Abstract
This study was the first report about complete chloroplast genome of Aster hypoleucus (Asteraceae, Astereae), an endemic species in Xizang (China). The circular whole cp genome of A. hypoleucus was 152,300 bp in length, contained a large single-copy (LSC) region of 84,031 bp and a small single-copy (SSC) region of 18,269 bp. These two regions were separated by a pair of inverted repeat regions (IRa and IRb), each of them 25,000 bp in length. A total of 134 functional genes were encoded, consisted of 89 protein-coding genes, 37 tRNA genes, and eight rRNA genes. The overall GC content of the chloroplast genome sequence was 37.3%, and the GC contents of the LSC, SSC, and IR regions were 35.2%, 31.2%, and 43.0%, respectively.
Keywords: Aster hypoleucus, chloroplast genome, phylogenomics analysis
The genus Aster is one of the most diverse genera in the tribe Astereae, family Asteraceae, including about 152 species (Nesom 1994). The species of Aster hypoleucus Hand.-Mazz. is the shrubby endemic species of the genus Aster, and it is narrowly distributed in Xizang Autonomous Region, China. It has ecological and medicinal value in western China. So far, the complete chloroplast genome of A. hypoleucus has not yet been published.
Fresh leaves of A. hypoleucus were collected from Lang county, Xizang Autonomous Region, China and deposited in Herbarium, Sichuan Normal University, SCNU (specimen no.: Z.X. Fu 1475). High quality total genomic DNA was extracted from ca. 6 cm2 sections of the silica-dried leaf using improved Tiangen Plant Genomic DNA Kits, add the 4 μl RNAseA and 20 μl Proteinase K after incubated (65 °C). Total DNA was directly constructed short-insert of 150 bp in length libraries and sequenced on the Illumina Genome Analyzer (Hiseq 2000) based the manufacturer’s protocol (Illumina, San Diego, CA, USA) by ORI-GENE, Beijing. Generally, more than 6 Gb of data was obtained for complete cp genome of A. hypoleucus. De novo assembled in CLC Genomic Workbench v11 (CLC Bio, Aarhus, Denmark) and consensus sequence in Geneious R11.1.5 (Biomatters Ltd., Auckland, New Zealand) with referenced chloroplast genome sequence of Conyza bonariensis (Accession: KX792499) and Lagenophora cuchumatanica (Accession: Accession: KX063879). The chloroplast genome was annotated using a web-based annotation program GeSeq (https://chlorobox.mpimp-golm.mpg.de/geseq.html) and editing by manual and imagining with OGDraw v1.2 (Lohse et al. 2013).
The complete chloroplast genome of A. hypoleucus (GenBank Accession No. MK290824) was 152,300 bp in length and a typical circular structure, which comprising a pair of inverted repeat (IR) of 25,000 bp divided by a large single copy (LSC) region of 84,031 bp and a small single copy (SSC) region of 18,269 bp (Figure 1). The general G + C content was 37.3% in the whole sequence, while corresponding values of 35.2%, 31.2%, and 43.0% in the LSC, SSC, and IR regions. The whole genome contained 134 genes, including 89 protein-coding genes, eight ribosomal RNA genes and 37 tRNA genes, nevertheless, 114 unique genes, 20 genes duplicated in the IRs. In addition, among the annotated chloroplast genomic sequence, 15 genes possessed only single intron, two genes (ycf3 and clpP) possessed two introns.
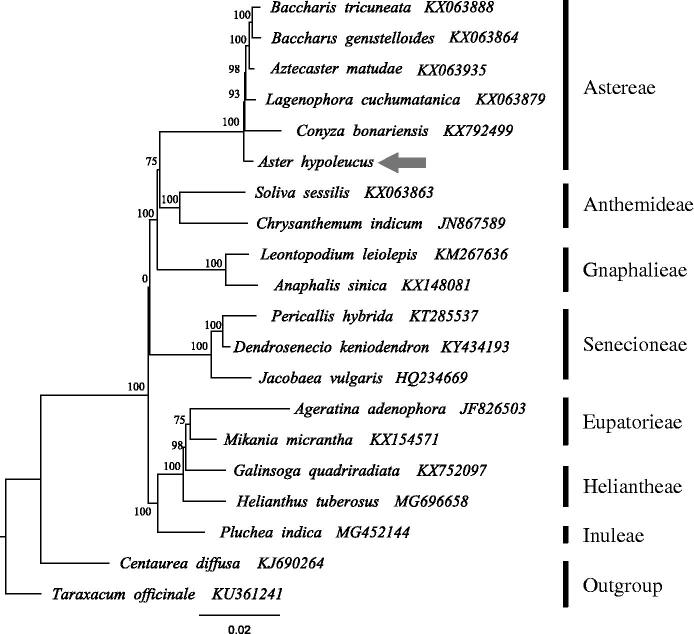
Figure 1.
The best Maximum-likelihood (ML) phylogram inferred from 20 chloroplast genomes in Asteraceae (bootstrap value are indicated on the branches). The gene’s accession number is list in the figure.
To construct the phylogenetic tree, all of the cp genome sequences were aligned in MAFFT (Katoh and Standley 2013). A maximum likelihood analysis based on the GTRGAMMA model was performed with RaxML v7.2.8 on the CIPRES (Stamatakis et al. 2008; Miller et al. 2010) using 1000 bootstrap replicates with Centaurea diffusa Lam. and Taraxacum officinale as outgroup. Phylogenetic analysis shows a well-supported sister-clade relationship A. hypoleucus and other members of Astereae (e.g. Conyza bonariensis, Lagenophora cuchumatanica, and Baccharis tricuneata) (Bootstrap support = 100, Figure 1).
The complete plastome sequence of A. hypoleucus will provide a useful resource for the conservation genetics of this species as well as for the phylogenetic studies for Asteraceae.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. Organellar genome DRAW—a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucl Acids Res. 41:575–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES science gateway for inference of large phylogenetic trees. Proceedings of the gateway computing environments workshop (GCE). New Orleans (LA): IEEE; p. 1–8. [Google Scholar]
- Nesom GL. 1994. Review of the taxonomy of Aster sensu lato (Asteraceae: Astereae), emphasizing the New World species. Phytologia. 77:14–297. [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol. 57:758–771. [DOI] [PubMed] [Google Scholar]