Abstract
Scutellaria indica var. coccinea (Lamiaceae) is an endemic taxon in Jeju Island, Korea. We present the complete chloroplast genome sequence of S. indica var. coccinea. Its genome size is 151,956 bp in length comprising a large single copy of 83,951 bp, a small single copy of 17,539 bp, and a pair of inverted repeats measuring 25,233 bp. The complete genome contains 115 genes including 80 protein-coding genes, four rRNAs, and 31 tRNAs. Phylogenetic analysis of this species with 10 published chloroplast genomes in Lamiaceae showed that S. insignis is a sister to S. indica var. coccinea.
Keywords: Chloroplast genome, genome skimming, endemic species, Korea, Scutellaria indica var. coccinea
Scutellaria L. is the second largest genus in the Lamiaceae with approximately 470 recognized species that are mainly distributed in temperate regions. Plants are annual or perennial herbs and rarely subshurubs or aquatic species. Eighteen taxa of Scutellaria are distributed in Korea (Kim and Lee 1995; Yang 2004; Kim et al. 2011). Three of them are endemic species in Korea: S. indica var. coccinea S. Kim & S. Lee, S. insignis Nakai, and S. pekinensis var. maxima S. T. Kim & S. Lee. Especially, S. indica var. coccinea is found only in a small volcanic crater (Sangumburi) of Jeju Island, which is located to the south of the Korean peninsula (Kim and Lee 1995).
The recent classification system of Scutellaria includes subgen. Scutellaria and subgen. Apelthanthus containing seven sections (Paton 1990). In this system, 34 unranked morphological ‘species-groups’ have also been suggested under Scutellaria, which is the largest section in the genus (ca. 240 species). In this species-group category, S. indica L. is included under ‘S. violacea species-group’ of the section Scutellaria, which is characterized by nutlets with acuminate papillae terminating in hooks. In S. indica, S. indica var. coccinea is distinguished from other varieties having widely cordate leaves, conspicuously sunken veins in adaxial leaf surface and dark pink corolla color.
In this study, we report a complete sequence of cp genome of S. indica var. coccinea (GenBank accession: MN047312). Genomic DNA was extracted from leaves of a plant cultivated in Sungshin University, which is propagated from seeds of a wild plant collected in Jeju Island (N33°25′57.33″, E126°41′26.72″). Voucher specimen is deposited in Sungshin University herbarium (S. Kim 2015-0298). The whole genome sequencing was conducted with 100 bp paired-end reads on the BGISEQ-500 sequencer (BGI, Shenzhen, China). To obtain the chloroplast genome (cp) sequence, paired-end reads were mapped against a reference cp genome (GenBank accession: NC_028533) which is previously reported from Scutellaria insignis using Geneious (v9.0.5; Kearse et al. 2012). The quality of a consensus sequence and its mapping condition were examined visually. The cp genome was annotated using GeSeq (Tillich et al. 2017).
The complete cp genome of S. indica var. coccinea is 151,956 bp in length, containing a large single copy (LSC) of 83,951 bp, a small single copy (SSC) of 17,539 bp, and a pair of inverted repeats (IR) measuring 25,233 bp. It includes 115 genes comprising 80 protein coding genes, four rRNA genes and 31 tRNA genes. Among these genes, 16 genes carry a single intron, and two genes (clpP and ycf3) contain two introns, respectively.
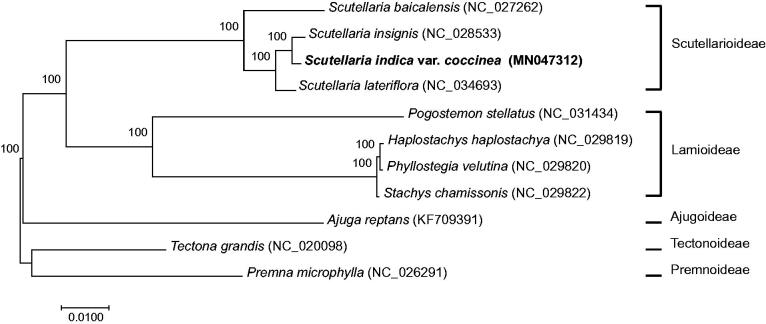
The phylogenetic tree was reconstructed with this cp genome and previously published cp genomes (three in Scutellaria and seven in other Lamiaceae; Figure 1). The maximum-likelihood analysis is performed using raxmlGUI (v.1.5; Silvestro and Michalak 2012). In the phylogenetic tree (Figure 1), S. indica var. coccinea is a sister to S. insignis. In this study, we report the cp genome of S. indica var. coccinea as an endemic taxon in Korea. The findings contribute to the conservation of this species and the phylogenetic study of Scutellaria.
Figure 1.
A maximum-likelihood tree based on ‘GTR + gamma + I’ model using the cp genome of S. indica var. coccinea and 10 selected cp genomes reported in Lamiaceae. Numbers above the node indicate bootstrap values.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. . 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CS, Kim SY, Byun GO. 2011. A new record for Korean flora: Scutellaria tuberifera C. Y. Wu & C. Chen (Lamiaceae). Kor J Plant Tax. 41:249–252. [Google Scholar]
- Kim ST, Lee ST. 1995. Taxonomy of the genus Scutellaria (Lamiaceae) in Korea. Korean J Pl Taxon. 25:71–102. (In Korean.) [Google Scholar]
- Paton A. 1990. A global taxonomic investigation of Scutellaria (Labiatae). Kew Bull. 45:399–450. [Google Scholar]
- Silvestro D, Michalak I. 2012. raxmlGUI: a graphical front-end for RAxML. Org Divers Evol. 12:335–337. [Google Scholar]
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq–versatile and accurate annotation of organelle genomes. Nucl Acid Res. 45:W6–W11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YH. 2004. One unrecorded species from: Scutellaria orthocalyx Hand.-Mazz. Korean J Plant Res. 17:41–42. (In Korean.) [Google Scholar]