In this study, Singh et al. investigated the functional diversification of mammalian Hox1 genes during evolution, using the Drosophila ortholog Labial to represent their ancestral functions. Using rescue experiments in Drosophila, as well as genome-wide transcription factor-binding experiments in differentiated mouse ES cells, the authors identified a novel six-amino-acid C-terminal motif (CTM) flanking the highly conserved homeodomain that serves as a major determinant of ancestral activity.
Keywords: Labial, Drosophila, mouse Hox genes, TALE proteins, gene duplication, functional diversification, protein evolution
Abstract
Gene duplication and divergence is a major driver in the emergence of evolutionary novelties. How variations in amino acid sequences lead to loss of ancestral activity and functional diversification of proteins is poorly understood. We used cross-species functional analysis of Drosophila Labial and its mouse HOX1 orthologs (HOXA1, HOXB1, and HOXD1) as a paradigm to address this issue. Mouse HOX1 proteins display low (30%) sequence similarity with Drosophila Labial. However, substituting endogenous Labial with the mouse proteins revealed that HOXA1 has retained essential ancestral functions of Labial, while HOXB1 and HOXD1 have diverged. Genome-wide analysis demonstrated similar DNA-binding patterns of HOXA1 and Labial in mouse cells, while HOXB1 binds to distinct targets. Compared with HOXB1, HOXA1 shows an enrichment in co-occupancy with PBX proteins on target sites and exists in the same complex with PBX on chromatin. Functional analysis of HOXA1–HOXB1 chimeric proteins uncovered a novel six-amino-acid C-terminal motif (CTM) flanking the homeodomain that serves as a major determinant of ancestral activity. In vitro DNA-binding experiments and structural prediction show that CTM provides an important domain for interaction of HOXA1 proteins with PBX. Our findings show that small changes outside of highly conserved DNA-binding regions can lead to profound changes in protein function.
The balance between maintaining ancestral roles and novel diversification of orthologous transcription factors (TFs) shapes the evolution of gene regulatory networks underlying morphological diversity. While we have some understanding of gene duplication and neofunctionalization events at the genomic level (Ohno 1970; Lundin 1993; Holland et al. 1994; Blomme et al. 2006), at protein level it is unclear how variations in amino acid sequence lead to functional diversification or retention of the ancestral function. Many TF families arose early in animal evolution and for >600 million years have maintained a relatively high degree of similarity in their in vitro DNA-binding specificities, contributing to the idea that divergence in TF DNA-binding specificity occurs relatively infrequently in evolution (Carroll 2005; Berger et al. 2008; Noyes et al. 2008; Nitta et al. 2015). This is in contrast to differences in their functional properties observed in vivo.
The Hox family of homeodomain containing TFs are central to the specification of structures along the anterior–posterior (AP) body axis in bilaterian animals and modifications in their expression and function have paralleled the emergence of diversity in body plans (Lewis 1978; McGinnis and Krumlauf 1992; Carroll 1995; Woltering et al. 2009; Arendt 2018). Despite a conserved genomic organization, collinear domains of Hox gene expression and roles in AP patterning (Lewis 1978; Duboule and Dollé 1989; Graham et al. 1989; Carroll 1995; Kmita and Duboule 2003), the evolution of Hox protein function is poorly understood. Hox proteins display nearly identical DNA-binding specificities in vitro (Carroll 2005; Berger et al. 2008; Noyes et al. 2008; Nitta et al. 2015), but the binding preferences for individual Hox homeodomains are difficult to identify because binding specificity is governed by pleiotropic interactions through multiple residues (Passner et al. 1999; Piper et al. 1999; Slattery et al. 2011; Zeiske et al. 2018). Furthermore, interactions with cofactors, such as the TALE (three-amino-acid loop extension) family members PBX/Exd and MEIS/Hth, can modify their binding preferences (Chan et al. 1994, 1997; Chang et al. 1995; Merabet and Mann 2016). HOX proteins are known to interact with PBX through a hexapeptide domain, but interactions may also occur in a context-dependent manner through a variety of other small motifs located in different regions of HOX proteins (Merabet et al. 2003; Merabet and Mann 2016; Dard et al. 2018). This makes it challenging to identify the amino acid differences that underly the functional diversity of Hox proteins in vivo. Here, we used in vivo cross-species functional analyses of the mouse HOX1 paralogous group TFs with their Drosophila ortholog Labial, as a paradigm to investigate the maintenance and diversification of ancestral protein function.
Drosophila has a single HOX1 gene, labial, while in mouse there are three paralogs, Hoxa1, Hoxb1, and Hoxd1, as Hoxc1 has been lost in mammalian lineages during vertebrate evolution (McGinnis and Krumlauf 1992). There is evidence suggesting Hoxd1 has neofunctionalized, with roles in new tissues (development of nociceptors of the mouse skin) due to changes in expression through evolution of regulatory elements (Guo et al. 2011; Pascual-Anaya et al. 2013). Mutations of Hoxa1 and Hoxb1 genes in humans and mice reveal that, like labial, they have essential roles in head and brain development (Merrill et al. 1989; Lufkin et al. 1991; Studer et al. 1996; Tischfield et al. 2005; Webb et al. 2012). However, it is unknown whether they possess ancestral functions of labial or have diversified and evolved new activities during evolution.
Here, we have used cross-species functional analysis of Drosophila Labial and its three mouse HOX1 orthologs, separated by 600 million years of evolution, as a paradigm to address protein evolution following gene duplication. Despite similar degrees of sequence divergence from Labial, only HOXA1 retains essential ancestral functions of Labial and displays similar DNA-binding properties. We used a series of chimeric proteins to map the determinants governing ancestral activity and found that small differences in multiple domains underlie functional diversification of mouse HOX1 paralogs. We identified a six-amino-acid motif (CTM) located immediately C-terminal to the highly conserved homeodomain that is a major determinant of ancestral activity. Our findings reveal that small changes can lead to profound differences in function and illustrates that overall similarity or divergence is not always predictive of protein function.
Results
High evolutionary divergence among mouse HOX1 paralogous proteins and Labial
Protein sequence alignments revealed that the three-mouse paralogous HOX1 proteins (HOXA1, HOXB1, and HOXD1) and Drosophila Labial display low (∼30%) overall similarity (Fig. 1A; Supplemental Fig. S1A). Most of the similarity (>50%) among HOX1 proteins is contributed by the DNA-binding region (homeodomain), and a hexapeptide motif, known to interact with the HOX cofactors, such as the TALE (three-amino-acid loop extension) family members PBX/Exd and MEIS/Hth (Chan et al. 1994; Chang et al. 1995). Alignment of the orthologous arthropod Labial and vertebrate HOX1 proteins also reveals that sequence similarity is primarily concentrated in these same domains while other regions are more variable (Supplemental Figs. S1, S2). Moreover, Labial in arthropods widely vary in size and Drosophila Labial is almost twice the size of the mouse HOX1 proteins, indicating that there have been global changes during evolution (Fig. 1A; Supplemental Fig. S2). Analyses of the vertebrate HOX1 proteins revealed multiple mutations in hexapeptide and homeodomain of the HOXD1, suggesting rapid evolution after gene duplication compared with the other paralogs (Supplemental Fig. S1D). The high degree of conservation within the homeodomain and large differences between HOX1 proteins in other regions make it challenging to identify sequence determinants that underlie ancestral or novel functional properties.
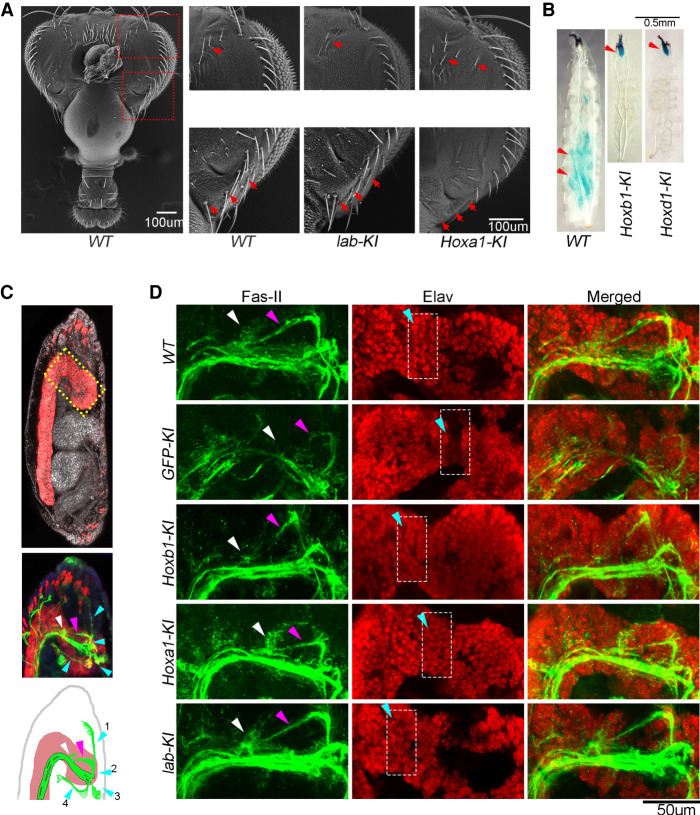
Figure 1.
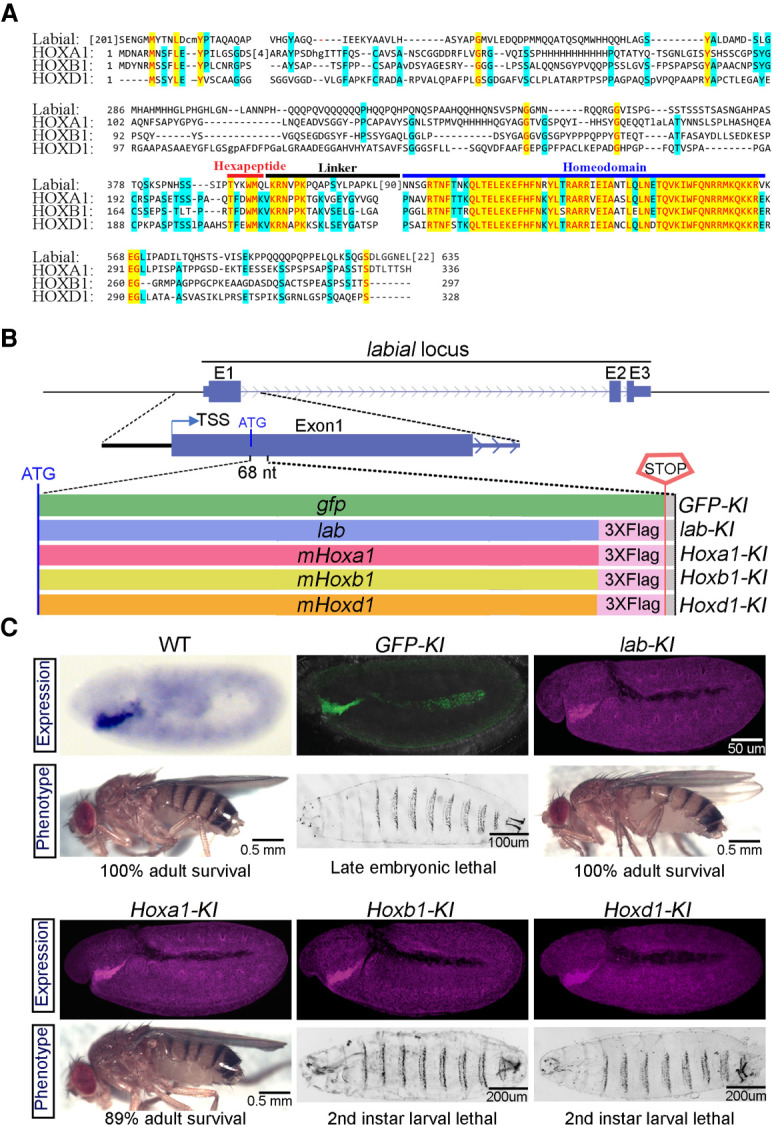
Mouse HOXA1 retains the essential functions of Drosophila Labial. (A) Protein sequence alignment of Drosophila Labial with mouse HOXA1, HOXB1, and HOXD1. Colored blocks indicate conserved amino acids in three (light blue) or all four (yellow) of the proteins. Horizontal bars above the alignment indicate specific domains in the proteins: hexapeptide (red), linker (black), and homeodomain (blue). (B) The top panel shows a schematic of the labial gene with introns (arrowed line) and exons (E1–E3 in blue blocks). The bottom panel shows five DNA cassettes independently inserted at the ATG codon in-frame with the endogenous labial open reading frame that encode GFP (GFP-KI), Drosophila Labial (lab-KI), and mouse HOXA1 (Hoxa1-KI), HOXB1 (Hoxb1-KI), and HOXD1 (Hoxd1-KI). Multiple stop codons in the cassettes prevent the translation of the endogenous labial gene. Labial and HOX proteins are epitope-tagged at the C-terminal with 3xFLAG. (C) Expression pattern (top) in 5- to 6-h-old embryos and corresponding larval/adult phenotypes (bottom) of WT and knock-in lines, with genotypes listed above the panels. The WT panel shows endogenous lab RNA expression. Expression of knocked in proteins is indicated by green for GFP expression and purple for immunostaining of the FLAG tag.
Mouse HOXA1 is the only paralog to have retained the essential ancestral functions of Labial
To functionally test whether the mouse HOX1 paralogs have retained ancestral activities of Labial, we used a CRISPR/CAS9-based approach to knock-in sequences encoding 3xFLAG-tagged versions of these proteins into the endogenous labial locus (Fig. 1B). The insertions are designed to express the mouse proteins under control of the endogenous regulatory elements of labial and prevent labial translation, creating a loss-of-function allele. This allowed us to simultaneously assess the consequence of loss of Labial function and its rescue by the orthologous proteins. The coding regions of the labial and gfp genes were also knocked-in in a similar manner as positive and negative controls, respectively. The inserted genes expressed the respective proteins with the expected spatial and temporal patterns displayed by endogenous Labial in embryos and in larval brain and eye-antennal imaginal discs (Fig. 1C; Supplemental Fig. S3A,B; Diederich et al. 1989). We quantified the relative levels of these proteins, along with another Drosophila Hox protein (Abd-A) as a control and found that the mouse HOX proteins are expressed at a comparable level in embryos (Supplemental Fig. S3C,D).
We analyzed the phenotypes of homozygous insertion lines and, as expected, the knock-in of gfp (GFP-KI) did not complement the loss of labial function, resulting in the well-characterized labial-null mutant phenotypes and late embryonic lethality (Fig. 1C; Supplemental Fig. S3E; Merrill et al. 1989). Conversely, lines with a knock-in of labial (lab-KI) displayed 100% viability, with no observable phenotypes, validating our experimental approach. Remarkably, Hoxa1-KI flies hatched, with 89% survival (Fig. 1C) with only subtle bristle pattern defects in adult head, indicating nearly complete functional complementation of labial mutant by mouse HOXA1 (Fig. 2A). In contrast, Hoxb1-KI and Hoxd1-KI lines were lethal at the second instar stage. Developmentally arrested larvae survived up to 5 d (Hoxd1-KI) or 15 d (Hoxb1-KI), but they never progressed to third instar stage (Fig. 1C). These larvae are smaller in size, malnourished and have food in the pharynx but not in the gut, suggesting a defect in feeding (Fig. 2B; Beadle et al. 1938; Bakker 1959). We investigated feeding behavior in Hoxb1-KI larvae and found a large reduction (∼65%) in the rate of mouth extensions and observed that many other larvae barely move their mouth hooks (Supplemental Movies S1–S3). This rescue assay reveals that HOXA1 has retained the major ancestral functions of Labial, while HOXB1 and HOXD1 have lost most of this activity. However, the partial survival of Hoxb1-KI and Hoxd1-KI lines up to the second instar larval stage suggests they may have retained some aspect of early Labial function in embryos.
Figure 2.
Hoxb1-KI embryos show defects in the development of stomatogastric nervous system. (A) Scanning electron micrographs of the posterior side of an adult head. Arrows (red) note bristles that are missing in Hoxa1-KI as compared with WT and lab-KI. (B) Colored food (blue) is present in the gut of WT larvae but remains in the pharynx of homozygous Hoxb1-KI and Hoxd1-KI (red arrows). (C) The top panel shows 15- to 16-h-old embryo (anterior up, dorsal right) immunostained with anti-Elav (red) to mark CNS, and DAPI (white) to mark nuclei. Middle panel shows anterior half of the embryo with anti-Fasciclin-II (FAS-II) immunostaining (green) that marks SNS (arrowheads). The botttom panel, schematic of SNS connection with tritocerebrum neurons (white arrow), through antennal nerve (pink arrow). SNS anteriorly connects to the pharynx by the frontal nerve (cyan arrow 1) and posteriorly connects with renal gland and gut by the recurrent nerve (arrows 2–4). (D) High magnification images of boxed region in C of WT, GFP-KI, Hoxb1-KI, Hoxa1-KI, and lab-KI embryos. The left panels show Fas-II expression in tritocerebrum neurons (white arrow) and antennal nerve (pink arrow). The middle panels show Elav expression in tritocerebrum region (box with cyan arrow), and the right panels show the merged images.
To better understand which aspects of labial functions are preserved or lost by Hoxb1, we performed a detailed analysis of Hoxb1-KI flies. The mouth hook develops normally in Hoxb1-KI embryos, compared with the deformities observed in labial mutants, indicating that other functions of Labial in feeding behavior are affected (19) (Supplemental Fig. S3E). The labial gene is expressed in the posterior tritocerebrum region of the embryonic fly central nervous system and is required for differentiation of neuroblasts and axonal pathfinding in this region (Hirth et al. 1998, 2001). The stomatogastric nervous system (SNS) is a set of ganglia connected to the tritocerebrum and innervates diverse muscles that control food uptake and transport (Fig. 2C; Hartenstein 1997; Schoofs et al. 2014). Immunostaining for neuronal (Elav) and neurite (Fasciclin-II) markers in late embryos (15–16 h), revealed defects in neuronal differentiation and projections of motor nerves in labial (GFP-KI) mutants (Fig. 2D). Both defects are rescued in the lab-KI and Hoxa1-KI lines. However, in Hoxb1-KI embryos, neuronal differentiation appears normal, but the axonal projections are not fully restored. Therefore, the larval lethality in the Hoxb1-KI line is likely due to a role of labial in patterning the SNS and control of feeding behaviors. Together, these complementation experiments in Drosophila demonstrate that for >600 million years of evolution HOXA1 has retained the key ancestral functions of Labial while HOXB1 and HOXD1 have diversified.
Genome-wide binding profiles of HOXA1 are different from HOXB1 and overlap with those of Drosophila Labial
The rescue experiments in Drosophila indicated that the properties of HOXB1 and HOXD1 proteins have functionally diverged from HOXA1. Next, we investigated whether these functional differences extend to mammalian cells by analyzing genome-wide DNA-binding properties and downstream targets using a well-characterized assay based on programmed differentiation of mouse ES cells into neural fates (De Kumar et al. 2015, 2017a,b,c). We showed previously that retinoid (RA) treatment of mouse ES cells induces the sequential expression of genes from the Hoxa and Hoxb clusters, including Hoxa1 and Hoxb1, in a manner similar to neural tissue of developing mouse embryos (De Kumar et al. 2015). However, endogenous Hoxd1 is not induced in this system, consistent with its lack of expression in the mouse neural tube. Hence, we only examined the DNA-binding properties of HOXA1 and HOXB1 in this system. Independent mouse ES cell lines carrying cDNAs encoding epitope-tagged (3xFLAG) versions of HOXA1 and HOXB1 proteins were inserted at a specific locus under tight inducible Dox control (Beard et al. 2006). We optimized the expression of the inserted cDNAs in response to Dox and RA programmed differentiation so that levels were comparable with that of the endogenous Hox gene. The genome-wide binding profiles of the HOX1 proteins, PBX and MEIS cofactors, and other TFs were then characterized by chromatin immunoprecipitation followed by deep sequencing (ChIP-seq) (Fig. 3A,C). The binding profiles were also compared with histone modifications associated with enhancers (H3K4me1 and H3K27Ac), occupancy of coactivators (p300) and chromatin accessibility (ATAC-seq) (Fig. 3B).
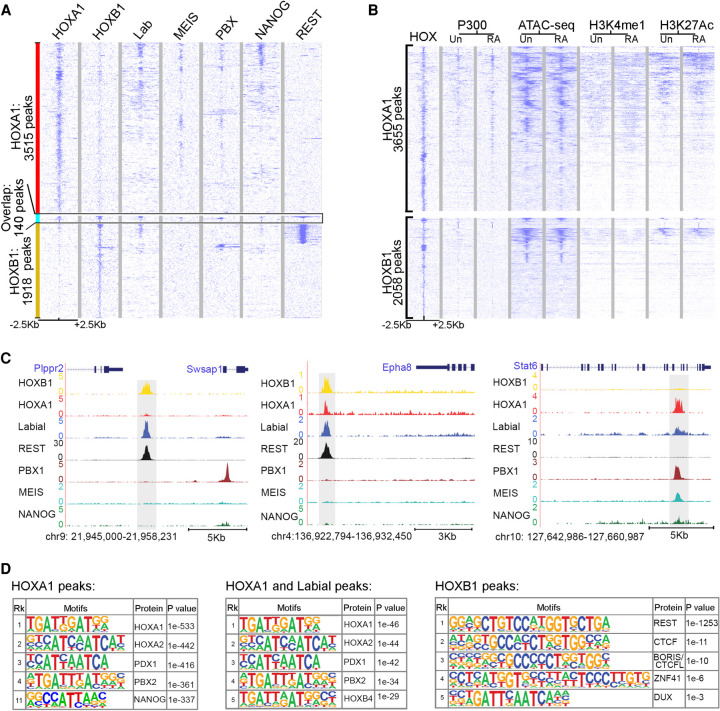
Figure 3.
HOXA1-bound regions overlap with Labial and differ from HOXB1 in mES cells. (A) Heat maps showing genome-wide binding peaks of HOXA1 and HOXB1 (±2.5 kb) and the occupancy of Labial, PBX, MEIS, NANOG, and REST at the corresponding sites in differentiated ES cells. Heat map is sorted first on HOXA1, second HOXA1-B1 cobound, and then HOXB1 peaks. (B) Genome-wide binding of HOXA1 and HOXB1 is compared with ATAC-seq, P300, H3K4me, and H3K27Ac profiles in undifferentiated (Un) and 24-h retinoic acid (RA) differentiated ES cells. Heat map is sorted on HOX binding peaks, respectively. (C) Genome browser screenshots of regions predominantly bound by HOXB1 (left), by HOXA1 (right) and by both HOXA1 and HOXB1 (middle). Corresponding binding profiles of Labial, REST, PBX, MEIS, and NANOG are also shown in these panels. Regions of interest are highlighted in gray. Nearby genes are displayed at the top and genomic coordinates are indicated at the bottom. (D) Tables show enriched motifs in HOXA1, HOXB1, and shared HOXA1/Labial binding peaks. Motifs in the table are sorted by their rank (Rk) and P-values.
We observed very little overlap (only140 peaks) between the genome-wide DNA-binding peaks of HOXB1 (2058) and HOXA1 (3655). Gene ontology analysis for adjacent genes reveal that HOXB1-binding peaks are enriched for genes required for neurogenesis and behavior, while HOXA1 peaks are enriched for genes involved in differentiation, morphogenesis, and development (Supplemental Fig. S4). Furthermore, analysis of transcription factor binding motifs reveals that HOXA1 bound peaks are enriched for consensus TF motifs, including HOXA1, PBX, MEIS, and NANOG (Fig. 3D; Merabet and Mann 2016; De Kumar et al. 2017a,b). In contrast, HOXB1 peaks are enriched for REST and CTCF consensus motifs. REST is a transcriptional repressor that interacts with SIN3A and is required for repression of neural genes in nonneural cells (Ballas and Mandel 2005). We validated the motif discovery analysis by demonstrating that there is also enriched occupancy of PBX, MEIS, and NANOG on HOXA1-bound regions and REST on HOXB1-binding peaks (Fig. 3A,C; Supplemental Fig. S5). In contrast to HOXA1, HOXB1-binding peaks display very little overlap with occupancy of P300 and only small subsets correlate with open chromatin or H3K4me1 and H3K27Ac histone marks (Fig. 3B; De Kumar et al. 2017b,c).
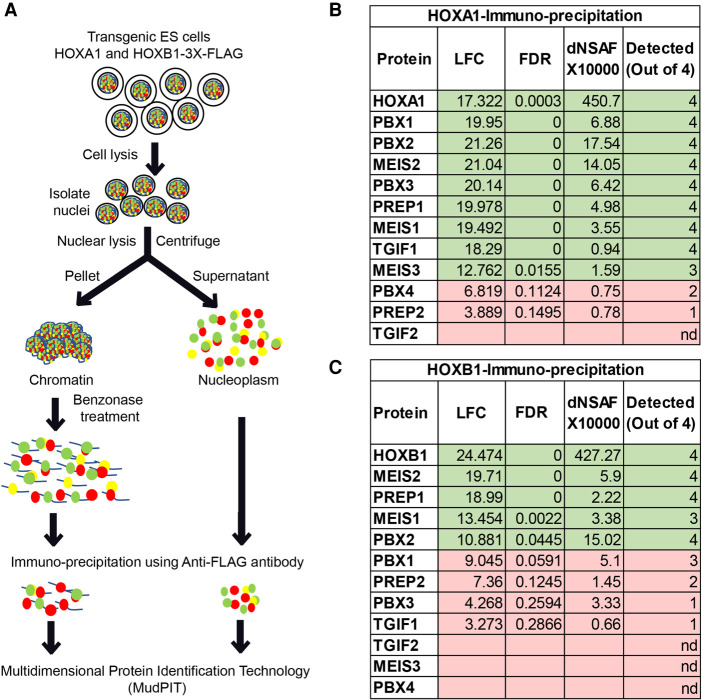
To explore the differences in genome-wide association of HOXA1 and HOXB1 with PBX and MEIS proteins, we performed immunoprecipitation of HOXA1 and HOXB1 in these ES cell lines followed by MudPIT. There was high enrichment of PBX and other TALE proteins in HOXA1 protein complexes as compared with HOXB1 (Fig. 4). This suggests that HOXA1 and PBX proteins exist in the same complex on chromatin and further support the observed enrichment in genome-wide co-occupancy of HOXA1 with TALE proteins (Fig. 4B; De Kumar et al. 2017b).
Figure 4.
Identification of proteins interacting with HOXA1 and HOXB1. (A) The schematic shows an outline of the experimental process used to prepare chromatin extract followed by immunoprecipitation (IP) and MudPIT. (B,C) Three-amino-acid loop extension (TALE) proteins enriched in IP-MudPIT experiments of HOXA1 (B) and HOXB1 (C) are listed with their QPROT-calculated LFC (log2 fold change) values, dNSAF values and sorted using QPROT-FDR (false discovery rate). Statistically significant interactions are highlighted in green. These proteins were detected in at least three out of four experimental replicates, as listed. TALE proteins with FDR value >0.05 or not detected (n.d.) in this experiment are highlighted in red.
To investigate whether the binding properties of these proteins are associated with ancestral functions, we performed similar experiments using a mouse ES cell line expressing Drosophila Labial. Regions bound by Labial show extensive overlap with HOXA1 binding peaks compared with those of HOXB1 (Fig. 3A). However, HOXB1 peaks (267) that are cobound with Labial also overlap with REST, indicating an ancestrally conserved association of HOX and REST (Fig. 3C). Motif analysis of the common binding peaks between HOXA1 and Labial revealed an enrichment of HOX, PBX, and MEIS consensus motifs, similar to those observed for HOXA1 alone (Fig. 3D). These analyses demonstrate that the HOXA1 and HOXB1 proteins have distinct binding properties in vivo and their occupancy correlates with downstream target genes involved in different biological processes. This functional difference between HOXA1 and HOXB1 is associated with their differential interaction with PBX. The unique genome-wide binding of HOXB1 proteins and associated genes required for neurogenesis and behavior suggests that HOXB1 has neofunctionalized by regulating a new set of the genes in genome. Together, these functional experiments in Drosophila and mammalian cells reveal that HOXA1 has retained ancestral properties of Labial while HOXB1 has diverged. This functional difference between HOXA1 and HOXB1 is associated with their differential interaction with PBX.
The unique genome-wide binding of HOXB1 proteins and associated genes required for neurogenesis and behavior suggests that HOXB1 has neofunctionalized by regulating a new set of the genes in genome. This is also supported by functional and mutational analyses of Hoxb1 in a number of vertebrate species, from zebrafish to humans, that have revealed a highly conserved role for HOXB1 in cranial nerve patterning (Goddard et al. 1996; Studer et al. 1996; McClintock et al. 2002; Webb et al. 2012).
Small differences in multiple domains underlie functional diversification of HOX1 proteins
Despite similar degrees of sequence divergence between HOX1 and Labial proteins, it is striking that only HOXA1 has retained the DNA-binding properties and ancestral function of Labial. The main sequence conservation of these proteins lies within their homeodomain and hexapeptide regions. HOXA1 and HOXB1 have exactly the same hexapeptide and 88% similarity in the homeodomain. Except for the two amino acids, all other changes in the homeodomain are conservative. This raises a question about the differences that underlie functional diversification between the HOXA1 and HOXB1 with respect to ancestral activities.
To identify the region(s) that underlie this functional diversification, we expressed a series of chimeric proteins from the labial locus, where different regions of HOXB1 were replaced by equivalent regions of HOXA1 and scored for their ability to rescue labial (Fig. 5A; Supplemental Table S1). HOX proteins have an N-terminal transcriptional activation/repression region, a hexapeptide motif (HP) that interacts with PBX/Exd cofactors and the homeodomain (HD) required for DNA binding (Merabet and Mann 2016). Roles of the linker (LK) region between HP and HD and the region C-terminal to HD are unclear. While most divergence lies in the N-terminal region (Fig. 1A; Supplemental Fig. S1), substituting HOXB1- N-terminal with HOXA1 (Chimera-1) did not alter its inability to rescue labial (Fig. 5A).
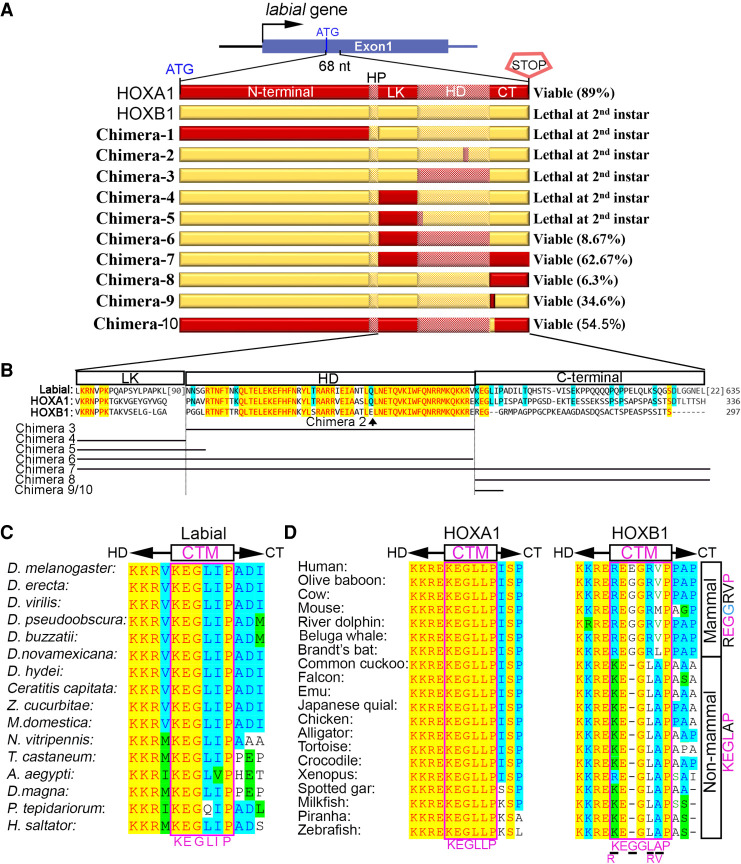
Figure 5.
Small differences in multiple regions functionally separate HOXA1 from HOXB1. (A) Schematic showing the first exon of the labial gene (top) along with diagrams of chimeric mouse HOX proteins (bottom) generated by replacing different regions of HOXB1 (yellow) with those of HOXA1 (red) proteins. Domains of the HOX proteins are indicated. (–) N-terminal, (HP) hexapeptide, (LK) linker, (HD) homeodomain, (CT) C-terminal. Survival rates of these genetic lines or genotypes are listed on the right. (B) Amino acid sequence alignment of LK, HD, and CT regions of Labial, HOXA1, and HOXB1. The line diagrams below represent position of regions replaced in each of the chimeras. (C) Protein sequence alignment around the CTM region among arthropods Labial (listed at the left). (D) Protein sequence alignment around the CTM region among vertebrate (listed at the left) HOXA1 and HOXB1. The CTM region among nonmammalian HOXB1 shows a high conservation of KEGLAP sequence but in mammals there is an insertion of a glycine (G) amino acid (blue) and conservation (pink) of only three amino acids.
Since the most divergent region did not account for the functional difference between HOXA1 and HOXB1, and the HP motif is identical between these proteins, we focused on small differences in the highly conserved LK to HD regions (Fig. 5A,B). Reverting E to Q at 39th position of HOXB1-HD, replacing the whole HD, replacing the LK region alone or in combination with the first four amino acids of the HD (Chimera-2 to Chimera-5) failed to rescue (Fig. 5A,B). The first evidence for rescue arose from Chimera-6 (8.7%), which had a combination of both the LK and HD regions. Furthermore, combining the C-terminal region with the LK-HD domains (Chimera-7) significantly enhanced the efficiency of rescue (62.7%) (Fig. 5A,B). Replacing only the C-terminal region in HOXB1 (Chimera-8) also produced viable flies at a low level (6.3%). These results imply that cooperation among the LK, HD, and C-terminal regions plays a major role in conferring ancestral function to HOXA1. While Chimera-7 rescues lethality with 62.7% efficiency, this is lower than the full-length HOXA1 protein (89%), revealing that the N-terminal domain may also have some contribution to ancestral activity. Together, these data indicate that small amino acid differences in multiple regions contribute to the loss of ancestral activity of HOXB1 during evolution.
A six-amino-acid C-terminal motif (CTM) is a major determinant of ancestral activity
Our chimeric analyses indicate that changes outside of the highly conserved DNA-binding regions can have profound effects on function. It is interesting that replacing only the C-terminal region of HOXB1 with that of HOXA1 (Chimera-8) is sufficient to facilitate a low level of rescue. The functional role of C-terminal regions of HOX proteins and their impact on DNA-binding properties has not been well characterized. Therefore, we examined the C-terminal region of HOX1 proteins to explore its role in ancestral activity. Alignment of the C-terminal domains of Labial in arthropods and HOXA1 and HOXB1 in vertebrates uncovered a six-amino-acid motif located immediately C-terminal to the HD, which is highly conserved in arthropod Labial and vertebrate HOXA1 proteins but has diverged in HOXB1 (Fig. 5C; Supplemental Figs. S2, S6). We named this sequence the C-terminal motif (CTM). The CTM motif is identical in all vertebrate HOXA1 proteins examined (KEGLLP), but in HOXB1 proteins it varies between nonmammalian (KEGLAP) and mammalian (REGGRVP) vertebrates (Fig. 5C).
To functionally assess the role of this motif, we replaced the CTM in HOXB1 with that of HOXA1 (Chimera-9). Remarkably, this small change was sufficient to rescue labial function with relatively high efficiency (34.6%) (Fig. 5A,B). Furthermore, to determine whether the CTM is required for ancestral activity of HOXA1, we replaced this domain in HOXA1 with the HOXB1 sequence (Chimera-10) and found that it reduced the ability of HOXA1 to rescue labial (89%–55%). These results confirm that the CTM region is a major determinant of ancestral activity in HOXA1 and amino acid differences in this region of HOXB1 lead to functional diversification. It is interesting that the level of rescue observed by replacing the whole C-terminal region (Chimera-8) is lower than that found by replacing only the six amino acids of the CTM (Chimera-9), 6.3% versus 34.6%, respectively. This suggests that interactions with other sequences in the C-terminal region have the potential to impact the role of the CTM or other domains in HOX1 proteins (Fig. 5A).
The CTM region modulates HOX1 interactions with PBX
The amino acid differences between HOXA1 and HOXB1 could directly alter interactions with DNA or modify protein–protein interactions and indirectly impact DNA-binding specificity (Treisman et al. 1989; Brickman et al. 2001; Merabet and Mann 2016). To begin to investigate the role of CTM region we compared the DNA-binding properties of Labial, HOXA1, HOXB1, Chimera-9, and Chimera-10 in an in vitro electrophoretic mobility shift assay (EMSA). We used a previously characterized HOXA1 target site identified downstream from Meis3 (De Kumar et al. 2017b). In vivo, the Meis3 site shows enriched occupancy of Labial, HOXA1, PBX, and Meis, but HOXB1 shows only weak occupancy (Fig. 6A). In contrast, in vitro, we observed binding of HOXA1, HOXB1, and Labial along with PBX on the Meis3 oligo containing the bipartite PBX-HOX motif (Supplemental Fig. S7A). To investigate the role of the CTM in DNA-binding properties of HOX1 proteins we used a comparative EMSA approach that quantified the binding affinity/stability of HOX-PBX-DNA ternary complexes on this site (Fig. 6B,C). We found that replacing the CTM of HOXA1 with that of HOXB1 (Chimera-10) reduces the affinity of interaction with PBX from 100% to 77%. Conversely, replacing the CTM of HOXB1 with that of HOXA1 (Chimera-9) enhances binding with PBX from 100% to 166%. To further generalize this observation to other loci in the genome, we repeated same experiment with a previously characterized binding site (R3) for HOX1 proteins on an auto/cross-regulatory element upstream of mouse Hoxb1 (Pöpperl et al. 1995; Ferretti et al. 2005) and observed similar results (Fig. 6C; Supplemental Fig. S7C). This shows that the CTM region of HOXA1 enhances the affinity/stability of HOX–PBX–DNA ternary complexes. To explore whether the interaction of the CTM region with PBX is evolutionarily conserved, we performed similar experiments with Exd, a Drosophila homolog of mouse PBX (Supplemental Fig. S7B). We observed a similar role for the CTM of HOXA1 in enhancing affinity/stability of ternary complexes. These data further support the functional in vivo rescue results in Drosophila, where presence of the CTM region of HOXA1 improves the rescue efficiency.
Figure 6.
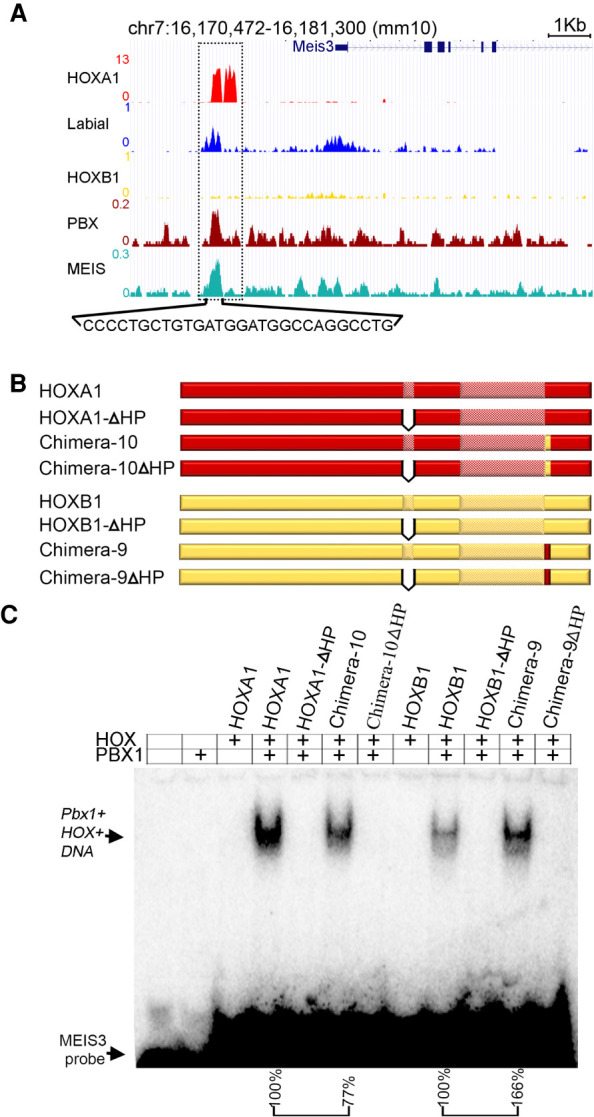
Comparing DNA-binding properties of HOX protein in vivo and in vitro. (A) An enhancer region downstream from Meis3 gene (dotted box) is occupied by HOXA1 (red) and Labial (blue), MEIS (Jade) and PBX (Maroon); however, HOXB1 binding is very weak in in vivo ChIP-seq experiments. (B) Full-length protein of HOXA1, HOXB1, Chimera-9, and Chimera-10 along with hexapeptide deletion mutant versions (ΔHP) were prepared in vitro. (C) EMSA in vitro assay with bipartite motif (bold sequence below the genome browser shot) in Meis3 enhancer show unbound probe at the bottom while PBX1–HOX–DNA ternary complex runs slow in the gel. Template is added in all the wells while PBX1 and different HOX proteins are mentioned above the gel. Same amounts of template, PBX and different versions of HOX proteins were added in all the wells. Quantification of intensity of PBX1–HOX–DNA ternary complex is indicated below the gel. Band intensity of Chimera-10 (77%) is normalized to that of full-length HOXA1 + PBX lane (100%). Similarly, intensity of the band in Chimera-9 (166%) is normalized to that in HOXB1 + PBX lane (100%).
Figure 7.
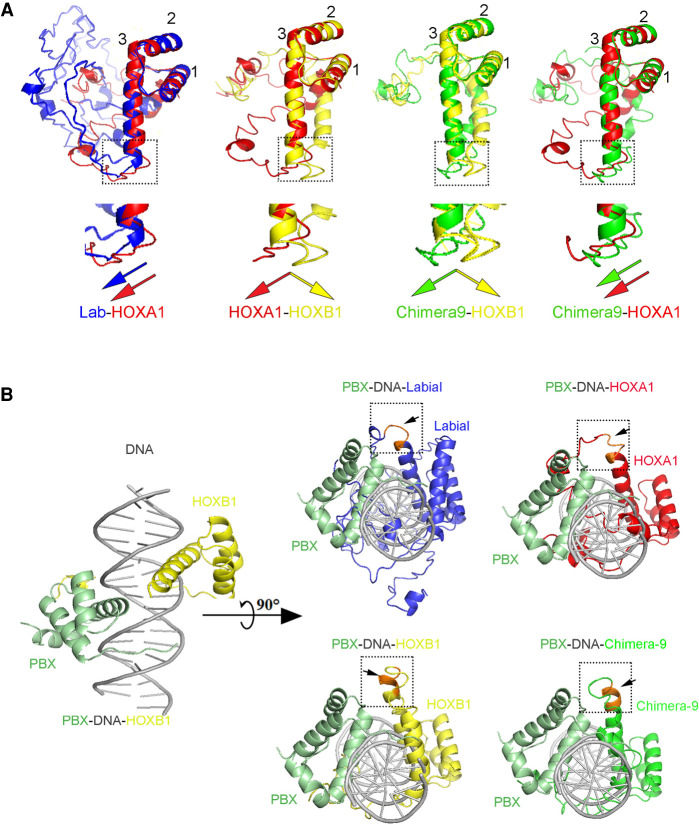
Structural alignment of predicted HOX1 orthologs with Pbx and DNA. (A) The top panels show structural alignments of predicted protein models of HP through C-terminal regions of Labial (blue), HOXA1 (red), HOXB1 (yellow), and Chimera-9 (green). The bottom panels show zoomed-in views of respective boxed areas to visualize the bending patterns of the CTM region represented by color-matched arrows. Known crystal structure of human HOXB1 (PDB ID: 1B72A) is shown at the left. (B) Predicted 3D structure of HP to end of the Labial, HOXA1, HOXB1, and Chimera-9 were then aligned to human HOXB1 in Pbx–HOXB1–DNA ternary complex to examine the position of CTM region (orange) with respect to PBX. In Labial (blue), HOXA1 (red), and Chimera-9 (green) CTM region bends toward PBX (olive green) protein but HOXB1 (yellow) displays bending in opposite direction (boxed with black arrow).
While these results indicate that the CTM region can modulate the binding affinity of HOX1 interactions PBX on DBA we next explored whether it is sufficient for interacting with PBX. In most HOX proteins, the hexapeptide domain is the primary region involved in direct physical interactions with PBX, but this domain is identical in HOXA1 and HOXB1. Some HOX–PBX interactions can also occur through other regions of HOX proteins (Merabet et al. 2003; Merabet and Mann 2016; Dard et al. 2018). Therefore, we compared the in vitro binding affinity of HOXA1, HOXB1, Chimera-9, and Chimera-10 proteins with and without the hexapeptide regions. In all cases, we observed a complete absence of binding in these proteins on the Meis3 and R3 probes when the hexapeptide was deleted (Fig. 6B,C; Supplemental Fig. S7B,C). These data reveal that the CTM of HOXA1 is not sufficient for mediating interactions with PBX. Our in vitro and in vivo binding properties of HOXA1 and HOXB1 proteins suggest that CTM region of HOXA1 has a conserved ability to enhance binding affinity/stability of HOX1/PBX complexes on DNA target sites.
To explore potential interactions between the CTM and PBX we used an in silico bioinformatics method (I-TASSER [iterative threading assembly refinement]) for predicting three-dimensional structure using a previously identified structure for the HP-HD region of human HOXB1 as a template (Piper et al. 1999; Roy et al. 2010). Overlay of the predicted 3D structures show similar conformations of the HD regions. However, the CTM regions in Labial, HOXA1, and Chimera-9 are similar to each other, but they are different from that in HOXB1 (Fig. 7A). We extended this analysis to include PBX, through alignments based on the structure of a ternary complex containing a human HOXB1–PBX1 heterodimer bound to DNA (Fig. 7B; Piper et al. 1999). The structural alignments predict bending of the CTM regions of Labial, HOXA1, and Chimera-9 toward PBX, while the CTM of HOXB1 bends in the opposite direction. This suggests that the CTM region may contribute to differences in physical interactions of HOXA1 and HOXB1 with PBX in vivo. This is consistent with our proteomic data, functional analyses and in vivo and in vitro binding properties demonstrating that, in contrast to HOXB1, HOXA1 and PBX proteins exist in the same complex on chromatin and there is enrichment in genome-wide co-occupancy of HOXA1 with TALE proteins (Figs. 3, 4). Hence, in addition to the HP region, the CTM appears to provide an important domain for interaction of HOXA1 proteins with PBX, which can modulate their genome-wide binding properties and function.
Discussion
The availability of large-scale genome sequencing and gene expression profiling has significantly improved our ability to understand how evolutionary forces act at regulatory regions of the genome for the divergence of the ancestral genes (Brookfield and Sharp 1994; Nurminsky et al. 1998; Sandve et al. 2018). However, how amino acid changes impact protein function are poorly understood (Soskine and Tawfik 2010). In this study we performed in vivo cross-species functional analyses of three mouse HOX1 paralogs with their Drosophila ortholog Labial to investigate the maintenance and diversification of ancestral protein function at the molecular and organism level. The functional analyses of chimeric proteins inserted into the Drosophila labial locus to show that a novel six-amino-acid CTM region is critical in mediating differences in the ancestral activity of HOXA1 and HOXB1. Our analyses indicate that small amino acid changes played a major role in the diversification of HOX1 protein function in development and evolution. These findings raise a number of issues relevant for understanding the evolution of proteins function.
Ancestral activity and neofunctionalization of mouse HOX1 paralogs
The process of gene duplication during vertebrate evolution provides substrates for functional divergence and emergence of evolutionary novelties. Gene duplication relaxes the functional constraint on an essential gene, and a random evolutionary process is expected to lead to subfunctionalization of ancestral activities among paralogs (Force et al. 1999). However, a striking finding from our analyses is that HOXA1 has selectively retained ancestral functions of Drosophila Labial despite extensive sequence divergence of these proteins over 600 million years. While HOXA1 and HOXB1 were thought to be functionally redundant in mouse (Tvrdik and Capecchi 2006), our data demonstrate that HOXB1 has lost ancestral activity and displays genome-wide binding properties and targets that are distinct from those of HOXA1. The occupancy of HOXB1 and HOXA1 correlates with downstream target genes involved in different biological processes (Supplemental Fig. S5) and functional differences in their roles in mouse development and fitness (Tvrdik and Capecchi 2006; Ruff et al. 2017). These observations suggest neo-functionalization of HOXB1 and HOXD1 during the duplication and divergence of HOX1 genes in evolution of vertebrates. Our functional analysis of HOX1 proteins is consistent with the classical neofunctionalization model where HOXA1 maintains ancestral functions while the other paralogs have evolved new functions or been lost (Ohno 1970).
Sequence similarity does not always reflect conserved protein function
All three mouse HOX1 paralogs (HOXA1, HOXB1, and HOXD1) display similar low levels of sequence similarity with Drosophila Labial. However, surprisingly we found that the most divergent regions do not account for the functional differences between these proteins associated with ancestral activity. Instead, small differences in multiple domains of mammalian HOX1 proteins have a profound impact in protein function and underlie the in vivo differences in their ability to fulfill the ancestral functions of Labial. The key functional differences in amino acids lay outside of the highly conserved hexapeptide and homeodomain, indicating that other domains impact specificity and function. Our analysis uncovered a six-amino-acid motif (CTM) flanking the homeodomain, that serves as a major determinant for conservation of ancestral Labial activity in HOXA1 protein. Generally, sequence similarity between two proteins is assumed to be the best predictor of functional conservation, but our work illustrates this simple formulation may not reveal the full spectrum of protein evolution and functional divergence.
HOX1 and PBX interactions
Our genomic and proteomic analyses of HOXA1 and HOXB1 in differentiated mouse ES cells revealed key differences in their genome-wide interactions with PBX in vivo. HOXA1, but not HOXB1, shows an enrichment in genome-wide co-occupancy with PBX proteins on target sites and HOXA1 and PBX exist in the same complex on chromatin (Figs. 3, 4). In vivo analysis in Drosophila identified the CTM region as a key determinant of the functional differences between HOXA1 and HOXB1. The in vitro biochemical data revealed that the CTM region of HOXA1 enhances its ability to directly interact with PBX on target sites (Fig. 6C; Supplemental Fig. S7C). Evolutionary divergence of the CTM region in HOXB1 reduces its ability to interact with PBX, and this can be restored by replacing the CTM of HOXB1 with that of HOXA1. These findings highlight the importance of HOX–PBX interactions in functional specificity of HOX proteins.
The hexapeptide domain of HOX proteins is considered as the primary region involved in direct physical interactions with cofactor PBX (Chang et al. 1995; Chan et al. 1996; Merabet and Mann 2016). However, recent studies have provided evidence for additional HOX–PBX interaction domains in several Drosophila and vertebrate HOX proteins. These domains are highly variable in terms of their location and sequences along the HOX proteins (Merabet et al. 2003; Merabet and Mann 2016; Dard et al. 2018). It is interesting that by analogy to the CTM of HOXA1, a nine-amino-acid paralog-specific UBDA motif has been identified immediately C-terminal to the HD in Drosophila Ubx and Abd-A HOX proteins (Chan and Mann 1993; Merabet et al. 2007; Saadaoui et al. 2011). The UBDA motif was further characterized biochemically and structurally and found to be a flexible extension in DNA recognition by the homeodomain and provide an alternative interaction mode with Exd/PBX proteins (Foos et al. 2015). This is in contrast to the CTM, which is not sufficient to mediate interactions with PBX independent of the hexapeptide domain. Intriguingly, the in silico structure prediction shows that the CTM region has different conformations between HOXA1 and HOXB1 that alter its relative proximity to PBX. The conformation in HOXA1 is similar to that of the motif in Labial and, in both cases, brings them closer to PBX, while HOXB1 bends in the opposite direction. This is consistent with the idea that the CTM represents an additional domain for HOX-PBX interaction and changes in this region modulate this interaction thereby altering HOX1 function. This suggests that differences in protein–protein interactions through CTM region are likely to impact DNA-binding specificity of HOXA1 and HOXB1. These data suggest that loss of CTM region in mouse HOXB1 have contributed to its diversion from ancestral activity.
Our functional and biochemical analyses, together with other studies, show that there are diverse types of HOX–PBX/Exd interactions, which are mediated through multiple determinants spread over the proteins (Merabet and Mann 2016). The presence of a variety of HOX–PBX interaction domains in Drosophila and mouse HOX proteins provides a means for diverse inputs that could be utilized for the diversification of protein function during evolution. These interaction domains appear to provide a commonly used target for diversification of paralog-specific HOX protein functions during evolution, as our functional studies have shown for the CTM region of HOX1proteins.
Materials and methods
Protein sequence alignments
Protein sequences were downloaded from NCBI protein database and aligned using Vector NTI software using default settings. Multiple alignment tool of NCBI was used with default settings to align Drosophila Labial with mouse HOXA1 and HOXB1 proteins because vector NTI software was inefficient in aligning highly diverged regions outside the homeodomain (Altschul et al. 1997).
Drosophila melanogaster culture
Drosophila culture was done using standard protocol and all experiments were performed at 25°C. The labial gene mutant (Lab4) was obtained from Bloomington Drosophila stock center. All the fly lines generated in this study are listed in Supplemental Table S1 to explain their name, genotype, and expressed protein.
Generation of Drosophila knock-in lines
Two guide RNAs (gRNAs) separated by 68 nt were designed and cloned in pCFD3-dU6:3gRNA (Addgene plasmid # 49410) to target labial gene close to start codon (ATG) in the germline (Port et al. 2014). Targeting cassettes consisted of the homology sequences and gene of interest with 3xFLAG-1XMyc epitope tag to insert it at the start codon of labial gene (Supplemental Table S2). These cassettes also had flanking Gypsy insulator to minimize positional effect due to site of integration and Ubiquitin promoter-mCherry as a transformant marker with flanking loxP sequences enabled the removal of these sequences in the presence of Cre protein. All the sequences were assembled in pUC19 vector using Gibson assembly. In the case of labial targeting cassette, the gRNA target regions were modified without changing amino acid sequences to avoid targeting of gRNAs on the cassette (Supplemental Table S2). Plasmid DNAs of targeting cassette and gRNA were coinjected in nos-Cas9 fly embryos. Injected embryos were grown at 25°C and adult hatched flies were crossed to flies with third chromosome balancer and progeny were screened for mCherry expression. These lines were further crossed with Cre expressing flies to remove “loxP-Gypsy insulator-Ubiquitin promoter-mCherry-Gypsy insulator-loxP” cassette and screen for loss of mCherry expression. The mCherry negative flies had only the inserted gene of interest with 3xFLAG tag and a loxP. Insertion of the required gene was confirmed by sequencing and only these lines were used for phenotypic analysis. Primer sequences used for gRNA and Gibson assembly of gfp, labial, Hoxa1, Hoxb1, and Chimeras are listed in Supplemental Table S2.
Whole-mount RNA in situ hybridization in embryos
The labial RNA in situ was performed to assess the expression of the genes in 5- to 6-h-old wild-type embryos. We followed the published protocol except that the experiments were done in microfuge tubes instead of 96-well plates (Weiszmann et al. 2009). A 442-bp-long fragment from 5′ end of labial gene was amplified using the following primers: Lab-F (CTGTGTGGCAAGTGAAGGGT) and SP6 promoter sequence contained reverse primer SP6-labR (ATTTAGGTGACACTATAGAGAGTGGCTACTAGGATGGA). The amplified fragment was used for making the probe using in vitro transcription.
Immunostaining
Drosophila embryos, larval eye-antennal and brain disc were immunostained using 1:80 diluted anti-FLAG antibody (Sigma F1804), 1:20 anti-Fas-II (DSHB 1D4), 1:40 anti-Elav (DSHB 7E8A10), and anti-Abd-A antibody (1:50 dilution; Santa Cruz Biotechnology SC-27063). In brief, 5- to 6-h-old embryos were fixed in 4% formaldehyde for 30 min, blocked for 2 h in PBSTB (1× PBS + 0.1% Triton + 1% BSA + 1% donkey serum) and incubated overnight in PBSTB with primary antibody. Embryos were washed in PBSTB and reincubated in 1:300 diluted fluorescence tagged secondary antibody. Finally, embryos were washed and mounted in VectaShield antifade mounting medium with DAPI for imaging. Imaginal discs were processed similarly for immunostaining except that we fixed dissected imaginal discs from third instar larvae in 4% paraformaldehyde. All images were taken using Zeiss confocal microscope and images were processed and analyzed using Zen software from Zeiss and ImageJ software. Adobe Photoshop was further used to arrange all the images in final figures without any additional processing.
Preparation of larval cuticle
Larval cuticle was prepared for phenotypic analysis of mouth hook from WT, labial mutant (lab4), Hoxa1, and Hoxb1 knock-in lines. Larvae were washed and mounted in lactic acid:H2O (3:1) medium and incubated for 72 h at 60°C to digest the internal parts (Stern and Sucena 2011). Cleared larvae were imaged using stereo microscope in light and dark field modes.
Larval feeding behavior
Food intake was tracked by feeding larvae with fly food containing food dye (Melcher and Pankratz 2005). Hoxb1-KI heterozygous, and homozygous embryos were sorted using GFP-expressing balancer chromosome. The embryos were then transferred to fly food with 1% blue food dye. After 48 h, larvae were collected from the food and imaged for localization of the food. Mouth hook extension of the second instar larvae of heterozygous and homozygous Hoxb1-KI larvae was analyzed in 2% baker's yeast medium (Bhatt and Neckameyer 2013). Frequency of mouth hook contraction was counted manually for 20 WT and Hoxb1-KI larvae under microscope. For calculating average contraction rate, we omitted data of Hoxb1-KI homozygous larvae that show very few or no contractions per minute.
Electron microscopy
Adult heads were imaged using Hitachi TM1000 table-top scanning electron microscope. To avoid any change in bristle pattern of the head, the heads were detached from the body and immediately imaged without any processing. A darker color contrast is used in the images for better visualization of the bristle phenotypes.
Mouse embryonic stem cell culture
Feeder free culture was used to grow transgenic KH2 cells using N2B27 + 2i medium supplemented with 2000 units/mL ESGRO (Millipore) (De Kumar et al. 2017a). Cells were differentiated in neurponal fate on a gelatinized plate using differentiation media (DMEM + 10% [v/v] serum + nonessential amino acids + 3.3 µM RA). ES cells were harvested at 80%–90% confluency for the experiments. We generated single-copy transgenic mouse KH2 ES cell line (Open Biosystems MES 4304) by inserting 3XFLAG-Myc-tagged Hoxa1, Hoxb1, and labial at modified Col1A1 locus that encodes the type I collagen protein under tight control of a doxycycline (Dox)-inducible promoter (Beard et al. 2006). All cell lines were tested for karyotype stability. A FACSCalibur was used to analyze DNA context and measure indirect inference karyotype stability. The conditions for Dox induction and programmed differentiation of the ES cells were optimized so that the expression levels of the epitope-tagged versions were at levels comparable with endogenous the Hox gene (De Kumar et al. 2015, 2017b). To assess the genome-wide DNA-binding properties by ChIP-seq we differentiated mouse ES cells for 24 h using retinoic acid (RA) and simultaneously induced epitope tagged HOX protein expression using optimized levels of doxycycline (1 mg/mL).
ATAC and ChIP-seq
We observed previously that program differentiation of ES cells into neuronal fates using retinoic acid (RA) leads to sequential activation of Hox genes in a manner mimicking their induction during embryonic development (Simeone et al. 1990, 1991; Papalopulu et al. 1991; Mazzoni et al. 2013; Sheikh et al. 2014; De Kumar et al. 2015). The expression of the epitope-tagged version of HOX proteins in response to RA is optimized to express as comparable with that of the endogenous gene (De Kumar et al. 2017b). ChIP-seq of HOXA1, HOXB1 and Labial proteins were done using anti-FLAG antibody in 24 h RA differentiated KH2 ES cells. ChIP was performed as per the Upstate protocol as described in Smith et al. (2010). ATAC-seq was done following the protocol of Buenrostro et al. (2015) using ∼50,000 feeder-free uninduced and 24-h differentiated ES cells. ATAC-seq and ChIP-seq for PBX, MEIS, REST, NANOG were performed on 24-h RA differentiated unmodified KH2 ES cells. The following antibodies were used for ChIP-seq experiments: anti-FLAG (Sigma-Aldrich F1804), anti-PBX (Santa Cruz Biotechnology SC-888), anti-MEIS (Santa Cruz Biotechnology SC-25412), anti-P300 (Santa Cruz Biotechnology SC-585X), anti-H3K27Ac (Abcam ab4729), anti-H3K4me3 (Abcam ab8895), and anti-NANOG (Cell Signaling Technology D1G10). All raw data files for ChIP-seq, ATAC-seq data have been submitted to the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/sra/?term=PRJNA503882, PRJNA341679, and PRJNA335616) and Sequence Read Archive under accession numbers SRX4980243 to SRX4980246. All original source data are also deposited in the Stowers Institute Original Data Repository and available online at http://odr.stowers.org/research/publications/libpb-1565.
Genomic analysis
ATAC-seq and ChIP-seq reads were aligned to mm10 with bowtie2 default parameters (Langmead and Salzberg 2012). For ATAC-seq processing, we removed duplicates and retained fragments with max insert size 500 bp and concordant pairs with MAPping quality (MAPQ) values > = 10. We used ataqv software package for ATAC-seq quality control (QC) (Orchard et al. 2020). ChIP-seq peaks were called with MACS2 parameters “-p 0.25 -m 5 50” to ensure sufficient peaks for IDR analysis (Zhang et al. 2008). MACS2 peak coordinates were trimmed back to meet the actual IP signal. For all the duplicates, the top 100,000 peaks by P-value were sent through IDR 2.0.7, and paired peaks with MACS2 fold change ≥ 4, q-value ≤ 0.05, and IDR global P ≤ 0.1 were collected (Supplemental Table S3). HOXA1 and HOXB1 ChIPs were performed multiple times independently (Supplemental Table S3). To generate a common peak list, we analyzed biological replicates done at same and different time points separately. For the replicates done at different time points, the top 30,000 peaks by P-value peaks were also subset at greater than or equal to fourfold change. To further reduce noise from the five HOXB1 single-replicate samples, all five peak lists were compared, and only peaks found in at least two replicates were retained.
Binding peaks of HOXA1 and HOXB1 were analyzed with binding peaks of Labial, PBX, MEIS, REST, and NANOG in 24-h RA differentiated mouse ES cells to find the overlapping/co-occupied regions in the genome. Heat maps were generated using Bioconductor CoverageView package for R. Heat map values are produced by normalizing reads to per million and taking log fold change (RPM-LFC) between IP and input and only positive values are plotted using hierarchical clustering. HOXB1 heat map is generated using highest RPM-LFC data among the HOXB1 experiments using common peak list. Transcription factor-binding motifs in the HOXA1- and HOXB1-binding peaks were analyzed using HOMER motif analysis tool (Heinz et al. 2010).
Gene ontology (GO) analysis of the HOXA1 and HOXB1 target genes
Protein-coding genes nearest to the HOXA1 and/or HOXB1 bound peaks were extracted (version Ensembl 91) and queried for enriched GO terms using the GeneOntology.org May 2018 database release (The Gene Ontology Consortium 2015; Huntley et al. 2015). For each term, counts were taken as all hits to the term itself or any of its child terms. Terms for gene lists were compared with the remaining genes in the database using a one-tailed Fisher's exact test, BH-adjusted P-value ≤ 0.05. Only significant terms with three or more genes in the list were retained or if >25% of all genes in the database with that term were also in the gene list.
Immunoprecipitation followed by MudPIT
Immunoprecipitations were performed for HOXA1 and HOXB1 proteins by anti-FLAG antibody Agarose beads (Sigma Aldrich A2220) using chromatin extract of retinoic acid differentiated mouse ES cells (Aygun et al. 2008; De Kumar et al. 2017b). Protein complexes associated with HOXA1 and HOXB1 were identified using MudPIT (multidimensional protein identification technology) on a LTQ linear ion trap mass spectrometer (Thermo Scientific) coupled to a Quaternary Agilent 1100 series HPLC pump and nano-LC electrospray ionization source. Tandem mass (MS/MS) spectra were interpreted using ProluCID (Cai et al. 2015) against a database consisting of 78014 nonredundant Mus musculus proteins (NCBI June 23, 2016, release), 193 usual contaminants (human keratins, IgGs, and proteolytic enzymes). DTASelect (Laurikkala et al. 2002) and swallow, an in-house-developed software (v. 0.0.1, https://github.com/tzw-wen/kite), were used to filter ProLuCID search results at given FDRs at the spectrum, peptide, and protein levels. Here all controlled FDRs were <5%. All 16 data sets (four IP replicates of HOXA1 and HOXB1 and four control IP replicates of HOXA1 and HOXB1) were contrasted against their merged data set using Contrast v1.9 and in-house-developed sandmartin v0.0.1. Our in-house-developed software, NSAF7 v0.0.1, was used to generate spectral count-based label-free quantitation results (dNASF) (Zhang et al. 2010). QPROT was used to assess the significant enrichment or depletion of affinity-purified proteins compared with negative controls (Choi et al. 2008). Mass spectrometry data files are available from Massive at ftp://MSV000085127@massive.ucsd.edu (password NPS01146) and ProteomeXchange (PXD018127).
Structure prediction and alignment
Three dimensional structures of HOXA1, HOXB1, Labial, and Chimera-9 proteins from hexapeptide to end of the protein were predicted using online software I-TASSER (iterative threading assembly refinement) using the known structure of ahuman HOXB1 protein (PDB ID-1B72) as a template (Roy et al. 2010). These structures were further aligned using PyMOL (The PyMOL molecular graphics system, version 2.0, Schrödinger, LLC) to understand the implication of structural changes on interaction with Pbx and DNA.
Electrophoretic mobility shift assays (EMSA)
In vitro binding properties of HOXA1, HOXB1, and their variants were tested on PBX-HOX bipartite motif oligos of Meis3 (CCCCTGCTGTGATGGATGGCCAGGCCTG) and Hoxb1 (GGGGTGATGGATGGGCGCTG) enhancers. Binding reaction was performed in 1× binding buffer (10 mM Tris-Cl at pH 7.5, 75 mM NaCl, 1 mM EDTA, 6% glycerol, 3 mM spermidine, 1 mM DTT, 1 µg of poly[dIdC], protease inhibitor) with in vitro translated protein (Tnt Promega kit) and 0.2 pmol of P-32 labeled oligonucleotides for 30 min on ice. In all these reactions labeled probe was added after all other components to start the reaction at similar time. In the cold competition reactions, the Cold competitor were added after 30 min of the reaction with hot probe and the reaction was further incubated for another 30 min. A mutant version of the Meis3 oligo (CCCCAGTCACCCCTGCTG) was used to assess the binding at the bipartite motif. Monoclonal anti-FLAG (Sigma F1804) was used in the reaction for supershift experiments. All reactions were run on 6% PAGE gels with 0.5× TBE buffer. For comparing affinity/stability of HOXA1 with its variants (HOXA1-ΔHP, Chimera-10, and Chimera-10ΔHP) and HOXB1 with its variants (HOXB1-ΔHP, Chimera-9, and Chimera-9ΔHP) we quantified in vitro translated proteins using western blot. Equal amount of HOXA1 and HOXB1, and variant proteins were used in the EMSA experiments. Quantification of shifted band in HOXA1, Chimera-10, HOXB1 and Chimera-9 samples were done using image quant software from GE healthcare.
Data and material availability
All original source data are deposited in the Stowers Institute Original Data Repository and are available online at http://odr.stowers.org/research/publications/libpb-1565. Drosophila lines generated and/or analyzed during this study are available on request. All software used in this study is published and publicly available: Vector NTI (https://www.thermofisher.com/us/en/home/life-science/cloning/vector-nti-software.html), Bowtie 2 (http://bowtie-bio.sourceforge.net/bowtie2/index.shtml), MACS2 (https://taoliu.github.io/MACS), CoverageView (https://bioconductor.org/packages/release/bioc/html/CoverageView.html), MEME (http://meme-suite.org), GO term (http://geneontology.org), and I-TASSER (https://zhanglab.ccmb.med.umich.edu/I-TASSER). All the parameters used for analyses of our data using this software is mentioned in the Materials and Methods.
Supplementary Material
Acknowledgments
We thank the Bloomington Drosophila Stock Center for providing the lab4 labial mutant; Takuya Akiyama for providing construct for loxP-Gypsy-mcherry cassette and Cas9-expressing flies; members of the Molecular Biology, Media Preparation, Tissue Culture, Bioinformatics, and Fly Stock Center facilities at the Stowers Institute for Medical Research (SIMR) for their support in this study. We also thank other members of the Krumlauf group for many valuable discussions and advice on this project. This work was supported by funds from the Stowers Institute for Medical Research (grant no. 1001) to R.K.
Author contributions: N.P.S., B.D.K., and R.K. conceived this research program. N.P.S. performed all fly and in vitro DNA mobility assays experiments. B.D.K. performed mouse ES cell ChIP-seq and ATAC-seq genomic experiments with support from M.E.P., A.P., B.D.K., and N.P.S. analyzed mouse ES cell genomic data. N.P.S. and M.E.P. performed proteomic immunoprecipitation experiments and L.F. and Y.Z. performed MudPIT runs and data analysis. J.W.C. provided advice on protocols and analysis of data from MudPIT and in vitro DNA mobility assays. K.S. provided laboratory space, equipment, and support for fly experiments. N.P.S., K.S., and R.K. analyzed the data, discussed the ideas and interpretations, and wrote the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.342329.120.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendt D. 2018. Hox genes and body segmentation. Science 361: 1310–1311. 10.1126/science.aav0692 [DOI] [PubMed] [Google Scholar]
- Aygun O, Svejstrup J, Liu Y. 2008. A RECQ5-RNA polymerase II association identified by targeted proteomic analysis of human chromatin. Proc Natl Acad Sci 105: 8580–8584. 10.1073/pnas.0804424105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker K. 1959. Feeding period, growth, and pupation in larvae of Drosophila melanogaster. Ent Exp Appl 2: 171–186. 10.1111/j.1570-7458.1959.tb00432.x [DOI] [Google Scholar]
- Ballas N, Mandel G. 2005. The many faces of REST oversee epigenetic programming of neuronal genes. Curr Opin Neurobiol 15: 500–506. 10.1016/j.conb.2005.08.015 [DOI] [PubMed] [Google Scholar]
- Beadle GW, Tatum EL, Clancy CW. 1938. Food level in relation to rate of development and Eye pigmentation in Drosophila melanogaster. Biol Bull 75: 447–462. 10.2307/1537573 [DOI] [Google Scholar]
- Beard C, Hochedlinger K, Plath K, Wutz A, Jaenisch R. 2006. Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis 44: 23–28. 10.1002/gene.20180 [DOI] [PubMed] [Google Scholar]
- Berger MF, Badis G, Gehrke AR, Talukder S, Philippakis AA, Peña-Castillo L, Alleyne TM, Mnaimneh S, Botvinnik OB, Chan ET, et al. 2008. Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell 133: 1266–1276. 10.1016/j.cell.2008.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt PK, Neckameyer WS. 2013. Functional analysis of the larval feeding circuit in Drosophila. J Vis Exp e51062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomme T, Vandepoele K, De Bodt S, Simillion C, Maere S, Van de Peer Y. 2006. The gain and loss of genes during 600 million years of vertebrate evolution. Genome Biol 7: R43 10.1186/gb-2006-7-5-r43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickman JM, Clements M, Tyrell R, McNay D, Woods K, Warner J, Stewart A, Beddington RS, Dattani M. 2001. Molecular effects of novel mutations in Hesx1/HESX1 associated with human pituitary disorders. Development 128: 5189–5199. [DOI] [PubMed] [Google Scholar]
- Brookfield JF, Sharp PM. 1994. Neutralism and selectionism face up to DNA data. Trends Genet 10: 109–111. 10.1016/0168-9525(94)90201-1 [DOI] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. 2015. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr Protoc Mol Biol 109: 21 29 21-29 10.1002/0471142727.mb2129s109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai MC, Xu Q, Pan YJ, Pan W, Ji N, Li YB, Jin HJ, Liu K, Ji ZL. 2015. ADReCS: an ontology database for aiding standardization and hierarchical classification of adverse drug reaction terms. Nucleic Acids Res 43: D907–D913. 10.1093/nar/gku1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB. 1995. Homeotic genes and the evolution of arthropods and chordates. Nature 376: 479–485. 10.1038/376479a0 [DOI] [PubMed] [Google Scholar]
- Carroll SB. 2005. Evolution at two levels: on genes and form. PLoS Biol 3: e245 10.1371/journal.pbio.0030245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SK, Mann RS. 1993. The segment identity functions of ultrabithorax are contained within its homeo domain and carboxy-terminal sequences. Genes Dev 7: 796–811. 10.1101/gad.7.5.796 [DOI] [PubMed] [Google Scholar]
- Chan SK, Jaffe L, Capovilla M, Botas J, Mann RS. 1994. The DNA binding specificity of Ultrabithorax is modulated by cooperative interactions with extradenticle, another homeoprotein. Cell 78: 603–615. 10.1016/0092-8674(94)90525-8 [DOI] [PubMed] [Google Scholar]
- Chan SK, Pöpperl H, Krumlauf R, Mann RS. 1996. An extradenticle-induced conformational change in a HOX protein overcomes an inhibitory function of the conserved hexapeptide motif. EMBO J 15: 2476–2487. 10.1002/j.1460-2075.1996.tb00605.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SK, Ryoo HD, Gould A, Krumlauf R, Mann RS. 1997. Switching the in vivo specificity of a minimal HOX-responsive element. Development 124: 2007–2014. [DOI] [PubMed] [Google Scholar]
- Chang CP, Shen WF, Rozenfeld S, Lawrence HJ, Largman C, Cleary ML. 1995. Pbx proteins display hexapeptide-dependent cooperative DNA binding with a subset of Hox proteins. Genes Dev 9: 663–674. 10.1101/gad.9.6.663 [DOI] [PubMed] [Google Scholar]
- Choi H, Fermin D, Nesvizhskii AI. 2008. Significance analysis of spectral count data in label-free shotgun proteomics. Mol Cell Proteomics 7: 2373–2385. 10.1074/mcp.M800203-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dard A, Reboulet J, Jia Y, Bleicher F, Duffraisse M, Vanaker JM, Forcet C, Merabet S. 2018. Human HOX proteins use diverse and context-dependent motifs to interact with TALE class cofactors. Cell Rep 22: 3058–3071. 10.1016/j.celrep.2018.02.070 [DOI] [PubMed] [Google Scholar]
- De Kumar B, Parrish ME, Slaughter BD, Unruh JR, Gogol M, Seidel C, Paulson A, Li H, Gaudenz K, Peak A, et al. 2015. Analysis of dynamic changes in retinoid-induced transcription and epigenetic profiles of murine Hox clusters in ES cells. Genome Res 25: 1229–1243. 10.1101/gr.184978.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Kumar B, Parker HJ, Parrish ME, Lange JJ, Slaughter BD, Unruh JR, Paulson A, Krumlauf R. 2017a. Dynamic regulation of Nanog and stem cell-signaling pathways by Hoxa1 during early neuro-ectodermal differentiation of ES cells. Proc Natl Acad Sci 114: 5838–5845. 10.1073/pnas.1610612114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Kumar B, Parker HJ, Paulson A, Parrish ME, Pushel I, Singh NP, Zhang Y, Slaughter BD, Unruh JR, Florens L, et al. 2017b. HOXA1 and TALE proteins display cross-regulatory interactions and form a combinatorial binding code on HOXA1 targets. Genome Res 27: 1501–1512. 10.1101/gr.219386.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Kumar B, Parker HJ, Paulson A, Parrish ME, Zeitlinger J, Krumlauf R. 2017c. Hoxa1 targets signaling pathways during neural differentiation of ES cells and mouse embryogenesis. Dev Biol 432: 151–164. 10.1016/j.ydbio.2017.09.033 [DOI] [PubMed] [Google Scholar]
- Diederich RJ, Merrill VK, Pultz MA, Kaufman TC. 1989. Isolation, structure, and expression of labial, a homeotic gene of the antennapedia complex involved in Drosophila head development. Genes Dev 3: 399–414. 10.1101/gad.3.3.399 [DOI] [PubMed] [Google Scholar]
- Duboule D, Dollé P. 1989. The structural and functional organization of the murine HOX gene family resembles that of Drosophila homeotic genes. EMBO J 8: 1497–1505. 10.1002/j.1460-2075.1989.tb03534.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti E, Cambronero F, Tümpel S, Longobardi E, Wiedemann LM, Blasi F, Krumlauf R. 2005. Hoxb1 enhancer and control of rhombomere 4 expression: complex interplay between PREP1–PBX1–HOXB1 binding sites. Mol Cell Biol 25: 8541–8552. 10.1128/MCB.25.19.8541-8552.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foos N, Maurel-Zaffran C, Maté MJ, Vincentelli R, Hainaut M, Berenger H, Pradel J, Saurin AJ, Ortiz-Lombardia M, Graba Y. 2015. A flexible extension of the Drosophila ultrabithorax homeodomain defines a novel Hox/PBC interaction mode. Structure 23: 270–279. 10.1016/j.str.2014.12.011 [DOI] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J. 1999. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151: 1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium. 2015. Gene ontology consortium: going forward. Nucleic Acids Res 43: D1049–D1056. 10.1093/nar/gku1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard J, Rossel M, Manley N, Capecchi M. 1996. Mice with targeted disruption of Hoxb1 fail to form the motor nucleus of the VIIth nerve. Development 122: 3217–3228. [DOI] [PubMed] [Google Scholar]
- Graham A, Papalopulu N, Krumlauf R. 1989. The murine and Drosophila homeobox gene complexes have common features of organization and expression. Cell 57: 367–378. 10.1016/0092-8674(89)90912-4 [DOI] [PubMed] [Google Scholar]
- Guo T, Mandai K, Condie BG, Wickramasinghe SR, Capecchi MR, Ginty DD. 2011. An evolving NGF–Hoxd1 signaling pathway mediates development of divergent neural circuits in vertebrates. Nat Neurosci 14: 31–36. 10.1038/nn.2710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartenstein V. 1997. Development of the insect stomatogastric nervous system. Trends Neurosci 20: 421–427. 10.1016/S0166-2236(97)01066-7 [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirth F, Hartmann B, Reichert H. 1998. Homeotic gene action in embryonic brain development of Drosophila. Development 125: 1579–1589. [DOI] [PubMed] [Google Scholar]
- Hirth F, Loop T, Egger B, Miller DF, Kaufman TC, Reichert H. 2001. Functional equivalence of Hox gene products in the specification of the tritocerebrum during embryonic brain development of Drosophila. Development 128: 4781–4788. [DOI] [PubMed] [Google Scholar]
- Holland PW, Garcia-Fernandez J, Williams NA, Sidow A. 1994. Gene duplications and the origins of vertebrate development. Dev Suppl 125–133. [PubMed] [Google Scholar]
- Huntley RP, Sawford T, Mutowo-Meullenet P, Shypitsyna A, Bonilla C, Martin MJ, O'Donovan C. 2015. The GOA database: gene ontology annotation updates for 2015. Nucleic Acids Res 43: D1057–D1063. 10.1093/nar/gku1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kmita M, Duboule D. 2003. Organizing axes in time and space; 25 years of colinear tinkering. Science 301: 331–333. 10.1126/science.1085753 [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with bowtie 2. Nat Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurikkala J, Pispa J, Jung HS, Nieminen P, Mikkola M, Wang X, Saarialho-Kere U, Galceran J, Grosschedl R, Thesleff I. 2002. Regulation of hair follicle development by the TNF signal ectodysplasin and its receptor Edar. Development 129: 2541–2553. [DOI] [PubMed] [Google Scholar]
- Lewis EB. 1978. A gene complex controlling segmentation in Drosophila. Nature 276: 565–570. 10.1038/276565a0 [DOI] [PubMed] [Google Scholar]
- Lufkin T, Dierich A, LeMeur M, Mark M, Chambon P. 1991. Disruption of the Hox-1.6 homeobox gene results in defects in a region corresponding to its rostral domain of expression. Cell 66: 1105–1119. 10.1016/0092-8674(91)90034-V [DOI] [PubMed] [Google Scholar]
- Lundin LG. 1993. Evolution of the vertebrate genome as reflected in paralogous chromosomal regions in man and the house mouse. Genomics 16: 1–19. 10.1006/geno.1993.1133 [DOI] [PubMed] [Google Scholar]
- Mazzoni EO, Mahony S, Peljto M, Patel T, Thornton SR, McCuine S, Reeder C, Boyer LA, Young RA, Gifford DK, et al. 2013. Saltatory remodeling of Hox chromatin in response to rostrocaudal patterning signals. Nat Neurosci 16: 1191–1198. 10.1038/nn.3490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock JM, Kheirbek MA, Prince VE. 2002. Knockdown of duplicated zebrafish hoxb1 genes reveals distinct roles in hindbrain patterning and a novel mechanism of duplicate gene retention. Development 129: 2339–2354. [DOI] [PubMed] [Google Scholar]
- McGinnis W, Krumlauf R. 1992. Homeobox genes and axial patterning. Cell 68: 283–302. 10.1016/0092-8674(92)90471-N [DOI] [PubMed] [Google Scholar]
- Melcher C, Pankratz MJ. 2005. Candidate gustatory interneurons modulating feeding behavior in the Drosophila brain. PLoS Biol 3: e305 10.1371/journal.pbio.0030305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merabet S, Mann RS. 2016. To be specific or not: the critical relationship between Hox and TALE roteins. Trends Genet 32: 334–347. 10.1016/j.tig.2016.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merabet S, Kambris Z, Capovilla M, Bérenger H, Pradel J, Graba Y. 2003. The hexapeptide and linker regions of the AbdA Hox protein regulate its activating and repressive functions. Dev Cell 4: 761–768. 10.1016/S1534-5807(03)00126-6 [DOI] [PubMed] [Google Scholar]
- Merabet S, Saadaoui M, Sambrani N, Hudry B, Pradel J, Affolter M, Graba Y. 2007. A unique extradenticle recruitment mode in the Drosophila Hox protein ultrabithorax. Proc Natl Acad Sci 104: 16946–16951. 10.1073/pnas.0705832104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill VK, Diederich RJ, Turner FR, Kaufman TC. 1989. A genetic and developmental analysis of mutations in labial, a gene necessary for proper head formation in Drosophila melanogaster. Dev Biol 135: 376–391. 10.1016/0012-1606(89)90187-5 [DOI] [PubMed] [Google Scholar]
- Nitta KR, Jolma A, Yin Y, Morgunova E, Kivioja T, Akhtar J, Hens K, Toivonen J, Deplancke B, Furlong EE, et al. 2015. Conservation of transcription factor binding specificities across 600 million years of bilateria evolution. Elife 4: e04837 10.7554/eLife.04837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noyes MB, Christensen RG, Wakabayashi A, Stormo GD, Brodsky MH, Wolfe SA. 2008. Analysis of homeodomain specificities allows the family-wide prediction of preferred recognition sites. Cell 133: 1277–1289. 10.1016/j.cell.2008.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurminsky DI, Nurminskaya MV, De Aguiar D, Hartl DL. 1998. Selective sweep of a newly evolved sperm-specific gene in Drosophila. Nature 396: 572–575. 10.1038/25126 [DOI] [PubMed] [Google Scholar]
- Ohno S. 1970. Evolution by gene duplication. Springer-Verlag, Berlin. [Google Scholar]
- Orchard P, Kyono Y, Hensley J, Kitzman JO, Parker SCJ. 2020. Quantification, dynamic visualization, and validation of bias in ATAC-seq data with ataqv. Cell Syst 10: 298–306.e4. 10.1016/j.cels.2020.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papalopulu N, Lovel-Badage R, Krumlauf R. 1991. The expression of murine Hox-2 genes is dependent on the differentiation pathway and displays a collinear sensitivity to retinoic acid in F9 cells and Xenopus embryos. Nucleic Acids Res 19: 5497–5506. 10.1093/nar/19.20.5497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual-Anaya J, D'Aniello S, Kuratani S, Garcia-Fernàndez J. 2013. Evolution of Hox gene clusters in deuterostomes. BMC Dev Biol 13: 26 10.1186/1471-213X-13-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passner J, Ryoo H, Shen L, Mann R, Aggarwal A. 1999. Structure of a DNA-bound ultrabithorax-extradenticle homeodomain complex. Nature 397: 714–719. 10.1038/17833 [DOI] [PubMed] [Google Scholar]
- Piper DE, Batchelor AH, Chang CP, Cleary ML, Wolberger C. 1999. Structure of a HoxB1–Pbx1 heterodimer bound to DNA: role of the hexapeptide and a fourth homeodomain helix in complex formation. Cell 96: 587–597. 10.1016/S0092-8674(00)80662-5 [DOI] [PubMed] [Google Scholar]
- Pöpperl H, Bienz M, Studer M, Chan SK, Aparicio S, Brenner S, Mann RS, Krumlauf R. 1995. Segmental expression of Hoxb-1 is controlled by a highly conserved autoregulatory loop dependent upon exd/pbx. Cell 81: 1031–1042. 10.1016/S0092-8674(05)80008-X [DOI] [PubMed] [Google Scholar]
- Port F, Chen HM, Lee T, Bullock SL. 2014. Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc Natl Acad Sci 111: E2967–E2976. 10.1073/pnas.1405500111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Kucukural A, Zhang Y. 2010. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 5: 725–738. 10.1038/nprot.2010.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruff JS, Saffarini RB, Ramoz LL, Morrison LC, Baker S, Laverty SM, Tvrdik P, Capecchi MR, Potts WK. 2017. Mouse fitness measures reveal incomplete functional redundancy of Hox paralogous group 1 proteins. PLoS One 12: e0174975 10.1371/journal.pone.0174975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saadaoui M, Merabet S, Litim-Mecheri I, Arbeille E, Sambrani N, Damen W, Brena C, Pradel J, Graba Y. 2011. Selection of distinct Hox–extradenticle interaction modes fine-tunes Hox protein activity. Proc Natl Acad Sci 108: 2276–2281. 10.1073/pnas.1006964108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandve SR, Rohlfs RV, Hvidsten TR. 2018. Subfunctionalization versus neofunctionalization after whole-genome duplication. Nat Genet 50: 908–909. 10.1038/s41588-018-0162-4 [DOI] [PubMed] [Google Scholar]
- Schoofs A, Hückesfeld S, Surendran S, Pankratz MJ. 2014. Serotonergic pathways in the Drosophila larval enteric nervous system. J Insect Physiol 69: 118–125. 10.1016/j.jinsphys.2014.05.022 [DOI] [PubMed] [Google Scholar]
- Sheikh BN, Downer NL, Kueh AJ, Thomas T, Voss AK. 2014. Excessive versus physiologically relevant levels of retinoic acid in embryonic stem cell differentiation. Stem Cells 32: 1451–1458. 10.1002/stem.1604 [DOI] [PubMed] [Google Scholar]
- Simeone A, Acampora D, Arcioni L, Andrews PW, Boncinelli E, Mavilio F. 1990. Sequential activation of HOX2 homeobox genes by retinoic acid in human embryonal carcinoma cells. Nature 346: 763–766. 10.1038/346763a0 [DOI] [PubMed] [Google Scholar]
- Simeone A, Acampora D, Nigro V, Faiella A, D'Esposito M, Stornaiuolo A, Mavilio F, Boncinelli E. 1991. Differential regulation by retinoic acid of the homeobox genes of the four HOX loci in human embryonal carcinoma cells. Mech Dev 33: 215–227. 10.1016/0925-4773(91)90029-6 [DOI] [PubMed] [Google Scholar]
- Slattery M, Riley T, Liu P, Abe N, Gomez-Alcala P, Dror I, Zhou T, Rohs R, Honig B, Bussemaker HJ, et al. 2011. Cofactor binding evokes latent differences in DNA binding specificity between Hox proteins. Cell 147: 1270–1282. 10.1016/j.cell.2011.10.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KT, Martin-Brown SA, Florens L, Washburn MP, Workman JL. 2010. Deacetylase inhibitors dissociate the histone-targeting ING2 subunit from the Sin3 complex. Chem Biol 17: 65–74. 10.1016/j.chembiol.2009.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soskine M, Tawfik DS. 2010. Mutational effects and the evolution of new protein functions. Nat Rev Genet 11: 572–582. 10.1038/nrg2808 [DOI] [PubMed] [Google Scholar]
- Stern DL, Sucena E. 2011. Preparation of cuticles from feeding Drosophila larvae. Cold Spring Harb Protoc 2011: 1394–1398. [DOI] [PubMed] [Google Scholar]
- Studer M, Lumsden A, Ariza-McNaughton L, Bradley A, Krumlauf R. 1996. Altered segmental identity and abnormal migration of motor neurons in mice lacking Hoxb-1. Nature 384: 630–634. 10.1038/384630a0 [DOI] [PubMed] [Google Scholar]
- Tischfield MA, Bosley TM, Salih MA, Alorainy IA, Sener EC, Nester MJ, Oystreck DT, Chan WM, Andrews C, Erickson RP, et al. 2005. Homozygous HOXA1 mutations disrupt human brainstem, inner ear, cardiovascular and cognitive development. Nat Genet 37: 1035–1037. 10.1038/ng1636 [DOI] [PubMed] [Google Scholar]
- Treisman J, Gonczy P, Vashishtha M, Harris E, Desplan C. 1989. A single amino acid can determine the DNA binding specificity of homeodomain proteins. Cell 59: 553–562. 10.1016/0092-8674(89)90038-X [DOI] [PubMed] [Google Scholar]
- Tvrdik P, Capecchi MR. 2006. Reversal of hox1 gene subfunctionalization in the mouse. Dev Cell 11: 239–250. 10.1016/j.devcel.2006.06.016 [DOI] [PubMed] [Google Scholar]
- Webb BD, Shaaban S, Gaspar H, Cunha LF, Schubert CR, Hao K, Robson CD, Chan WM, Andrews C, MacKinnon S, et al. 2012. HOXB1 founder mutation in humans recapitulates the phenotype of Hoxb1−/− mice. Am J Hum Genet 91: 171–179. 10.1016/j.ajhg.2012.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiszmann R, Hammonds AS, Celniker SE. 2009. Determination of gene expression patterns using high-throughput RNA in situ hybridization to whole-mount Drosophila embryos. Nat Protoc 4: 605–618. 10.1038/nprot.2009.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woltering JM, Vonk FJ, Müller H, Bardine N, Tuduce IL, de Bakker MA, Knöchel W, Sirbu IO, Durston AJ, Richardson MK. 2009. Axial patterning in snakes and caecilians: evidence for an alternative interpretation of the Hox code. Dev Biol 332: 82–89. 10.1016/j.ydbio.2009.04.031 [DOI] [PubMed] [Google Scholar]
- Zeiske T, Baburajendran N, Kaczynska A, Brasch J, Palmer AG III, Shapiro L, Honig B, Mann RS. 2018. Intrinsic DNA shape accounts for affinity differences between Hox-cofactor binding sites. Cell Rep 24: 2221–2230. 10.1016/j.celrep.2018.07.100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based analysis of ChIP-seq (MACS). Genome Biol 9: R137 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wen Z, Washburn MP, Florens L. 2010. Refinements to label free proteome quantitation: how to deal with peptides shared by multiple proteins. Anal Chem 82: 2272–2281. 10.1021/ac9023999 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.