Abstract
The advances of large-scale genomics studies have enabled compilation of cell type–specific, genome-wide DNA functional elements at high resolution. With the growing volume of functional annotation data and sequencing variants, existing variant annotation algorithms lack the efficiency and scalability to process big genomic data, particularly when annotating whole-genome sequencing variants against a huge database with billions of genomic features. Here, we develop VarNote to rapidly annotate genome-scale variants in large and complex functional annotation resources. Equipped with a novel index system and a parallel random-sweep searching algorithm, VarNote shows substantial performance improvements (two to three orders of magnitude) over existing algorithms at different scales. It supports both region-based and allele-specific annotations and introduces advanced functions for the flexible extraction of annotations. By integrating massive base-wise and context-dependent annotations in the VarNote framework, we introduce three efficient and accurate pipelines to prioritize the causal regulatory variants for common diseases, Mendelian disorders, and cancers.
Variant annotation is a common procedure in human genome studies for interpreting the biological function and disease relevance of given genetic variants or somatic mutations (MacArthur et al. 2014). It greatly aids prioritization analysis regarding certain genetic hypotheses and facilitates functional follow-up on selected variants. As the growing volume of large-scale genome sequencing of human populations, such as the UK Biobank study (Bycroft et al. 2018), the National Heart, Lung, and Blood Institute's Trans-Omics for Precision Medicine (TOPMed) program (Brody et al. 2017), and functional genomics data, such as The Encyclopedia of DNA Elements (ENCODE) Project (The ENCODE Project Consortium 2012; Davis et al. 2018) and the International Human Epigenome Consortium (IHEC) project (Bujold et al. 2016), efficient interpretation of genome variants is profoundly affected by the unprecedented scale of genomic features and the resources used for their annotation. For example, a widely used metric for mutation deleteriousness, Combined Annotation Dependent Depletion (CADD) (Kircher et al. 2014), integrates more than 100 annotations for all 8.6 billion possible substitutions and 48 million short indels in the human reference genome, and it is archived in a compressed file of >300 GB. Based on the whole CADD annotation database, it may take 5–100 h to finish a personal genome annotation (around 5 million single-nucleotide variants [SNVs] and indels) using present state-of-the-art variant annotation tools. Another resource, CistromeDB (Zheng et al. 2019), aggregates 360 million genomic intervals for more than 6000 human tissue-/cell type–specific epigenomic profiles; such cumulative region-based annotations will ultimately pose a challenge to the efficient interpretation of noncoding regulatory variants. Therefore, the development of fast, scalable, and versatile annotation retrieval strategies is crucial for the broad use of big genomic features in genetic study and precision medicine.
To retrieve relevant information from annotation databases, computational methods need first to identify annotation records that overlap query variants and then extract specified annotation fields. Such processing is usually done by sequential chromosome sweeping between query variants and the annotation database—using such as BEDTools (Quinlan and Hall 2010), BEDOPS (Neph et al. 2012), ANNOVAR (Wang et al. 2010), and BCFtools (Li 2011a)—or independent random access from a whole annotation database using an index, such as the UCSC binning algorithm (Kent et al. 2002), Tabix (Li 2011b) and VEP (McLaren et al. 2016). The vcfanno annotator implements a parallel chrom-sweep algorithm based on a Tabix index and enables streaming query within defined chunks (Pedersen et al. 2016). Recently, GIGGLE introduced a temporal indexing scheme for the fast identification of shared genomic loci between query features and thousands of genome interval files (Layer et al. 2018). Despite their achievements, no algorithm can properly adapt to query variants and annotation databases with different data scales and levels of feature distribution. This unresolved bottleneck can be attributed to the high burden of disk reads from massive annotation records, which significantly hamper the running speed and scalability of existing tools. In addition, few tools have been developed to efficiently accommodate genome-scale queries and accurately prioritize disease-causal variants by leveraging large-scale functional genomics data.
Here, we present a novel index system and an ultrafast parallel intersection algorithm, called VarNote, to process variant annotations and genomic features at scale. VarNote fits different levels of data distribution and reduces the runtime of common variant annotation tasks by more than two to three orders of magnitude. It supports both region-based and allele-specific annotations for different file formats and introduces many advanced functions to improve its flexibility in use. To facilitate efficient and accurate prioritization of disease-causal regulatory variants for clinicians and biologists in different medical genetics fields, we also develop three online VarNote applications: (1) causal regulatory variants prioritization from GWAS results of common diseases; (2) pathogenic regulatory variants prioritization from genome sequencing of rare inherited diseases; and (3) driver regulatory variants prioritization from personal genome profile of cancers.
Results
VarNote index system and random-sweep algorithm
Functional genomics studies constantly produce unprecedentedly large amounts of data at genome-wide scale, which enables comprehensive annotation of genetic variants. A Tabix index together with an associated bgzip file is currently one of the most widely used storage formats for genomic annotation. Tabix combined the binning index and linear index to quickly retrieve features overlapping specified regions, but it was optimized for single independent queries. For tasks that involve the query of each of the whole-genome sequencing (WGS) variants over a huge annotation database (e.g., CADD contains more than 100 annotations for each of 8.6 billion possible substitutions), tools relying on a Tabix index need to repetitively decompress gzip blocks and parse chromosome positions from original annotation records, thus introducing many redundant operations that can be reduced by more efficient means.
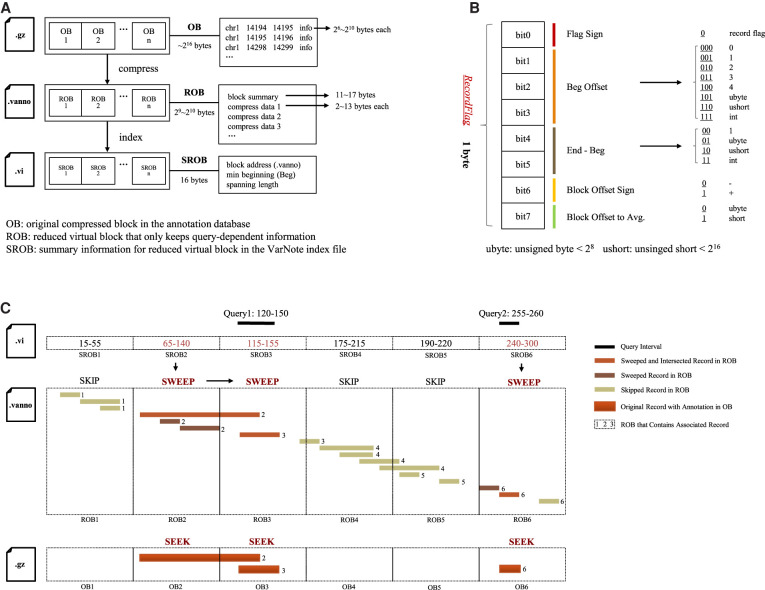
To maximally reduce time-consuming disk reads, we developed a novel index system for annotation databases. Given a bgzip-compressed annotation database, we created a positioning file that only keeps query-dependent information and an index file that allows fast retrieval of genomic position (Fig. 1A; Supplemental Fig. S1). Briefly, we first tailored the annotation metadata of each record in the original compressed block (OB) and used a reduced virtual block (ROB) that only stores block summary information together with chromosome position information for each record (Supplemental Fig. S2A). To further compress the record position information, we introduced an 8-bit “RecordFlag” to dynamically determine the exact storage volume of the chromosome position and block offset for sequential records (Fig. 1B). These strategies reduce the size of the original annotation database by a factor of approximately 100. For instance, the algorithm can convert 344 GB of CADD annotations for all possible SNVs into a 6-GB VarNote positioning file. Building on this positioning file with lossless compression of query-dependent information, we created a linear index that merely contains summary information of each ROB (SROB) (Supplemental Figs. S2B, S3), which ensures memory-efficient sweeping of chromosome positions. Taken together, our VarNote index system will significantly minimize disk reads during annotation and provide infrastructure for the fast retrieval of large numbers of query intervals (for details, see Methods).
Figure 1.
The key components of VarNote. (A) Architecture of VarNote index system. Bgzip-compressed annotation database (.bgz file) will be first converted to VarNote positioning file (.vanno file). The system tailors and encodes information of each original compressed block in the annotation database (OB) to generate a reduced virtual block that only keeps query-dependent information (ROB). The bgzip-compressed VarNote positioning file contains concatenated compressed block that stores ROB bytes. Then, summary information of the reduced virtual block (SROB) is linearly indexed to generate VarNote index file (.vi file). (B) Bit encoding of record position information. The system uses an 8-bit “RecordFlag” to encode position information of annotation record in corresponding OB. The first bit represents a sign of annotation record start; the second through fourth bits encode storage size of beginning (Beg) offset from the previous record; the fifth and sixth bits encode storage size of distance between End and Beg for current record; the seventh bit is the direction sign of the block offset; the eighth bit indicates the storage size of the block offset to average. (C) Workflow of random sweep. The algorithm accepts two dummy query intervals in the same chromosome (query 1 starts from 120 and ends in 150; query 2 starts from 255 and ends in 260) and efficiently executes the annotation intersection across a corresponding chromosome by leveraging the VarNote index system and original annotation database. The query 1 is first stream-compared with position information of each SROB in the VarNote index file (.vi file) to determine intersected ROBs (query 1 overlaps with ROB2 and ROB3). The algorithm directly skips unrelated ROBs and quickly locates the intersected ROBs using random access (ROB1 is completely skipped in the following searching). Because ROB only contains query-dependent information of annotation records, VarNote can sweep the ROB more efficiently with saved disk reads (query 1 intersects an annotation record in the ROB2, and intersects another annotation record in the ROB3). Once all intersected annotation records within a ROB are identified, the algorithm can instantly seek full annotation information of record hits from the corresponding OB (only needs to seek two records in the OB2 and OB3 of annotation database for query 1). Similarly, VarNote skips over ROB4 and ROB5, only sweeps across ROB6, and finally seeks an annotation record at OB6 for query 2.
By leveraging the VarNote index system, we also combined random-access and chromosome-sweep strategies to implement a unified and efficient searching approach for sorted query intervals/variants, called the random-sweep algorithm (Fig. 1C). Specifically, the algorithm first loads the VarNote small index and sequentially sweeps SROBs to locate intersected ROBs. Selected ROBs are random accessed from the VarNote positioning file through block summary information, while unassociated ROBs are directly skipped. As position information for each annotation record is bit-encoded within consecutive ROBs, decoding ROB content and applying the chromosome-sweep algorithm can identify all annotation record hits (Supplemental Fig. S4; Methods).
The gained speed and scalability of VarNote were attained for the following reasons. First, the intersection between query intervals and annotation records mostly relies on the VarNote positioning file instead of the original annotation database. The only step associated with the original annotation database is the extraction of annotation fields for final record hits using random access, hence excessive disk reads can be largely saved. Second, random-sweep searching is a coherent process in which the combination of a global linked list and a file pointer ensure straightforward intersections without returns; meanwhile, the algorithm can jump over unrelated data blocks. This strategy enables VarNote to be scalable to large data sets and still be efficient for small inputs, especially for sparse and unbalanced queries. Third, there is no repetitive decomposition of the same gzip blocks during the annotation process, thus further accelerating annotation searches.
Comparison of VarNote with existing tools for interval-level annotations
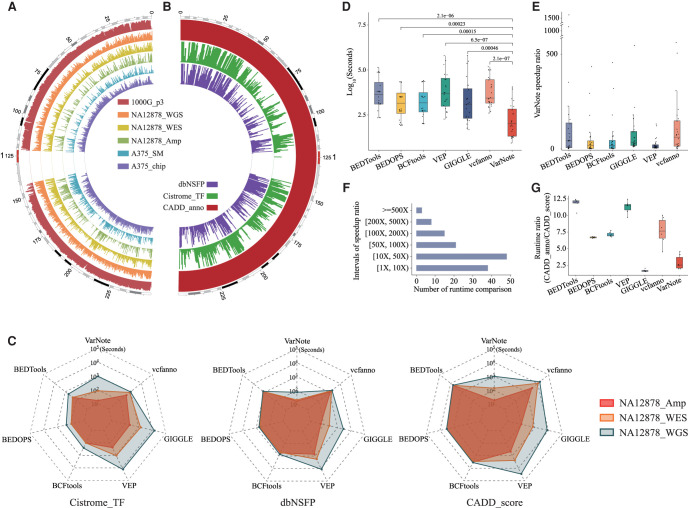
Genomic features intersection is a common part of many bioinformatics analyses such as variant annotation, yet existing tools rely strongly on the data scales and feature distributions of the query variants and annotation databases across the whole chromosome. To comprehensively evaluate VarNote in comparison with existing interval-level annotation tools, we first established a set of benchmark data sets encompassing various data scales and feature distributions (Supplemental Table S1). Six query variant data sets were generated from a genome-wide genotyping chip (A375_ chip), targeted sequencing (NA12878_Amp), whole-exome sequencing (WES) (NA12878_WES, A375_SM), whole-genome sequencing (WGS) (NA12878_WGS), and all known variants in The 1000 Genomes Project phase3 (1000G_p3); they ranged from highly unbalanced and sparse queries to highly balanced and dense queries (Fig. 2A; Supplemental Fig. S5). Several commonly used annotation databases were also prepared, including functional prediction of all potential nonsynonymous single-nucleotide variants from dbNSFP (Liu et al. 2016b), aggregated ChIP-seq peak calling results of human transcription factors from CistromeDB (Cistrome_TF) (Zheng et al. 2019), deleteriousness scores of all possible SNVs from CADD (CADD_score), and 114 related annotations (CADD_anno). These annotation databases represent distinct distributions of interspersed (dbNSFP), overlapped (Cistrome_TF), or tandem (CADD_score or CADD_anno) genomic features, respectively, which could serve as comprehensive benchmarks to evaluate the performance of VarNote and exiting algorithms (Fig. 2B; Supplemental Fig. S6).
Figure 2.
Comparison of VarNote with existing tools for interval-level annotations. (A) The genomic distribution of six query variant data sets across Chromosome 1 of the human reference genome, including variant call result of The 1000 Genomes Project phase3 (1000G_p3), variant call result of 10x Genomics Chromium whole-genome sequencing for NA12878 (NA12878_WGS), variant call result of Nextera Rapid Capture Exome and Expanded Exome whole-exome sequencing for NA12878 (NA12878_WES), variant call result of Ion AmpliSeq Exome capture sequencing data for NA12878 (NA12878_Amp), genotype calling result of Affymetrix Genome-Wide Human SNP Array 6.0 data for A375 cell line (A375_chip), and somatic mutation call result of whole-exome sequencing data for A375 cell line (A375_SM). These data sets span the highly unbalanced and sparse queries to the highly balanced and dense queries. (B) The genomic distribution of three annotation databases across Chromosome 1 of the human reference genome, including functional prediction and annotation of all potential nonsynonymous SNVs (dbNSFP), Cistrome aggregated ChIP-seq peak calling result of human transcription factors (Cistrome_TF), CADD deleteriousness score, and related annotation of all possible SNVs (CADD_anno). (C) The runtime comparisons among VarNote, BEDTools, BEDOPS, BCFtools, VEP, vcfanno, and GIGGLE for sequencing variants at different scales and commonly used annotation databases. (D) The runtime distribution of 24 combinatory tests for each algorithm. (E) The speed ratio of VarNote compared with other methods for each of 24 tests. (F) The number of runtime comparisons between VarNote and other methods within corresponding speed ratio intervals. (G) The runtime ratio distribution for processing long annotation database CADD_anno over short annotation database CADD_score.
Using the aforementioned six query data sets and four annotation databases, we constructed 24 combinatory tests for the following evaluations. We compared the performance of VarNote with five state-of-the-art bioinformatics tools for intersecting genomic features. These commonly used, representative tools can achieve interval-level overlap annotation based on distinct algorithms, including BEDTools, BEDOPS, BCFtools, VEP, GIGGLE, and vcfanno (Supplemental Table S2). Overall, VarNote runs more quickly than the previous methods and outperforms them in almost all combinatory tests in which the benchmark data sets hold various data scales and feature distributions (Supplemental Fig. S7). Specifically, VarNote shows significantly extended scalability and is well-adapted to increasing query intervals and annotation databases (Fig. 2C). In contrast, the runtime of random-access-derived methods, like VEP and GIGGLE, tightly correlates with the data scale of queries, reaching hours when annotating a WGS data set. The chromosome-sweep-derived methods (BEDTools, BEDOPS, and BCFtools) show low efficiency when annotating targeted sequencing and WES data, especially when the annotation database is huge. Although vcfanno introduces random access into the chromosome-sweep algorithm, its moderate performance largely depends on the feature distributions of the query and annotation database (Fig. 2C).
In addition to excellent scalability, VarNote shows ultrafast genomic feature intersection, being between 123 and 1514 times quicker than the other methods for the 24 benchmarks (Supplemental Fig. S7; Supplemental Table S3). VarNote finishes half of the tests within 100 sec, whereas the median runtimes of other tools are ∼1000–4000 sec for all the designed tests. The distribution of VarNote's runtimes significantly deviate from those of the other methods (Mann–Whitney U test) (Fig. 2D). VarNote runs more quickly than the other tools in 135 out of 144 (93.75%) comparisons but is slightly inferior in some evaluations involving a small annotation database and huge query. However, the second best tool, BEDOPS, only wins 69.44% of comparisons, revealing that VarNote can adapt to the full range of data scales and feature distributions while maintaining stably superior performance (Supplemental Fig. S8). Among the preceding runtimes, 65.98%, 32.64%, and 18.06% of VarNote's were more than 10, 50, and 100 times quicker, respectively, than those of the other algorithms, representing a powerful improvement (Fig. 2E,F). In addition, VarNote and GIGGLE have similar runtimes when processing the short annotation database CADD_score and the long annotation database CADD_anno, whereas the other tools show ∼10 times longer runtimes, which implies that unnecessary disk reads can be largely saved by using an auxiliary positioning file (Fig. 2G).
Fast and parallelizable variant-level annotations
Efficient and accurate extraction of genomic features from annotation databases facilitates variant interpretation, particularly for the increasingly high volumes of DNA sequencing data gathered in the precision medicine era. However, current variant-level annotation tools coupled with different intersection algorithms either can barely process WGS variants or show limited efficiency for huge annotation databases. The majority of intersection algorithms do not support multithreading, which leads current variant annotation tools to implement split-and-join parallelization only by Tabix index and random access. To investigate the ability of VarNote for all-around variant-level annotations and multithreading tasks, we selected three variant call results as query data sets (targeted sequencing, WES, and WGS) and extracted designed feature fields across three genome-wide annotation databases: dbNSFP, gnomAD (Karczewski et al. 2020), and CADD_anno. We also compared VarNote's performance against three frequently adopted and high-powered variant annotation methods, VEP, BCFtools, and vcfanno, which present relatively strong performance using random access, chromosome sweep, and mixed strategies, respectively (Supplemental Table S4).
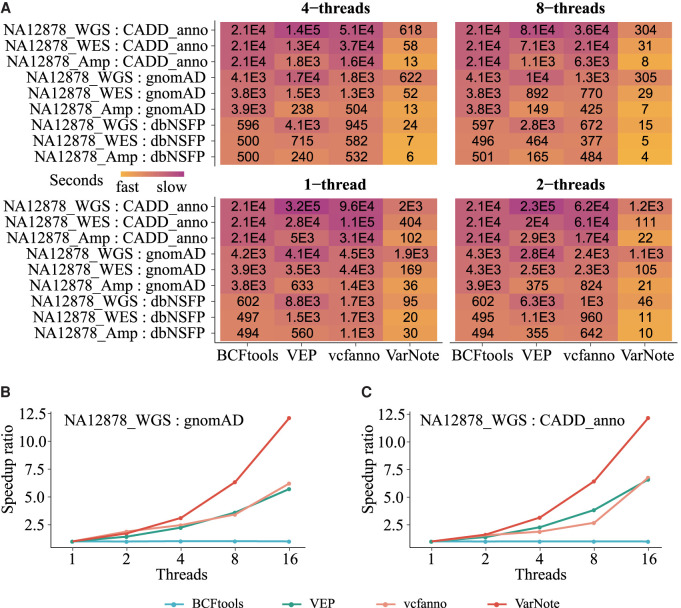
Generally, with a single thread, VarNote outperforms the other tools, running between ∼5 and 500 times more quickly in all the benchmarks. It takes <2 min to annotate NA12878_Amp targeted sequencing variants with any of the prepared annotation databases, whereas the other tools need up to hours for a large annotation database such as CADD_anno. In addition, VarNote completes common annotation tasks for WES data within 7 min. For example, it only takes 20 sec to annotate NA12878_WES variants by extracting dbNSFP nonsynonymous SNVs functional prediction scores, whereas the other tools require ∼500–1700 sec. Moreover, to annotate WGS data containing several million variants, VarNote performs the most time-consuming CADD_anno task within 35 min using a single thread. In contrast, BCFtools, vcfanno, and VEP require 6, 27, and 90 h to execute the same job, respectively, making them unsuitable for personal genome applications (Fig. 3A).
Figure 3.
Comparison of VarNote with existing tools for variant-level annotations. (A) The runtime comparisons of variant annotation using multithreading. Three variant call results as query data sets (NA12878_Amp, NA12878_WES, and NA12878_WGS) and three genome-wide annotation databases (dbNSFP, gnomAD, and CADD_anno) were used. (B) The parallelization efficiency comparisons for variant annotation task of NA12878_WGS querying on gnomAD. (C) The parallelization efficiency comparisons for variant annotation task of NA12878_WGS querying on CADD_anno.
We assessed multithreading performance by having selected tools perform the same evaluations using two to eight threads. Using more than four threads, VarNote can reduce the runtime of WGS annotation tasks to ∼10 min and lower the runtime of WES annotation tasks to nearly 1 min. When annotating targeted sequencing variants, it is more than 1000 times faster than chromosome-sweep-derived methods like BCFtools because of its restriction of parallelization (Fig. 3A). For the most time-consuming WGS annotation tasks, the speedup ratio curves of VarNote are generally sublinear and show better parallelization efficiency than other methods (Fig. 3B,C), further indicating that VarNote's index system can significantly eliminate input/output (I/O) bottlenecks for large-scale variant annotations. Because BCFtools only supports multithreading for output compression, increasing the number of cores barely improved the runtime.
Advanced functions of VarNote for versatile genomic feature intersection and variant annotation
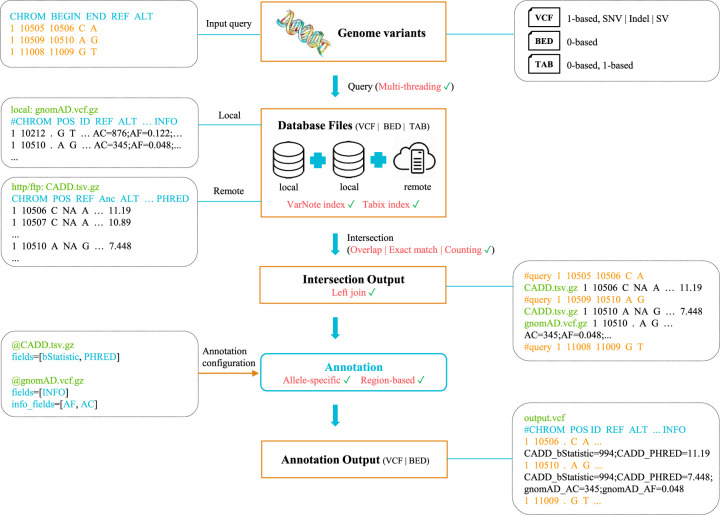
We designed several advanced features to improve the usability of VarNote in complex annotation tasks (Fig. 4). First, because most existing annotation databases are indexed by Tabix, VarNote also provides a random-sweep searching based on the Tabix index (Supplemental Fig. S9). By performing the same tests for variant-level annotation, we found that VarNote's Tabix mode still outperformed other tools, particularly when multithreading was applied (Supplemental Fig. S10). This implies that VarNote can faithfully process existing annotation resources without reindexing them. However, for large-scale and frequently used annotation data sets such as CADD, gnomAD, and dbNSFP, we strongly suggest using VarNote's index system to gain speed. By introducing extra file pointers, VarNote is able to support random sweep at multiple annotation databases. This is crucial to personal genome annotation tasks involving annotation resources spanning different contexts and biological domains, such as allele frequency, conservation, and functional prediction scores. In addition, VarNote allows remote annotation via FTP/HTTP, especially for big data sets that are time-consuming to download. Owing to the VarNote positioning file that only keeps query-dependent information, such remote queries differ from Tabix in having significantly reduced network data transmission load and allowing multithreading. Finally, VarNote also supports quick counting of the number of intersected features only based on the VarNote positioning file, which will be most efficient and suitable to prioritize context-dependent annotations through their relevance to the set of query variants, such as to identify the causal tissues/cell types for genome-wide association study (GWAS) variants (Huang et al. 2018) or to colocalize ChIP-seq binding events with particular transcription factors (Kanduri et al. 2019).
Figure 4.
The workflow and supported features of the VarNote annotation program. It accepts an input query (VCF, BED, or TAB format) and intersects annotation records from local or remote databases, then extracts annotation fields according to predefined annotation configuration, and finally outputs annotation results. It also supports many advanced features (shown in red).
To extend its flexibility, VarNote allows users to describe specified operations in a configuration file to process customizable features with complex data structures (Fig. 4). For example, compared with existing tools that usually require uniformly formatted annotations such as BED (0-based) or VCF (1-based) files, VarNote can extract annotation fields from any indexed tab-delimited annotation files. This could save time in format transformation and additional disk space for large annotation databases, such as CADD and dbNSFP. When multiple annotation databases are available, VarNote allows feature extraction using both interval-level overlap and variant-level exact matching. It also has an annotation mode supporting allele-specific variant annotation for SNV/indel and region-specific annotation for structure variations. Selected fields of intersected outputs can be extracted and filled to the same line of the input query according to the annotation configuration file. To facilitate the integration of VarNote into advanced genomic programs, we also provide an application programming interface for developers.
Applications of VarNote to genome-scale prioritization of functional and pathogenic regulatory variants
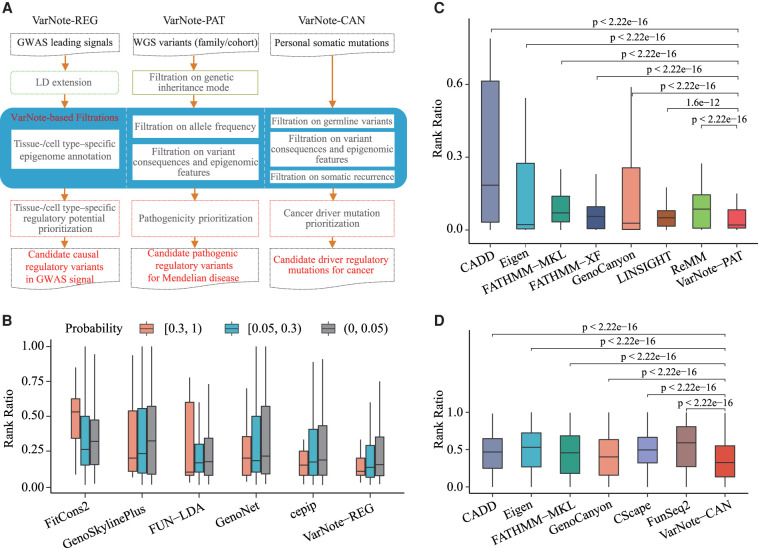
GWASs have identified many genetic variants associated with hundreds of medical traits and diseases, and most of these associations are suggested to be mediated by context-specific regulatory codes in the noncoding human genome (Li et al. 2016). For Mendelian diseases and cancer, WGS technologies are frequently incorporated into the exploration of noncoding pathogenic variants from patients’ entire genomes (Castel et al. 2018). However, very few computational tools have been developed to efficiently manipulate such genome-scale data and accurately prioritize the true disease-causal regulatory variants. To show the applicability and efficiency of VarNote for identifying functional and pathogenic regulatory variants at genome-scale, we integrated base-wise variant annotations and several state-of-the-art regulatory variant prediction methods to develop three online computational pipelines (http://mulinlab.org/varnote/application.html): (1) disease-causal regulatory variants prioritization for GWAS results; (2) pathogenic regulatory variants prioritization for rare inherited diseases; and (3) driver regulatory variants prioritization for cancers (Fig. 5A).
Figure 5.
The applied pipelines and performance evaluations of VarNote for disease-causal variant prioritization. (A) The function summary of three designed pipelines based on VarNote framework. (B) The box plot of rank ratio for each PICS causal variant among different PICS probability intervals; the rank ratio is measured by the rank of observed variant/total number of investigated variants (including extended highly linked variant in LD) in each GWAS signal. (C) The box plot of rank ratio for each spike-in pathogenic variant using simulated WGS variants; the rank ratio is measured by the rank of pathogenic variant/total number of qualified variants after filtrations in each simulated individual genome, tested by one-tailed Mann–Whitney U test. (D) The box plot of rank ratio for each spike-in somatic eQTL mutation using simulated cancer genome profiles; the rank ratio is measured by the rank of somatic eQTL mutation/total number of qualified mutations after filtrations in each simulated cancer genome, tested by one-tailed Mann–Whitney U test.
Efficient and accurate prioritization of GWAS causal regulatory variants by integrating VarNote and tissue-/cell type–specific epigenomes
Although statistical fine-mapping of GWAS summary data provides a valid avenue to identify causal variants, it usually fails to narrow down the likely causal variants with extremely high linkage disequilibrium (LD) in each credible set and cannot evaluate context-dependent effects for regulatory variants in the noncoding genome (Schaid et al. 2018; Wang et al. 2020). Recently, several tissue-/cell type–specific regulatory variant prediction methods have been developed based on large-scale epigenomic features (Rojano et al. 2019), but no computational tool can leverage these methods to prioritize the potential causal regulatory variants from GWAS signals. By incorporating 127 Roadmap tissue-/cell type–specific epigenomic features, 1000 Genomes LD information, and five recent context-dependent regulatory variant prediction models—cepip (Li et al. 2017b), GenoSkyline-Plus (Lu et al. 2017), FUN-LDA (Backenroth et al. 2018), GenoNet (He et al. 2018), and FitCons2 (Gulko and Siepel 2019)—we implemented a comprehensive and fast pipeline, called VarNote-REG, to accurately prioritize GWAS casual regulatory variants based on the VarNote variant annotation framework. Simplistically, given GWAS leading signals and a selected trait-matched tissue/cell type, VarNote-REG will first identify all associated variants in the defined LD block and extract epigenomic features for them. To prioritize the context-dependent regulatory potential for variants in each LD block, VarNote-REG introduces a combined rank score based on five state-of-the-art prediction scores (Methods).
VarNote-REG can complete prioritization jobs for common GWAS results, such as 161 fine-mapped loci for coronary artery disease (van der Harst and Verweij 2018) and 154 fine-mapped loci for inflammatory bowel disease (Huang et al. 2017a), within 2 min. We benchmarked the performance of VarNote-REG using PICS GWAS fine-mapped variants for 21 autoimmune diseases (Farh et al. 2015). We first filtered out LD blocks either associated with autoimmune diseases possibly driven by nonregulatory effects or containing limited highly linked (LD R2 ≥ 0.8) noncausal variants. For the remaining GWAS signals, we tested whether the variants with high PICS causal probability could be ranked above other highly linked variants in the matched tissue/cell type and compared the performances of various prediction methods (Methods). Using predication scores from 16 ENCODE cell types, we found that the ranking of causal variants within each causal LD block varied with both the cell type and the prediction method. The causal variants were generally higher ranking in the E116 lymphoblastoid cell line than in other cell types (Supplemental Fig. S11), indicating that selection of relevant cell types matching autoimmune disease may improve the prioritization of true causal regulatory variants. By selecting E116 lymphoblastoid-specific prediction and partitioning the fine-mapped causal variants into three separate groups, we also observed that variants in the ≥0.3 PICS probability group obtained higher ranks than those with low PICS probability. Particularly, our combined rank score showed better discriminatory ability than each separate method, that is, comparing the ≥0.3 PICS probability group with the [0.05 0.3) and <0.05 PICS probability groups (Fig. 5B). These results suggest that VarNote-REG could efficiently and accurately prioritize disease-causal regulatory variants.
Fast and whole-genome prioritization of pathogenic regulatory variants for rare inherited diseases
Rare noncoding variants can cause inherited disorders by affecting the function of regulatory elements, and recent genetic studies of Mendelian disease have applied WGS to identify pathogenic regulatory variants (Weedon et al. 2014; Marshall et al. 2020). Several computational methods have also been developed to prioritize the pathogenesis/deleteriousness of regulatory variants (Smedley et al. 2016). However, accurate identification of disease-causal regulatory variants from family-based WGS data requires time-consuming filtrations of genome-scale annotations as well as integration of pathogenic regulatory variant predictions. By introducing several unique filtration strategies and integrating base-wise pathogenic prediction scores, we developed a fast and accurate pipeline, VarNote-PAT, for geneticists and clinicians to prioritize likely pathogenic regulatory variants based on WGS data. Given a VCF file and matching pedigree file, VarNote-PAT can narrow down the candidate variants based on different filtrations, including variant quality, allele frequency, genetic inheritance mode, variant consequence, and tissue-/cell type–specific epigenomic features. In addition, VarNote-PAT combines seven recent prediction methods to improve the prioritization of pathogenic regulatory variants (Methods).
Building on the VarNote framework, VarNote-PAT can finish a complete prioritization task on trio-based WGS data (∼5 million variants) within 60 min, whereas an existing online program, Genomiser (Smedley et al. 2016), can only deal with 100,000 variants in each run. To evaluate the performance of pathogenic variant prioritization, we first curated 18 pathogenic regulatory variants of rare inherited diseases that have been experimentally validated by different functional assays (Supplemental Table S5). We then simulated WGS data of 503 subjects by spiking each validated pathogenic variant into individual genomes from The 1000 Genomes Project EUR population (Methods). By filtering and scoring WGS variants for each simulated genome with the VarNote-PAT pipeline, we found that the ranks of spike-in pathogenic variants averaged in the top 6.5%. Compared with existing prediction scores, such as ReMM (Smedley et al. 2016) and CADD, we observed that the validated pathogenic variants were ranked significantly higher using our combined scores (Mann–Whitney U test) (Fig. 5C). These evaluations show that VarNote-PAT can greatly improve the speed and accuracy of discovering WGS-based rare pathogenic regulatory variants.
Filtration and prioritization of regulatory somatic mutations in personal cancer genomes using VarNote
Recurrent somatic mutations in the noncoding regulatory region have been revealed as cancer drivers (Yang and Adli 2019). However, no computational pipeline has been specifically designed to screen and prioritize cancer driver regulatory mutations for a given individual cancer genome profile. Here, we encapsulated VarNote and several annotation databases to develop an online pipeline, called VarNote-CAN, for regulatory somatic mutation prioritization. VarNote-CAN first filters germline variants and nonrecurrent mutations and then annotates recurrent somatic mutations using tissue-/cell type–specific epigenomic features. It uses our cancer driver regulatory mutation prediction model, regBase-CAN (Zhang et al. 2019), to prioritize the remaining candidate mutations. We benchmarked the performance of VarNote-CAN using 1950 simulated personal cancer genomes based on 158 highly recurrent mutations in the somatic eQTL intervals identified by a large-scale pan-cancer whole-genome analysis (PCAWG Transcriptome Core Group et al. 2020) (Methods). Compared with existing prediction scores for cancer regulatory mutations, such as FunSeq2 (Fu et al. 2014) and CScape (Rogers et al. 2017), the overall rank of somatic eQTL SNVs in all cancer genomes was higher using regBase-CAN (Mann–Whitney U test) (Fig. 5D), indicating the applicability of VarNote-CAN to whole cancer genome analysis.
Discussion
Large-scale genomic sequencing together with constantly evolving biotechnologies in functional genomics have posed great challenges to the efficient annotation of variant functions as well as interpreting their causal links with diseases (Starita et al. 2017). An immediate challenge is the development of rapid computational methods that scale to the WGS variants and the vast amount of functional annotations across the entire genome. To address these efficiency and scalability issues, we have implemented a new index schema to tailor annotation databases. Building on this, a fast random-sweep search algorithm was designed with coherent support for multithreading. In contrast to previous strategies that merely operate original annotation databases or repetitively decompress gzip blocks, VarNote greatly saves excessive disk reads and significantly enhances scalability, thereby achieving rapid annotation for large-scale genomic features. The efficiency and scalability of VarNote make it potentially broadly applicable to large-scale genomic feature annotations. To further extend the convenience of human variant annotations for clinicians and biologists without the experience of command line operation, we designed three web tools that allow users to upload well-formatted variant lists (VCF, BED, or tab-delimited) and prioritize disease-causal regulatory variants in different scenarios. Using GWAS fine-mapped variants and simulated individual genomes, we have shown the excellent usability of the designed pipelines and demonstrated the efficient diagnosis potential afforded by VarNote's index system and ultrafast search algorithm.
Existing interval intersection and variant annotation algorithms either cannot scale to large data sets (e.g., random-access-derived methods) or lack efficiency on small and moderate queries (e.g., chromosome-sweep-derived methods). Although tools such as vcfanno use the Tabix index to implement streaming queries for sorted inputs, they usually introduce many repetitive block decompositions and unnecessary disk accesses. To the best of our knowledge, VarNote is the first tool that can adapt well to query variants and annotation databases with different levels of data scales and feature distributions. From highly unbalanced queries (e.g., variants from targeted sequencing) to extremely balanced 1000G variants, VarNote, in comparison to other existing methods, consistently shows superior performance to extract the target fields from large-scale annotations. Nevertheless, there are some rare worst-case situations in which the runtime of VarNote could slightly fall behind those of other methods; these include cases with genome features in both the query and annotation database being largely similar or the annotation database being small while the query is very large. Our previous tests show that VarNote works best when the query variants are moderate (like WES and WGS data) and the database is huge (like CADD with full annotations), which involves using exactly the same short slabs as both random-access-derived methods and chromosome-sweep-derived methods. Because all of the benchmarks in this study were performed on SAS hard drives, the bottleneck of disk I/O could be reached when using a large number of threads, particularly for those tools equipped with faster searching algorithms. We suggest using SSD hard drives for large-scale annotation tasks, which will not only reduce random access time but also allow better parallelization efficiency during annotation.
Many other sophisticated computational tools could be used to annotate variants in genetic and genomic studies, for example, ANNOVAR (Wang et al. 2010), SnpEff (Cingolani et al. 2012), KGGSeq (Li et al. 2017a), WGSA (Liu et al. 2016a), Bystro (Kotlar et al. 2018), AIList (Feng et al. 2019), and Oncotator (Ramos et al. 2015). Because current variant annotation methods usually adopt similar intersection and feature extraction algorithms, or barely support customized operations, we only considered representative and relatively efficient ones in our completely fair benchmarks. Unlike many gene-based and filter-oriented annotation tools, VarNote concentrates on region-based and allele-specific annotation tasks for which many exiting methods substantially lack scalability (such as running out of memory or extremely long running times) when dealing with large-scale genomic features. In conjunction with accurate gene-based and filter-oriented annotation tools, VarNote will significantly accelerate personal genome interpretation in the precision medicine era.
Methods
The VarNote index strategy
The VarNote index comprises two main constructing steps, including generations of VarNote positioning file and VarNote index file. VarNote accepts a position-sorted (first by sequence name and then by leftmost coordinate) and block-compressed gzip (bgzip) annotation database and converts it to a new bgzip file that only keeps query-dependent information (Supplemental Fig. S1). To maximally reduce the file size and significantly save the disk accesses, VarNote transforms each original compressed block (OB) into a reduced virtual block (ROB). Specifically, for each OB that contains at most 216 bytes, block summary information together with position information (including chromosome position and file block position) of each record are calculated and encoded to constitute ROB. The block summary information includes a unique 64-bit OB address (defined by bgzip), position information of first record, and average block offset of all records in the current OB. The position information of each record involves a record flag sign, position offset, and block offset to average. To further compress the position information of each record, an 8-bit “RecordFlag” is used to dynamically determine the exact storage volume of chromosome position offset and block offset for different records. Thus, this bit encoding strategy enables the algorithm to store consecutive base-wise chromosome positions with only 2 bytes, significantly lessening the overall storage space of ROB. In most situations, VarNote can transform each 216 bytes OB into ∼29–210 bytes ROB, which achieves an approximately 100 times size reduction of original annotation database. Finally, a bgzip byte stream of sequential ROBs is used to generate the VarNote positioning file (Supplemental Fig. S2A). To facilitate the efficient identification of associated ROBs intersecting with queries, VarNote creates the linear index file that contains summary information of each ROB (SROB). The SROB includes a unique 64-bit address of ROB start in the VarNote positioning file, initial beginning position of the current ROB, as well as spanning length of all records in the current ROB (Supplemental Figs. S2B, S3). The combination of the VarNote positioning file and the VarNote index file achieves fast retrieval of large numbers of query intervals.
Random-sweep searching algorithm
VarNote intellectively leverages random-access and chromosome sweep for efficient interval intersection. The same as an annotation database, query intervals/variants should be sorted first by sequence name and then by leftmost coordinate. The algorithm will first load the VarNote index and then sequentially sweep SROBs, by streaming comparison of the position information between query and each ROB, to find the first intersected ROB for initial query interval. The intersected ROB is random accessed from the VarNote positioning file through the 64-bit address of ROB start. Then the VarNote chromosome-sweep algorithm is used to decode ROB content and identify record hits. It uses a global linked list to cache position information of intersected records to make sure the sweeping is unreturned. In detail, VarNote will cache intersected records after finishing the sweeping procedure of a query interval, and then it checks whether there are any overlaps in the global linked list before it sweeps the following annotation records when the next query comes. To ensure memory-efficient searching, VarNote removes those records that no longer intersect with the following query intervals from the global linked list. In addition, VarNote uses file pointers to keep and synchronize chromosome position, and therefore makes sure the sweeping process continues. Also, the query interval may span multiple ROBs, therefore the sweeping will continue in the following ROBs until the end position of the query interval is less than the beginning position of the next ROB. Once the sweeping is finished for query intervals and the next ROB does not overlap with it, VarNote will directly seek and parse the annotation information of record hits from the corresponding OB in the annotation database (Supplemental Fig. S4). The VarNote random-sweep algorithm iteratively executes the above processes for all query intervals in a scalable manner. It also ensures that associated ROBs and OBs are decompressed only once, that is, using Intel Genomics Kernel Library (Guilford et al. 2017) for speeding up, during the whole job, which significantly save the most time-consuming disk access and block decompression (see the pseudocode of the random-sweep searching algorithm in Supplemental Table S6).
Random-sweep searching on Tabix index
To facilitate querying when only the Tabix index is available, we also implemented a random-sweep searching algorithm based on Tabix binning and linear indexes. Generally, each chromosome is partitioned into segments spanning 128-kb intervals, and query intervals belonging to separate segments will be grouped together for subsequent searching. First, for each query interval group, we used the Tabix algorithm to identify and merge associated bins, then assemble corresponding bgzip blocks. Second, the aforementioned chromosome-sweep algorithm is used to intersect annotation records across assembled blocks, where nonconsecutive or unassociated blocks can be randomly accessed or skipped (Supplemental Fig. S9). Iteratively, the algorithm finishes all of the query interval groups.
Parallelization and remote access
Owing to the inherent attribute of the VarNote random-sweep algorithm, we can easily implement a parallel version of VarNote using a MapReduce programming idea. More specifically, input query file is equally partitioned to N subsets (N is a given number of threads), and for each subset VarNote independently executes random-sweep searching against annotation databases. To ensure a sorted output, VarNote merges the annotation result of all query subsets by the original order. In addition, using network streaming connection, VarNote supports remote access when huge annotation databases locate in the remote FTP/HTTP site.
Benchmark data set and environment
To comprehensively evaluate the performance of VarNote at different scales of the query variant and annotation database, we downloaded and compiled commonly used data sets from several public repositories. For the query data set, variant calling results of the NA12878 genome for AmpliSeq targeted sequencing, whole-exome sequencing (WES), and whole-genome sequencing (WGS) were downloaded from GIAB FTP (Zook et al. 2016). Variant calling result of The 1000 Genomes Project phase3 (1000G) was downloaded from EBI FTP (The 1000 Genomes Project Consortium 2015). Somatic mutation calling and germline variant genotyping results of A375 cancer cell line were downloaded from GDSC (Iorio et al. 2016). For annotation databases, we downloaded files from gnomAD (known whole-genome variant allele frequency), dbNSFP (functional prediction and annotation of all potential nonsynonymous single-nucleotide variants), CADD (deleteriousness score and related annotation of all possible single-nucleotide variants), and Cistrome Human_TF (aggregated ChIP-seq peak calling result of human transcription factors). For a data set with separate chromosome or assay files, we merged them into one annotation database (see details in Supplemental Table S1). All tests in this study were performed on a server with Intel Xeon 2.60 GHz E5-2690 v4 CPU and RAID5 mode on eight 7200 RPM SAS hard drives.
Evaluation for interval-level annotations
We compared the performance of VarNote intersection function with several well-performed tools in each algorithm category, including BEDTools, BEDOPS, BCFtools, VEP, vcfanno, and GIGGLE. Using prepared VCF inputs (AmpliSeq, WES and WGS, 1000G) and BED databases (dbNSFP, Cistrome Human_TF, and CADD with or without related annotations) at different data sizes, we tested the runtime and scalability of each software for interval-level annotations. Because BEDTools, BEDOPS, BCFtools, and GIGGLE do not support multiple threads execution, all interval-level comparisons were based on single thread mode (see details in Supplemental Table S2). For VEP, the actual runtime of the intersection was calculated by subtracting the runtime of the gene-based annotation.
Evaluation for variant-level annotations
We evaluated the performance for VarNote and three representative variant annotation tools (BCFtools, VEP, and vcfanno), which use unique search algorithms, respectively. Three VCF queries (NA12878 AmpliSeq, WES, and WGS) were used to test the allele-specific annotations against one VCF database (gnomAD) and two BED databases (dbNSFP and CADD with full annotations). Besides, we also inspected the performance of these programs at multiple threads (see details in Supplemental Table S4).
Implementation of the VarNote-REG pipeline
Genotype data of different populations were retrieved from The 1000 Genomes Project phase 3 release, and LD was estimated by a correlation method. Consolidated and imputed epigenomes from 127 human tissues/cell lines were downloaded from the web portal of the NIH Roadmap Epigenomics project (Roadmap Epigenomics Consortium et al. 2015), which includes ChIP-seq narrow peaks for eight histone modifications (H3K4me1, H3K4me2, H3K4me3, H3K27ac, H3K27me3, H3K36me3, H3K79me2, and H3K9me3) and DNase-seq peaks. Jannovar (Jäger et al. 2014) was used to map variant consequences. Five tissue-/cell type–specific regulatory variants prediction methods—cepip (Li et al. 2017b), GenoSkyline-Plus (Lu et al. 2017), FUN-LDA (Backenroth et al. 2018), GenoNet (He et al. 2018), and FitCons2 (Gulko and Siepel 2019)—were used to prioritize likely causal variants in each GWAS signal. For each candidate variant in its LD block, the ranks of prediction scores were combined by calculating rank product. By encapsulating VarNote programming interfaces, the VarNote-REG pipeline introduces three main steps to prioritize regulatory variant in each input GWAS signal: (1) variant normalization and LD expansion; (2) tissue-/cell type–specific epigenomic features annotation; and (3) tissue-/cell type–specific regulatory potential prioritization. The online VarNote-REG pipeline and visualization is available at http://mulinlab.org/varnote/application.html#REG.
Evaluation of VarNote-REG for tissue-/cell type–specific regulatory variant prioritization
Candidate causal variants for 21 autoimmune diseases were downloaded from a PICS GWAS fine-mapping study (Farh et al. 2015). We first used Jannovar to annotate the variant consequence and excluded the signals (leading variant-associated LD block) in which the causal variant with the largest PICS probability having protein-coding or splicing-altering consequences. For each leading variant-associated GWAS signal, we extended the linked variants in this signal (R2 ≥ 0.8) and treated them as noncausal variants. We removed the GWAS signal once the extended noncausal variants are insufficient (less than five variants). We used VarNote to retrieve tissue-/cell type–specific regulatory variant prediction scores on 16 ENCODE cell types for five existing methods and calculated combined scores by rank product. We then tested whether the causal variants could be ranked higher than highly linked noncausal variants in different tissues/cell types and compared the performance among used prediction methods.
Implementation of the VarNote-PAT pipeline
Allele frequency information for different populations were downloaded from The 1000 Genomes Project phase 3 release and gnomAD v2.1 (Karczewski et al. 2020). Four genetic inheritance modes were supported in VarNote-PAT, including autosomal dominant/recessive and X-linked dominant/recessive inheritances. Variant filtration strategies were similar to our previous WES platform wKGGSeq (Li et al. 2015). We specially introduced tissue/cell-type-specific epigenomic feature annotation and filtration using 127 Roadmap epigenomic profiles. We combined the prediction scores of seven methods—CADD v1.4 (Kircher et al. 2014), ReMM (Smedley et al. 2016), Eigen (Ionita-Laza et al. 2016), GenoCanyon (Lu et al. 2015), FATHMM_MKL (Shihab et al. 2015), FATHMM-XF (Rogers et al. 2018), and LINSIGHT (Huang et al. 2017b)—for evaluating pathogenic noncoding variant by a rank product method. Overall, the VarNote-PAT pipeline incorporated four main steps to rank pathogenic regulatory variant from WGS variants: (1) filtration on allele frequency; (2) filtration on genetic inheritance mode; (3) filtration on variant consequences and epigenomic features; and (4) pathogenicity prioritization. The online VarNote-PAT pipeline and visualization is available at http:// mulinlab.org/varnote/application.html#PAT.
Evaluation of VarNote-PAT for rare pathogenic variant prioritization
Known rare pathogenic regulatory variants (minor allele frequency in EUR < 0.001) were collected from ClinVar (Landrum et al. 2020), Genomiser (Smedley et al. 2016), RegBase (Zhang et al. 2019), NCBoost (Caron et al. 2019), and CDTS (di Iulio et al. 2018). We used Jannovar to annotate the variant consequence and excluded the variants with protein-coding or splicing-altering attributes. Because some of these pathogenic variants were not confirmed by functional study, we manually inspected which pathogenic variants were validated by functional experiments (such as luciferase reporter assay of promoter/enhancer, electrophoretic mobility shift assay, chromatin conformation capture assay, etc.) from the original publication and other literature. We simulated WGS results of pathogenic variant carrier by spiking each validated variant into each individual genome (503 individuals in EUR population from The 1000 Genomes Project) according to dominant disease inheritance mode. We then tested whether the spike-in pathogenic variants could be top-ranked in the simulated individual genomes (filtering criteria: allele frequency: less than 0.001 in EUR; variant consequence: protein-coding and splicing-altering excluded; genome region: 10 kb upstream and downstream from the gene promoter) and compared the performance among our combined strategy and existing methods of pathogenic regulatory variant prediction.
Implementation of the VarNote-CAN pipeline
The used variant annotations for allele frequency and tissue/cell-type-specific epigenomic features are the same as those in the VarNote-PAT pipeline. Noncoding somatic mutations and their recurrence information were downloaded from COSMIC v90 (Tate et al. 2019). We used our recent regBase-CAN score (Zhang et al. 2019) to prioritize cancer driver regulatory variants. The VarNote-CAN pipeline incorporated four main steps to rank cancer driver regulatory variants for personal cancer genome: (1) filtration on germline variants; (2) filtration on variant consequences and epigenomic features; (3) filtration on somatic recurrence; and (4) cancer driver mutation prioritization. The online VarNote-CAN pipeline and visualization is available at http://mulinlab.org/varnote/application.html#CAN.
Evaluation of VarNote-CAN for regulatory somatic mutation prioritization
Somatic eGene and associated eQTL intervals were downloaded from a large-scale pan-cancer whole-genome analysis (PCAWG Transcriptome Core Group et al. 2020). In each eQTL interval, we identified the highly recurrent somatic mutations based on COSMIC v90 data and constructed a data set of candidate driver regulatory mutations. Because some intervals contain multiple best somatic mutations with the same recurrence, we only selected one representative by requiring a median pathogenic score on regBase-CAN. We simulated individual cancer mutation profiles by spiking the candidate driver regulatory mutation into somatic mutation calling results of each WGS cancer genome (1950 patients collected from ICGC). We then executed VarNote-CAN pipeline for all simulated personal cancer genomes (filtering criteria: variant consequence: protein-coding and splicing-altering excluded; genome region: 10 kb upstream and downstream from the gene promoter) and compared the ranks of the spike-in cancer driver mutations among our regBase-CAN score and existing somatic regulatory mutation prediction methods, including FunSeq2 v2.1.6 (Fu et al. 2014), CScape (Rogers et al. 2017), CADD v1.4 (Kircher et al. 2014), Eigen (Ionita-Laza et al. 2016), GenoCanyon (Lu et al. 2015), and FATHMM_MKL (Shihab et al. 2015).
Software availability
All source code and scripts for methods evaluation and manuscript results are available at GitHub (https://github.com/mulinlab/VarNote) and as Supplemental Code. Software, documentation, and VarNote online applications are available at http://mulinlab.org/varnote. VarNote web servers are running at qual-core mode and SAS hard disks for genomic feature intersection and variant annotation.
Competing interest statement
The authors declare no competing interests.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China 31871327 (M.J.L.), and Natural Science Foundation of Tianjin City 19JCJQJC63600, 18JCZDJC34700 (M.J.L.). We thank all tool and resource providers.
Author contributions: D.H. and M.J.L. implemented the algorithms. D.H., X.Y., and M.J.L. conducted the computational analysis described in the paper. D.H. and Y.Z. developed the database and web portal. M.J.L. conceived the study and wrote the paper. H.Y., H.X., JH.W., S.Z., W.N., and P.W. evaluated the computational tool, tested the web portal, and reviewed the manuscript. L.S., C.X., M.L., JW.W., W.L., and H.S.K. suggested the tool functions and reviewed the manuscript. P.C.S. and K.W. contributed to coordination and supervision of computational methodologies and reviewed the manuscript. All authors read and approved the final manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.267997.120.
References
- The 1000 Genomes Project Consortium. 2015. A global reference for human genetic variation. Nature 526: 68–74. 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backenroth D, He Z, Kiryluk K, Boeva V, Pethukova L, Khurana E, Christiano A, Buxbaum JD, Ionita-Laza I. 2018. FUN-LDA: a latent Dirichlet allocation model for predicting tissue-specific functional effects of noncoding variation: methods and applications. Am J Hum Genet 102: 920–942. 10.1016/j.ajhg.2018.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody JA, Morrison AC, Bis JC, O'Connell JR, Brown MR, Huffman JE, Ames DC, Carroll A, Conomos MP, Gabriel S, et al. 2017. Analysis commons, a team approach to discovery in a big-data environment for genetic epidemiology. Nat Genet 49: 1560–1563. 10.1038/ng.3968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujold D, Morais DAL, Gauthier C, Côté C, Caron M, Kwan T, Chen KC, Laperle J, Markovits AN, Pastinen T, et al. 2016. The International Human Epigenome Consortium data portal. Cell Syst 3: 496–499.e2. 10.1016/j.cels.2016.10.019 [DOI] [PubMed] [Google Scholar]
- Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, Motyer A, Vukcevic D, Delaneau O, O'Connell J, et al. 2018. The UK Biobank resource with deep phenotyping and genomic data. Nature 562: 203–209. 10.1038/s41586-018-0579-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron B, Luo Y, Rausell A. 2019. NCBoost classifies pathogenic non-coding variants in mendelian diseases through supervised learning on purifying selection signals in humans. Genome Biol 20: 32 10.1186/s13059-019-1634-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castel SE, Cervera A, Mohammadi P, Aguet F, Reverter F, Wolman A, Guigo R, Iossifov I, Vasileva A, Lappalainen T. 2018. Modified penetrance of coding variants by cis-regulatory variation contributes to disease risk. Nat Genet 50: 1327–1334. 10.1038/s41588-018-0192-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6: 80–92. 10.4161/fly.19695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CA, Hitz BC, Sloan CA, Chan ET, Davidson JM, Gabdank I, Hilton JA, Jain K, Baymuradov UK, Narayanan AK, et al. 2018. The Encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res 46: D794–D801. 10.1093/nar/gkx1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- di Iulio J, Bartha I, Wong EHM, Yu HC, Lavrenko V, Yang D, Jung I, Hicks MA, Shah N, Kirkness EF, et al. 2018. The human noncoding genome defined by genetic diversity. Nat Genet 50: 333–337. 10.1038/s41588-018-0062-7 [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farh KK, Marson A, Zhu J, Kleinewietfeld M, Housley WJ, Beik S, Shoresh N, Whitton H, Ryan RJ, Shishkin AA, et al. 2015. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature 518: 337–343. 10.1038/nature13835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Ratan A, Sheffield NC. 2019. Augmented interval list: a novel data structure for efficient genomic interval search. Bioinformatics 35: 4907–4911. 10.1093/bioinformatics/btz407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Liu Z, Lou S, Bedford J, Mu XJ, Yip KY, Khurana E, Gerstein M. 2014. FunSeq2: a framework for prioritizing noncoding regulatory variants in cancer. Genome Biol 15: 480 10.1186/s13059-014-0480-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilford J, Bergelson L, Powley G, Tucker G, Lichtenstein L, Vaidya P, Roazen D. 2017. Accelerating the compression and decompression of genomics data using GKL provided by Intel. https://www.intel.com/content/dam/www/public/us/en/documents/white-papers/accelerating-genomics-data-gkl-white-paper.pdf.
- Gulko B, Siepel A. 2019. An evolutionary framework for measuring epigenomic information and estimating cell-type-specific fitness consequences. Nat Genet 51: 335–342. 10.1038/s41588-018-0300-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, Liu L, Wang K, Ionita-Laza I. 2018. A semi-supervised approach for predicting cell-type specific functional consequences of non-coding variation using MPRAs. Nat Commun 9: 5199 10.1038/s41467-018-07349-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Fang M, Jostins L, Umićević Mirkov M, Boucher G, Anderson CA, Andersen V, Cleynen I, Cortes A, Crins F, et al. 2017a. Fine-mapping inflammatory bowel disease loci to single-variant resolution. Nature 547: 173–178. 10.1038/nature22969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YF, Gulko B, Siepel A. 2017b. Fast, scalable prediction of deleterious noncoding variants from functional and population genomic data. Nat Genet 49: 618–624. 10.1038/ng.3810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D, Yi X, Zhang S, Zheng Z, Wang P, Xuan C, Sham PC, Wang J, Li MJ. 2018. GWAS4D: multidimensional analysis of context-specific regulatory variant for human complex diseases and traits. Nucleic Acids Res 46: W114–W120. 10.1093/nar/gky407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ionita-Laza I, McCallum K, Xu B, Buxbaum JD. 2016. A spectral approach integrating functional genomic annotations for coding and noncoding variants. Nat Genet 48: 214–220. 10.1038/ng.3477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio F, Knijnenburg TA, Vis DJ, Bignell GR, Menden MP, Schubert M, Aben N, Goncalves E, Barthorpe S, Lightfoot H, et al. 2016. A landscape of pharmacogenomic interactions in cancer. Cell 166: 740–754. 10.1016/j.cell.2016.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäger M, Wang K, Bauer S, Smedley D, Krawitz P, Robinson PN. 2014. Jannovar: a Java library for exome annotation. Hum Mutat 35: 548–555. 10.1002/humu.22531 [DOI] [PubMed] [Google Scholar]
- Kanduri C, Bock C, Gundersen S, Hovig E, Sandve GK. 2019. Colocalization analyses of genomic elements: approaches, recommendations and challenges. Bioinformatics 35: 1615–1624. 10.1093/bioinformatics/bty835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP, et al. 2020. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581: 434–443. 10.1038/s41586-020-2308-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. 2002. The human genome browser at UCSC. Genome Res 12: 996–1006. 10.1101/gr.229102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M, Witten DM, Jain P, O'Roak BJ, Cooper GM, Shendure J. 2014. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 46: 310–315. 10.1038/ng.2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotlar AV, Trevino CE, Zwick ME, Cutler DJ, Wingo TS. 2018. Bystro: rapid online variant annotation and natural-language filtering at whole-genome scale. Genome Biol 19: 14 10.1186/s13059-018-1387-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrum MJ, Chitipiralla S, Brown GR, Chen C, Gu B, Hart J, Hoffman D, Jang W, Kaur K, Liu C, et al. 2020. ClinVar: improvements to accessing data. Nucleic Acids Res 48: D835–D844. 10.1093/nar/gkz972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layer RM, Pedersen BS, DiSera T, Marth GT, Gertz J, Quinlan AR. 2018. GIGGLE: a search engine for large-scale integrated genome analysis. Nat Methods 15: 123–126. 10.1038/nmeth.4556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2011a. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27: 2987–2993. 10.1093/bioinformatics/btr509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2011b. Tabix: fast retrieval of sequence features from generic TAB-delimited files. Bioinformatics 27: 718–719. 10.1093/bioinformatics/btq671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MJ, Deng J, Wang P, Yang W, Ho SL, Sham PC, Wang J, Li M. 2015. wKGGSeq: a comprehensive strategy-based and disease-targeted online framework to facilitate exome sequencing studies of inherited disorders. Hum Mutat 36: 496–503. 10.1002/humu.22766 [DOI] [PubMed] [Google Scholar]
- Li MJ, Liu Z, Wang P, Wong MP, Nelson MR, Kocher JP, Yeager M, Sham PC, Chanock SJ, Xia Z, et al. 2016. GWASdb v2: an update database for human genetic variants identified by genome-wide association studies. Nucleic Acids Res 44: D869–D876. 10.1093/nar/gkv1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Li J, Li MJ, Pan Z, Hsu JS, Liu DJ, Zhan X, Wang J, Song Y, Sham PC. 2017a. Robust and rapid algorithms facilitate large-scale whole genome sequencing downstream analysis in an integrative framework. Nucleic Acids Res 45: e75 10.1093/nar/gkx019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MJ, Li M, Liu Z, Yan B, Pan Z, Huang D, Liang Q, Ying D, Xu F, Yao H, et al. 2017b. cepip: context-dependent epigenomic weighting for prioritization of regulatory variants and disease-associated genes. Genome Biol 18: 52 10.1186/s13059-017-1177-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, White S, Peng B, Johnson AD, Brody JA, Li AH, Huang Z, Carroll A, Wei P, Gibbs R, et al. 2016a. WGSA: an annotation pipeline for human genome sequencing studies. J Med Genet 53: 111–112. 10.1136/jmedgenet-2015-103423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Wu C, Li C, Boerwinkle E. 2016b. dbNSFP v3.0: a one-stop database of functional predictions and annotations for human nonsynonymous and splice-site SNVs. Hum Mutat 37: 235–241. 10.1002/humu.22932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Hu Y, Sun J, Cheng Y, Cheung KH, Zhao H. 2015. A statistical framework to predict functional non-coding regions in the human genome through integrated analysis of annotation data. Sci Rep 5: 10576 10.1038/srep10576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Powles RL, Abdallah S, Ou D, Wang Q, Hu Y, Lu Y, Liu W, Li B, Mukherjee S, et al. 2017. Systematic tissue-specific functional annotation of the human genome highlights immune-related DNA elements for late-onset Alzheimer's disease. PLoS Genet 13: e1006933 10.1371/journal.pgen.1006933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur DG, Manolio TA, Dimmock DP, Rehm HL, Shendure J, Abecasis GR, Adams DR, Altman RB, Antonarakis SE, Ashley EA, et al. 2014. Guidelines for investigating causality of sequence variants in human disease. Nature 508: 469–476. 10.1038/nature13127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CR, Bick D, Belmont JW, Taylor SL, Ashley E, Dimmock D, Jobanputra V, Kearney HM, Kulkarni S, Rehm H, et al. 2020. The medical genome initiative: moving whole-genome sequencing for rare disease diagnosis to the clinic. Genome Med 12: 48 10.1186/s13073-020-00748-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, Flicek P, Cunningham F. 2016. The Ensembl Variant Effect Predictor. Genome Biol 17: 122 10.1186/s13059-016-0974-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neph S, Kuehn MS, Reynolds AP, Haugen E, Thurman RE, Johnson AK, Rynes E, Maurano MT, Vierstra J, Thomas S, et al. 2012. BEDOPS: high-performance genomic feature operations. Bioinformatics 28: 1919–1920. 10.1093/bioinformatics/bts277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- PCAWG Transcriptome Core Group, Calabrese C, Davidson NR, Demircioğlu D, Fonseca NA, He Y, Kahles A, Lehmann KV, Liu F, Shiraishi Y, et al. 2020. Genomic basis for RNA alterations in cancer. Nature 578: 129–136. 10.1038/s41586-020-1970-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen BS, Layer RM, Quinlan AR. 2016. Vcfanno: fast, flexible annotation of genetic variants. Genome Biol 17: 118 10.1186/s13059-016-0973-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos AH, Lichtenstein L, Gupta M, Lawrence MS, Pugh TJ, Saksena G, Meyerson M, Getz G. 2015. Oncotator: cancer variant annotation tool. Hum Mutat 36: E2423–E2429. 10.1002/humu.22771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Roadmap Epigenomics Consortium, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, et al. 2015. Integrative analysis of 111 reference human epigenomes. Nature 518: 317–330. 10.1038/nature14248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers MF, Shihab HA, Gaunt TR, Campbell C. 2017. CScape: a tool for predicting oncogenic single-point mutations in the cancer genome. Sci Rep 7: 11597 10.1038/s41598-017-11746-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers MF, Shihab HA, Mort M, Cooper DN, Gaunt TR, Campbell C. 2018. FATHMM-XF: accurate prediction of pathogenic point mutations via extended features. Bioinformatics 34: 511–513. 10.1093/bioinformatics/btx536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojano E, Seoane P, Ranea JAG, Perkins JR. 2019. Regulatory variants: from detection to predicting impact. Brief Bioinform 20: 1639–1654. 10.1093/bib/bby039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaid DJ, Chen W, Larson NB. 2018. From genome-wide associations to candidate causal variants by statistical fine-mapping. Nat Rev Genet 19: 491–504. 10.1038/s41576-018-0016-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shihab HA, Rogers MF, Gough J, Mort M, Cooper DN, Day IN, Gaunt TR, Campbell C. 2015. An integrative approach to predicting the functional effects of non-coding and coding sequence variation. Bioinformatics 31: 1536–1543. 10.1093/bioinformatics/btv009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smedley D, Schubach M, Jacobsen JOB, Kohler S, Zemojtel T, Spielmann M, Jager M, Hochheiser H, Washington NL, McMurry JA, et al. 2016. A whole-genome analysis framework for effective identification of pathogenic regulatory variants in mendelian disease. Am J Hum Genet 99: 595–606. 10.1016/j.ajhg.2016.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starita LM, Ahituv N, Dunham MJ, Kitzman JO, Roth FP, Seelig G, Shendure J, Fowler DM. 2017. Variant interpretation: functional assays to the rescue. Am J Hum Genet 101: 315–325. 10.1016/j.ajhg.2017.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, Boutselakis H, Cole CG, Creatore C, Dawson E, et al. 2019. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res 47: D941–D947. 10.1093/nar/gky1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Harst P, Verweij N. 2018. Identification of 64 novel genetic loci provides an expanded view on the genetic architecture of coronary artery disease. Circ Res 122: 433–443. 10.1161/CIRCRESAHA.117.312086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. 2010. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38: e164 10.1093/nar/gkq603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Huang D, Zhou Y, Yao H, Liu H, Zhai S, Wu C, Zheng Z, Zhao K, Wang Z, et al. 2020. CAUSALdb: a database for disease/trait causal variants identified using summary statistics of genome-wide association studies. Nucleic Acids Res 48: D807–D816. 10.1093/nar/gkaa807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weedon MN, Cebola I, Patch AM, Flanagan SE, De Franco E, Caswell R, Rodríguez-Segui SA, Shaw-Smith C, Cho CH, Allen HL, et al. 2014. Recessive mutations in a distal PTF1A enhancer cause isolated pancreatic agenesis. Nat Genet 46: 61–64. 10.1038/ng.2826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Adli M. 2019. Mapping and making sense of noncoding mutations in the genome. Cancer Res 79: 4309–4314. 10.1158/0008-5472.CAN-19-0905 [DOI] [PubMed] [Google Scholar]
- Zhang S, He Y, Liu H, Zhai H, Huang D, Yi X, Dong X, Wang Z, Zhao K, Zhou Y, et al. 2019. regBase: whole genome base-wise aggregation and functional prediction for human non-coding regulatory variants. Nucleic Acids Res 47: e134 10.1093/nar/gkz774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng R, Wan C, Mei S, Qin Q, Wu Q, Sun H, Chen CH, Brown M, Zhang X, Meyer CA, et al. 2019. Cistrome Data Browser: expanded datasets and new tools for gene regulatory analysis. Nucleic Acids Res 47: D729–D735. 10.1093/nar/gky1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zook JM, Catoe D, McDaniel J, Vang L, Spies N, Sidow A, Weng Z, Liu Y, Mason CE, Alexander N, et al. 2016. Extensive sequencing of seven human genomes to characterize benchmark reference materials. Sci Data 3: 160025 10.1038/sdata.2016.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.