Abstract
Quercus fenchengsis is a rare Chinese oak species sporadically recorded in Fengcheng, Liaoning, Yuntai Mountain, Henan, and Qinling Mountains, Shaanxi. Here, the complete chloroplast (cp) genome of Q. fenchengensis was first reported. The cp genome is 161,296 bp in length with a GC content of 36.81%, and encodes 134 genes (86 protein-coding genes, 40 tRNA genes and eight rRNA genes). The phylogenetic tree based on the cp genome confirmed that Q. fenchengensis has a close relationship with Quercus aliena acutiserrata and Quercus dentata, consistent with the previous morphology-based suggestion that it would be a hybrid of Q. aliena acutiserrata and Q. dentata.
Keywords: Quercus fenchengensis, chloroplast genome, phylogenetic relationship
Quercus fenchengensis is a Chinese oak first recorded in Fengcheng, Liaoning (Jen et al. 1984), and then found in Yuntai Mountain, Henan and Qinling Mountains, Shaanxi. Morphologically, the nut and shell look like the Quercus dentata, but with short bracteoles; the branches and leaves resemble the Quercus aliena acutiserrata, but with sparse stelatehairs on the underside of the leaf (Jen et al. 1984). It has been suggested that it may be the hybrid of Q. aliena acutiserrata and Q. dentata (Xu and Ren 1998; Huang et al. 1999). So far, the molecular information regarding this rare oak species is severely limited. In this study, we report the complete chloroplast (cp) genome of the species to provide useful genetic information for this species.
The leaves of Q. fenchengensis was collected from a single individual tree at the oak field (N40°28′13.69″; E123°59′17.07″) of Sericultural Institute of Liaoning Province, Fengcheng, China. The specimen was stored with the collection number of OAK_FEN_01 at Department of Sericulture, Shenyang Agricultural University, China. Chloroplast DNA extraction and next-generation sequencing were conducted with an Illumina Hiseq 2500 platform by Personal Biotechnology (Shanghai, China). A reference-guided genome assembly was performed with the cp genome of Q. aliena (KU240007; Yang et al. 2016) as the reference. We used the program DOGMA to annotate the cp genome (Wyman et al. 2004) and compared them with the published cp genomes of Quercus species for correction manually. The cp genome sequence has been deposited at Genbank with accession number MN095295.
The cp genome of Q. fenchengensis is 161,296 in size, forming a typical quadripartite structure as known oaks. The circular cp genome is composed of two single-copy regions separated by a pair of inverted repeats [large single-copy region (LSC): 90,584 bp; small single-copy region (SSC): 19,058 bp; inverted repeats (IRs): 25,827 bp]. The cp genome encodes 134 genes, including 86 protein-coding genes, 40 tRNA genes, and 8 rRNA genes. There are 22 intron-containing genes in this cp genome, including 14 protein-coding genes and 8 tRNA genes. The two ycf1 genes stretch across the IR/SSC borders.
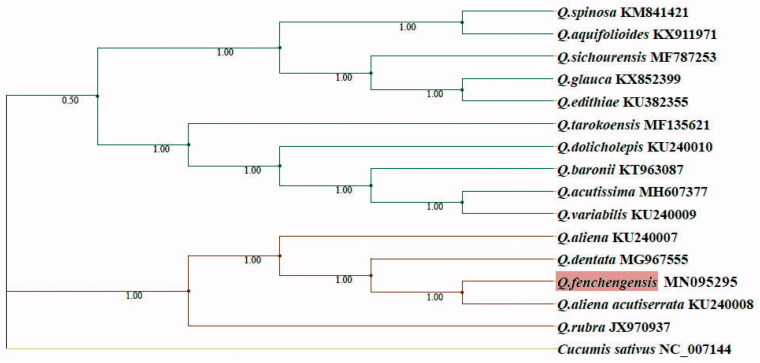
The cp genome sequence of Q. fenchengensis together with other 14 Quercus species was used to reconstruct the phylogenetic relationships of oaks with Bayesian inference (BI) method under the GTR + G + I model by MrBayes (Huelsenbeck and Ronquist 2001). In the phylogenetic tree (Figure 1), the majority of phylogenetic branches are supported by the high posterior probabilities. The phylogenetic analysis based on the cp genome confirmed that Q. fenchengensis has a close relationship with Q. aliena acutiserrata followed by Q.dentata. This result is in line with the previous morphological characters-based suggestion that Q. fenchengensis would be a hybrid of Q. aliena acutiserrata and Q. dentata (Xu and Ren 1998; Huang et al. 1999). However, comparison of the cp genome sequence between Q. fenchengensis and Q. aliena acutiserrata revealed 290 single nucleotide variants (SNVs). Thus, to address the evolutionary origin of Q. fenchengensis, more samplings of oak species and nuclear gene markers need to be covered.
Figure 1.
Phylogeny of 15 Quercus species based on whole chloroplast genomes. BI tree was built with Bayesian inference method under the model of GTR + G + I. The accession numbers are listed following species name. Bayesian posterior probability is given at each node.
Disclosure statement
The authors declare no competing financial interest. The authors alone are responsible for the content and writing of the paper.
References
- Huang CJ, Zhang YT, Barthlomew B. 1999. Fagaceae In: Wu Z-y, Raven PH, editors. Flora of China (vol. 4). Beijing (China): Science Press; p. 370–380. [Google Scholar]
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17:754–755. [DOI] [PubMed] [Google Scholar]
- Jen HW, Wang LM, Gao RQ. 1984. Taxa nova Quercus. Bull Bot Res. 4:195–199. [Google Scholar]
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255. [DOI] [PubMed] [Google Scholar]
- Xu YC, Ren XW. 1998. Flora of China (vol. 22). Beijing (China): Science Press; p. 213–263. [Google Scholar]
- Yang Y, Zhou T, Duan D, Yang J, Feng L, Zhao G. 2016. Comparative analysis of the complete chloroplast genomes of five Quercus species. Front Plant Sci. 7:959. [DOI] [PMC free article] [PubMed] [Google Scholar]