10813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas NS, Apollonio B, Ramsay AG. 2016. Tumor microenvironment (TME)-driven immune suppression in B cell malignancy. Biochim Biophys Acta 1863: 471–482. 10.1016/j.bbamcr.2015.11.003 [DOI] [PubMed] [Google Scholar]
- Norrback KF, Hultdin M, Dahlenborg K, Osterman P, Carlsson R, Roos G. 2001. Telomerase regulation and telomere dynamics in germinal centers. Eur J Haematol 67: 309–317. 10.1034/j.1600-0609.2001.00588.x [DOI] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99: 247–257. 10.1016/S0092-8674(00)81656-6 [DOI] [PubMed] [Google Scholar]
- Okosun J, Bödör C, Wang J, Araf S, Yang CY, Pan C, Boller S, Cittaro D, Bozek M, Iqbal S, et al. 2014. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat Genet 46: 176–181. 10.1038/ng.2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega-Molina A, Boss IW, Canela A, Pan H, Jiang Y, Zhao C, Jiang M, Hu D, Agirre X, Niesvizky I, et al. 2015. The histone lysine methyltransferase KMT2D sustains a gene expression program that represses B cell lymphoma development. Nat Med 21: 1199–1208. 10.1038/nm.3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan H, Jiang Y, Boi M, Tabbò F, Redmond D, Nie K, Ladetto M, Chiappella A, Cerchietti L, Shaknovich R, et al. 2015. Epigenomic evolution in diffuse large B-cell lymphomas. Nat Commun 6: 6921 10.1038/ncomms7921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa I, Vinuesa CG. 2018. Synaptic interactions in germinal centers. Front Immunol 9: 1858 10.3389/fimmu.2018.01858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, Potter NE, Heuser M, Thol F, Bolli N, et al. 2016. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med 374: 2209–2221. 10.1056/NEJMoa1516192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschka P, Schlenk RF, Gaidzik VI, Habdank M, Krönke J, Bullinger L, Späth D, Kayser S, Zucknick M, Götze K, et al. 2010. IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. J Clin Oncol 28: 3636–3643. 10.1200/JCO.2010.28.3762 [DOI] [PubMed] [Google Scholar]
- Pasqualucci L, Dalla-Favera R. 2015. The genetic landscape of diffuse large B-cell lymphoma. Semin Hematol 52: 67–76. 10.1053/j.seminhematol.2015.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualucci L, Dominguez-Sola D, Chiarenza A, Fabbri G, Grunn A, Trifonov V, Kasper LH, Lerach S, Tang H, Ma J, et al. 2011. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature 471: 189–195. 10.1038/nature09730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel JP, Gönen M, Figueroa ME, Fernandez H, Sun Z, Racevskis J, Van Vlierberghe P, Dolgalev I, Thomas S, Aminova O, et al. 2012. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med 366: 1079–1089. 10.1056/NEJMoa1112304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer M, Grau M, Lenze D, Wenzel SS, Wolf A, Wollert-Wulf B, Dietze K, Nogai H, Storek B, Madle H, et al. 2013. PTEN loss defines a PI3K/AKT pathway-dependent germinal center subtype of diffuse large B-cell lymphoma. Proc Natl Acad Sci 110: 12420–12425. 10.1073/pnas.1305656110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan RT, Dalla-Favera R. 2004. The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature 432: 635–639. 10.1038/nature03147 [DOI] [PubMed] [Google Scholar]
- Phan RT, Saito M, Basso K, Niu H, Dalla-Favera R. 2005. BCL6 interacts with the transcription factor Miz-1 to suppress the cyclin-dependent kinase inhibitor p21 and cell cycle arrest in germinal center B cells. Nat Immunol 6: 1054–1060. 10.1038/ni1245 [DOI] [PubMed] [Google Scholar]
- Postow MA, Chesney J, Pavlick AC, Robert C, Grossmann K, McDermott D, Linette GP, Meyer N, Giguere JK, Agarwala SS, et al. 2015. Nivolumab and ipilimumab versus ipilimumab in untreated melanoma. N Engl J Med 372: 2006–2017. 10.1056/NEJMoa1414428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prebet T, Sun Z, Figueroa ME, Ketterling R, Melnick A, Greenberg PL, Herman J, Juckett M, Smith MR, Malick L, et al. 2014. Prolonged administration of azacitidine with or without entinostat for myelodysplastic syndrome and acute myeloid leukemia with myelodysplasia-related changes: results of the US Leukemia Intergroup trial E1905. J Clin Oncol 32: 1242–1248. 10.1200/JCO.2013.50.3102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quek L, David MD, Kennedy A, Metzner M, Amatangelo M, Shih A, Stoilova B, Quivoron C, Heiblig M, Willekens C, et al. 2018. Clonal heterogeneity of acute myeloid leukemia treated with the IDH2 inhibitor enasidenib. Nature Med 24: 1167–1177. 10.1038/s41591-018-0115-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quivoron C, Couronne L, Della Valle V, Lopez CK, Plo I, Wagner-Ballon O, Do Cruzeiro M, Delhommeau F, Arnulf B, Stern MH, et al. 2011. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell 20: 25–38. 10.1016/j.ccr.2011.06.003 [DOI] [PubMed] [Google Scholar]
- Rampal R, Alkalin A, Madzo J, Vasanthakumar A, Pronier E, Patel J, Li Y, Ahn J, Abdel-Wahab O, Shih A, et al. 2014. DNA hydroxymethylation profiling reveals that WT1 mutations result in loss of TET2 function in acute myeloid leukemia. Cell Rep 9: 1841–1855. 10.1016/j.celrep.2014.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranuncolo SM, Polo JM, Dierov J, Singer M, Kuo T, Greally J, Green R, Carroll M, Melnick A. 2007. Bcl-6 mediates the germinal center B cell phenotype and lymphomagenesis through transcriptional repression of the DNA-damage sensor ATR. Nat Immunol 8: 705–714. 10.1038/ni1478 [DOI] [PubMed] [Google Scholar]
- Ranuncolo SM, Polo JM, Melnick A. 2008. BCL6 represses CHEK1 and suppresses DNA damage pathways in normal and malignant B-cells. Blood Cells Mol Dis 41: 95–99. 10.1016/j.bcmd.2008.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen KD, Helin K. 2016. Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev 30: 733–750. 10.1101/gad.276568.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen KD, Jia G, Johansen JV, Pedersen MT, Rapin N, Bagger FO, Porse BT, Bernard OA, Christensen J, Helin K. 2015. Loss of TET2 in hematopoietic cells leads to DNA hypermethylation of active enhancers and induction of leukemogenesis. Genes Dev 29: 910–922. 10.1101/gad.260174.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen KD, Berest I, Kebetaler S, Nishimura K, Simon-Carrasco L, Vassiliou GS, Pedersen MT, Christensen J, Zaugg JB, Helin K. 2019. TET2 binding to enhancers facilitates transcription factor recruitment in hematopoietic cells. Genome Res 29: 564–575. 10.1101/gr.239277.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravandi F, Alattar ML, Grunwald MR, Rudek MA, Rajkhowa T, Richie MA, Pierce S, Daver N, Garcia-Manero G, Faderl S, et al. 2013. Phase 2 study of azacytidine plus sorafenib in patients with acute myeloid leukemia and FLT-3 internal tandem duplication mutation. Blood 121: 4655–4662. 10.1182/blood-2013-01-480228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy A, Zhang J, Davis NS, Moffitt AB, Love CL, Waldrop A, Leppa S, Pasanen A, Meriranta L, Karjalainen-Lindsberg ML, et al. 2017. Genetic and functional drivers of diffuse large B cell lymphoma. Cell 171: 481–494.e15. 10.1016/j.cell.2017.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas MA, Melnick AM. 2019. Role of chromosomal architecture in germinal center B cells and lymphomagenesis. Curr Opin Hematol 26: 294–302. 10.1097/MOH.0000000000000505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert MF, Morin S, Beaulieu N, Gauthier F, Chute IC, Barsalou A, MacLeod AR. 2003. DNMT1 is required to maintain CpG methylation and aberrant gene silencing in human cancer cells. Nat Genet 33: 61–65. 10.1038/ng1068 [DOI] [PubMed] [Google Scholar]
- Robert C, Schachter J, Long GV, Arance A, Grob JJ, Mortier L, Daud A, Carlino MS, McNeil C, Lotem M, et al. 2015. Pembrolizumab versus ipilimumab in advanced melanoma. N Engl J Med 372: 2521–2532. 10.1056/NEJMoa1503093 [DOI] [PubMed] [Google Scholar]
- Rosenwald A, Wright G, Chan WC, Connors JM, Campo E, Fisher RI, Gascoyne RD, Muller-Hermelink HK, Smeland EB, Giltnane JM, et al. 2002. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med 346: 1937–1947. 10.1056/NEJMoa012914 [DOI] [PubMed] [Google Scholar]
- Rossi D, Trifonov V, Fangazio M, Bruscaggin A, Rasi S, Spina V, Monti S, Vaisitti T, Arruga F, Famà R, et al. 2012. The coding genome of splenic marginal zone lymphoma: activation of NOTCH2 and other pathways regulating marginal zone development. J Exp Med 209: 1537–1551. 10.1084/jem.20120904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russler-Germain DA, Spencer DH, Young MA, Lamprecht TL, Miller CA, Fulton R, Meyer MR, Erdmann-Gilmore P, Townsend RR, Wilson RK, et al. 2014. The R882H DNMT3A mutation associated with AML dominantly inhibits wild-type DNMT3A by blocking its ability to form active tetramers. Cancer Cell 25: 442–454. 10.1016/j.ccr.2014.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxonov S, Berg P, Brutlag DL. 2006. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci 103: 1412–1417. 10.1073/pnas.0510310103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk T, Chen WC, Göllner S, Howell L, Jin L, Hebestreit K, Klein HU, Popescu AC, Burnett A, Mills K, et al. 2012. Inhibition of the LSD1 (KDM1A) demethylase reactivates the all-trans-retinoic acid differentiation pathway in acute myeloid leukemia. Nat Med 18: 605–611. 10.1038/nm.2661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schichman SA, Caligiuri MA, Gu Y, Strout MP, Canaani E, Bloomfield CD, Croce CM. 1994. ALL-1 partial duplication in acute leukemia. Proc Natl Acad Sci 91: 6236–6239. 10.1073/pnas.91.13.6236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz R, Wright GW, Huang DW, Johnson CA, Phelan JD, Wang JQ, Roulland S, Kasbekar M, Young RM, Shaffer AL, et al. 2018. Genetics and pathogenesis of diffuse large B-cell lymphoma. N Engl J Med 378: 1396–1407. 10.1056/NEJMoa1801445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sermer D, Pasqualucci L, Wendel HG, Melnick A, Younes A. 2019. Emerging epigenetic-modulating therapies in lymphoma. Nat Rev Clin Oncol 16: 494–507. 10.1038/s41571-019-0190-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaknovich R, Cerchietti L, Tsikitas L, Kormaksson M, De S, Figueroa ME, Ballon G, Yang SN, Weinhold N, Reimers M, et al. 2011. DNA methyltransferase 1 and DNA methylation patterning contribute to germinal center B-cell differentiation. Blood 118: 3559–3569. 10.1182/blood-2011-06-357996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Zhu YM, Fan X, Shi JY, Wang QR, Yan XJ, Gu ZH, Wang YY, Chen B, Jiang CL, et al. 2011. Gene mutation patterns and their prognostic impact in a cohort of 1185 patients with acute myeloid leukemia. Blood 118: 5593–5603. 10.1182/blood-2011-03-343988 [DOI] [PubMed] [Google Scholar]
- Shen L, Wu H, Diep D, Yamaguchi S, D'Alessio AC, Fung HL, Zhang K, Zhang Y. 2013. Genome-wide analysis reveals TET- and TDG-dependent 5-methylcytosine oxidation dynamics. Cell 153: 692–706. 10.1016/j.cell.2013.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shia W-J, Okumura AJ, Yan M, Sarkeshik A, Lo M-C, Matsuura S, Komeno Y, Zhao X, Nimer SD, Yates JR III, et al. 2012. PRMT1 interacts with AML1-ETO to promote its transcriptional activation and progenitor cell proliferative potential. Blood 119: 4953–4962. 10.1182/blood-2011-04-347476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih AH, Jiang Y, Meydan C, Shank K, Pandey S, Barreyro L, Antony-Debre I, Viale A, Socci N, Sun Y, et al. 2015. Mutational cooperativity linked to combinatorial epigenetic gain of function in acute myeloid leukemia. Cancer Cell 27: 502–515. 10.1016/j.ccell.2015.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih AH, Meydan C, Shank K, Garrett-Bakelman FE, Ward PS, Intlekofer AM, Nazir A, Stein EM, Knapp K, Glass J, et al. 2017. Combination targeted therapy to disrupt aberrant oncogenic signaling and reverse epigenetic dysfunction in IDH2- and TET2-mutant acute myeloid leukemia. Cancer Discov 7: 494–505. 10.1158/2159-8290.CD-16-1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlush LI, Zandi S, Mitchell A, Chen WC, Brandwein JM, Gupta V, Kennedy JA, Schimmer AD, Schuh AC, Yee KW, et al. 2014. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature 506: 328–333. 10.1038/nature13038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Meissner A. 2013. DNA methylation: roles in mammalian development. Nat Rev Genet 14: 204–220. 10.1038/nrg3354 [DOI] [PubMed] [Google Scholar]
- Sneeringer CJ, Scott MP, Kuntz KW, Knutson SK, Pollock RM, Richon VM, Copeland RA. 2010. Coordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas. Proc Natl Acad Sci 107: 20980–20985. 10.1073/pnas.1012525107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spina V, Khiabanian H, Messina M, Monti S, Cascione L, Bruscaggin A, Spaccarotella E, Holmes AB, Arcaini L, Lucioni M, et al. 2016. The genetics of nodal marginal zone lymphoma. Blood 128: 1362–1373. 10.1182/blood-2016-02-696757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein EM, DiNardo CD, Pollyea DA, Fathi AT, Roboz GJ, Altman JK, Stone RM, DeAngelo DJ, Levine RL, Flinn IW, et al. 2017. Enasidenib in mutant IDH2 relapsed or refractory acute myeloid leukemia. Blood 130: 722–731. 10.1182/blood-2017-04-779405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strati P, Kantarjian H, Ravandi F, Nazha A, Borthakur G, Daver N, Kadia T, Estrov Z, Garcia-Manero G, Konopleva M, et al. 2015. Phase I/II trial of the combination of midostaurin (PKC412) and 5-azacytidine for patients with acute myeloid leukemia and myelodysplastic syndrome. Am J Hematol 90: 276–281. 10.1002/ajh.23924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. 2009. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324: 930–935. 10.1126/science.1170116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarighat SS, Santhanam R, Frankhouser D, Radomska HS, Lai H, Anghelina M, Wang H, Huang X, Alinari L, Walker A, et al. 2016. The dual epigenetic role of PRMT5 in acute myeloid leukemia: gene activation and repression via histone arginine methylation. Leukemia 30: 789–799. 10.1038/leu.2015.308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teater M, Dominguez PM, Redmond D, Chen Z, Ennishi D, Scott DW, Cimmino L, Ghione P, Chaudhuri J, Gascoyne RD, et al. 2018. AICDA drives epigenetic heterogeneity and accelerates germinal center-derived lymphomagenesis. Nat Commun 9: 222 10.1038/s41467-017-02595-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada K, Yamaguchi H, Ueki T, Usuki K, Kobayashi Y, Tajika K, Gomi S, Kurosawa S, Saito R, Furuta Y, et al. 2018. Usefulness of BCOR gene mutation as a prognostic factor in acute myeloid leukemia with intermediate cytogenetic prognosis. Genes Chromosomes Canc 57: 401–408. 10.1002/gcc.22542 [DOI] [PubMed] [Google Scholar]
- Thol F, Bollin R, Gehlhaar M, Walter C, Dugas M, Suchanek KJ, Kirchner A, Huang L, Chaturvedi A, Wichmann M, et al. 2014. Mutations in the cohesin complex in acute myeloid leukemia: clinical and prognostic implications. Blood 123: 914–920. 10.1182/blood-2013-07-518746 [DOI] [PubMed] [Google Scholar]
- Thota S, Viny AD, Makishima H, Spitzer B, Radivoyevitch T, Przychodzen B, Sekeres MA, Levine RL, Maciejewski JP. 2014. Genetic alterations of the cohesin complex genes in myeloid malignancies. Blood 124: 1790–1798. 10.1182/blood-2014-04-567057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tothova Z, Krill-Burger JM, Popova KD, Landers CC, Sievers QL, Yudovich D, Belizaire R, Aster JC, Morgan EA, Tsherniak A, et al. 2017. Multiplex CRISPR/Cas9-based genome editing in human hematopoietic stem cells models clonal hematopoiesis and myeloid neoplasia. Cell Stem Cell 21: 547–555.e8. 10.1016/j.stem.2017.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CH, Hou HA, Tang JL, Kuo YY, Chiu YC, Lin CC, Liu CY, Tseng MH, Lin TY, Liu MC, et al. 2017. Prognostic impacts and dynamic changes of cohesin complex gene mutations in de novo acute myeloid leukemia. Blood Cancer J 7: 663 10.1038/s41408-017-0022-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao T, Shi Y, Kornblau S, Lu H, Konoplev S, Antony A, Ruvolo V, Qiu YH, Zhang N, Coombes KR, et al. 2012. Concomitant inhibition of DNA methyltransferase and BCL-2 protein function synergistically induce mitochondrial apoptosis in acute myelogenous leukemia cells. Ann Hematol 91: 1861–1870. 10.1007/s00277-012-1537-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velichutina I, Shaknovich R, Geng H, Johnson NA, Gascoyne RD, Melnick AM, Elemento O. 2010. EZH2-mediated epigenetic silencing in germinal center B cells contributes to proliferation and lymphomagenesis. Blood 116: 5247–5255. 10.1182/blood-2010-04-280149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victora GD, Dominguez-Sola D, Holmes AB, Deroubaix S, Dalla-Favera R, Nussenzweig MC. 2012. Identification of human germinal center light and dark zone cells and their relationship to human B-cell lymphomas. Blood 120: 2240–2248. 10.1182/blood-2012-03-415380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viny AD, Ott CJ, Spitzer B, Rivas M, Meydan C, Papalexi E, Yelin D, Shank K, Reyes J, Chiu A, et al. 2015. Dose-dependent role of the cohesin complex in normal and malignant hematopoiesis. J Exp Med 212: 1819–1832. 10.1084/jem.20151317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner EJ, Carpenter PB. 2012. Understanding the language of Lys36 methylation at histone H3. Nat Rev Mol Cell Biol 13: 115–126. 10.1038/nrm3274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Xiao M, Chen X, Chen L, Xu Y, Lv L, Wang P, Yang H, Ma S, Lin H, et al. 2015. WT1 recruits TET2 to regulate its target gene expression and suppress leukemia cell proliferation. Mol Cell 57: 662–673. 10.1016/j.molcel.2014.12.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward PS, Cross JR, Lu C, Weigert O, Abel-Wahab O, Levine RL, Weinstock DM, Sharp KA, Thompson CB. 2011. Identification of additional IDH mutations associated with oncometabolite R(-)-2-hydroxyglutarate production. Oncogene 31: 2491–2498. 10.1038/onc.2011.416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters LR, Ahsan FM, Wolf DM, Shirihai O, Teitell MA. 2018. Initial B cell activation induces metabolic reprogramming and mitochondrial remodeling. iScience 5: 99–109. 10.1016/j.isci.2018.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissmann S, Alpermann T, Grossmann V, Kowarsch A, Nadarajah N, Eder C, Dicker F, Fasan A, Haferlach C, Haferlach T, et al. 2012. Landscape of TET2 mutations in acute myeloid leukemia. Leukemia 26: 934–942. 10.1038/leu.2011.326 [DOI] [PubMed] [Google Scholar]
- Wu SY, Chiang CM. 2007. The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. J Biol Chem 282: 13141–13145. 10.1074/jbc.R700001200 [DOI] [PubMed] [Google Scholar]
- Xie W, Schultz MD, Lister R, Hou Z, Rajagopal N, Ray P, Whitaker JW, Tian S, Hawkins RD, Leung D, et al. 2013. Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell 153: 1134–1148. 10.1016/j.cell.2013.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Wang P, Xiao MT, et al. 2011. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell 19: 17–30. 10.1016/j.ccr.2010.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, On DM, Ma A, Parton T, Konze KD, Pattenden SG, Allison DF, Cai L, Rockowitz S, Liu S, et al. 2015. Selective inhibition of EZH2 and EZH1 enzymatic activity by a small molecule suppresses MLL-rearranged leukemia. Blood 125: 346–357. 10.1182/blood-2014-06-581082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Rau R, Goodell MA. 2015. DNMT3A in haematological malignancies. Nat Rev Cancer 15: 152–165. 10.1038/nrc3895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap DB, Chu J, Berg T, Schapira M, Cheng SW, Moradian A, Morin RD, Mungall AJ, Meissner B, Boyle M, et al. 2011. Somatic mutations at EZH2 Y641 act dominantly through a mechanism of selectively altered PRC2 catalytic activity, to increase H3K27 trimethylation. Blood 117: 2451–2459. 10.1182/blood-2010-11-321208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Jima D, Moffitt AB, Liu Q, Czader M, Hsi ED, Fedoriw Y, Dunphy CH, Richards KL, Gill JI, et al. 2014. The genomic landscape of mantle cell lymphoma is related to the epigenetically determined chromatin state of normal B cells. Blood 123: 2988–2996. 10.1182/blood-2013-07-517177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Dominguez-Sola D, Hussein S, Lee JE, Holmes AB, Bansal M, Vlasevska S, Mo T, Tang H, Basso K, et al. 2015. Disruption of KMT2D perturbs germinal center B cell development and promotes lymphomagenesis. Nat Med 21: 1190–1198. 10.1038/nm.3940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Vlasevska S, Wells VA, Nataraj S, Holmes AB, Duval R, Meyer SN, Mo T, Basso K, Brindle PK, et al. 2017. The CREBBP acetyltransferase is a haploinsufficient tumor suppressor in B-cell lymphoma. Cancer Discov 7: 322–337. 10.1158/2159-8290.CD-16-1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorko NA, Bernot KM, Whitman SP, Siebenaler RF, Ahmed EH, Marcucci GG, Yanes DA, McConnell KK, Mao C, Kalu C, et al. 2012. Mll partial tandem duplication and Flt3 internal tandem duplication in a double knock-in mouse recapitulates features of counterpart human acute myeloid leukemias. Blood 120: 1130–1136. 10.1182/blood-2012-03-415067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, et al. 2011. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 478: 524–528. 10.1038/nature10334 [DOI] [PMC free article] [PubMed] [Google Scholar]