Abstract
Regeneration is one of the great mysteries of biology. Planarians are flatworms capable of dramatic feats of regeneration, which have been studied for over two centuries. Recent findings identify key cellular and molecular principles underlying these feats. A stem cell population (neoblasts) generates new cells and is comprised of pluripotent stem cells (cNeoblasts) and fate-specified cells (specialized neoblasts). Positional information is constitutively active and harbored primarily in muscle, where it acts to guide stem-cell-mediated tissue turnover and regeneration. I describe here a model in which positional information and stem cells combine to enable regeneration.
Introduction
Regeneration involves the replacement of missing organs, appendages, or large body regions. Capacity for regeneration exists in representative species from almost every major animal phylum, including in chordates, platyhelminthes, ctenophores, arthropods, echinoderms, hemichordates, sponges, annelids, arthropods, nermerteans, and acoels. Interest in regeneration is centuries old and stems from the prominent and widespread existence of regeneration in biology, interest in regenerative medicine, and because the process easily captures the imagination: the regrowth of limbs, lower jaws, parts of the heart, spinal cord, and complete new heads ignite curiosity. How do highly regenerative animals do it and why can’t we?
Planarians are flatworms (phylum Platyhelminthes) found in freshwater bodies and their regenerative abilities have been documented for centuries (Pallas, 1766; Dalyell, 1814). Planarians can regenerate new heads, tails, sides, or entire organisms from small body fragments in a process taking days to weeks. Because of their ease of culture and robust regeneration, they have been popular subjects. For instance, planarian regeneration has caught the attention over the years (to differing degrees) of diverse investigators, such as Michael Faraday and T.H. Morgan (Faraday, 1833; Morgan, 1898). A razor blade, magnifying glass, and imagination are enough for experimentation.
Classical inquiry into planarian regeneration involved diverse injuries and transplantations. A suite of molecular and cellular tools have enabled a recent era of intensive molecular genetic inquiry into planarian regeneration (Umesono et al., 1997; Sánchez Alvarado and Newmark, 1999; Newmark and Sánchez Alvarado, 2000; Reddien et al., 2005a; Hayashi et al., 2006; Wagner et al., 2011; Wurtzel et al., 2015; An et al., 2018; Fincher et al., 2018; Grohme et al., 2018; Plass et al., 2018; Zeng et al., 2018). Much excellent and fascinating work on planarian biology will not be reviewed here, such as the role of a myriad of molecules that give planarian stem cells (neoblasts) their attributes, the molecular genetics of the planarian germline, organ formation and function, signaling pathway function and evolution, cilia, genome repair and protection, aging, epigenetics, regulatory RNAs, immune biology, and planarian embryogenesis. Instead, I aim to synthesize key recent results into a mechanistic model for planarian regeneration. After introducing planarian biology, there are four sections: First, I describe pluripotent stem cells (cNeoblasts) and fate-specified stem cells (specialized neoblasts) that provide the cellular basis for regeneration. Second, I describe positional information that is harbored in muscle and how it is re-set after injury. Third, I describe how the combination of positional information and its influence on stem cells (neoblasts) can explain the logic of regeneration. I describe how progenitor targeting by extrinsic cues and self-organization combine to determine where regenerative progenitors go. Finally, I synthesize these findings into pillar concepts that promote understanding of regeneration, tissue turnover, and growth.
Planarian biology and regeneration
Planarians have a complex anatomy including brain, eyes, musculature, intestine, protonephridia, and epidermis, all arranged in complex patterns (Hyman, 1951). The bilobed planarian brain is comprised of a myriad of different neuron types and glia, and connects to two ventral nerve cords. The body-wall musculature contains longitudinal, circular, and diagonal fibers. The epidermis produces mucous and is heavily ciliated ventrally for locomotion. A ciliated excretory system, the protonephridia, is distributed broadly for waste excretion and osmoregulation. A highly branched intestine distributes nutrients and connects to a muscular pharynx located centrally that serves as both mouth and anus. Surrounding internal organs is a mesenchymal tissue compartment (the parenchyma) that includes the only proliferative cells of the adult soma, the neoblasts. Extensive single-cell sequencing has generated a transcriptome atlas for essentially all cell types that comprise planarian anatomy, giving planarians a wealth of molecular resources for their study (Fincher et al., 2018; Plass et al., 2018). Because small body fragments can regenerate an entire planarian, there exist mechanisms in the adult for the production of all adult cell types and tissue patterns.
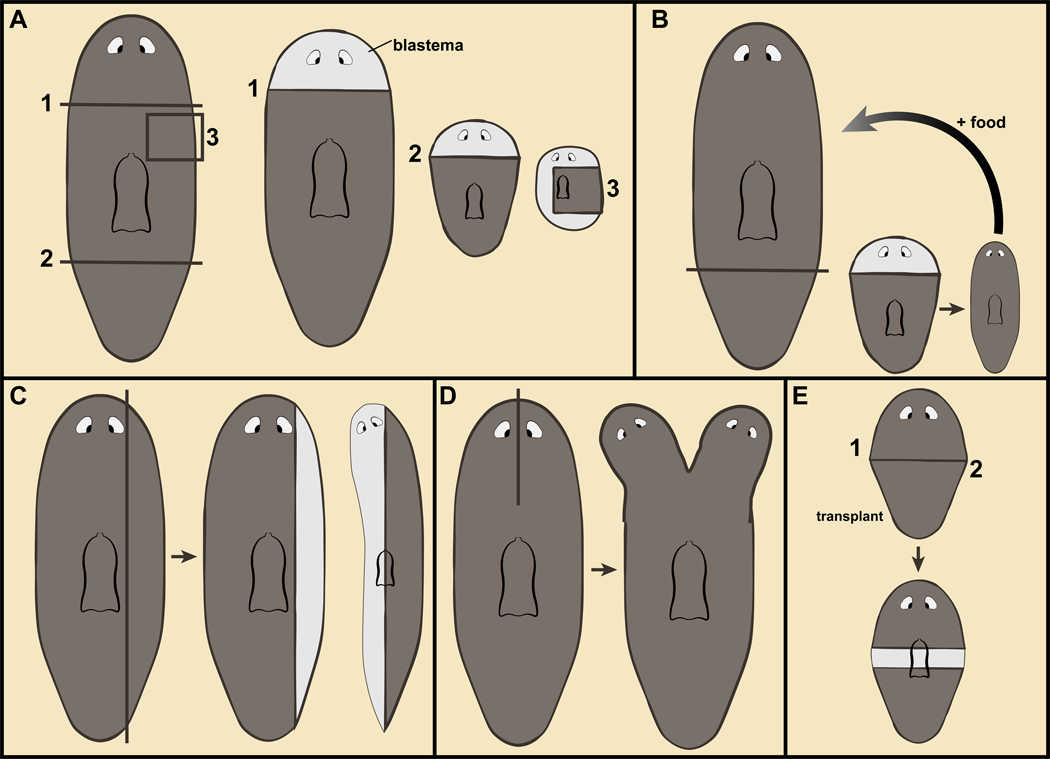
Planarian regeneration involves new tissue production in blastemas (Figure 1A). Because a small planarian body fragment cannot eat until suitable anatomy has been regenerated (including pharynx and brain), regeneration must occur with existing resources. Missing tissues thus cannot be regrown at their original scale. Consequently, blastema formation typically only regenerates some of the missing tissues (such as a head) and is coupled with changes in pre-existing body regions for the regeneration of other missing tissues (Figure 1A). Because the consequent animal will be smaller than the original, some tissues will initially be overabundant in the amputated fragment. Such tissues adjust their position and size relative to regenerating tissues (Figure 1B) (Morgan, 1898; Agata et al., 2003; Oviedo et al., 2003; Reddien and Sánchez Alvarado, 2004; Pellettieri et al., 2010; Forsthoefel et al., 2011; Hill and Petersen, 2015; Atabay et al., 2018; Hill and Petersen, 2018). These changes have been referred to as morphallaxis (Morgan, 1898; Reddien and Sánchez Alvarado, 2004). Morphallaxis is largely (if not entirely) the result of new cell production and cell loss, rather than changes in the differentiated state of cells.
Figure 1. Planarian regeneration.
A–E. Planarian fragments regenerate missing body parts in outgrowths at the wound called blastemas (lighter pigmentation) and through changes in pre-existing tissues (morphallaxis). B. Regeneration from a fragment results in a small animal that can eat and grow towards the original size. C. Parasagittal amputation is followed with ML axis regeneration. D. Incisions not allowed to seal result in duplications of body regions. E. Intercalary regeneration between juxtaposed body fragments.
Planarians can also regenerate missing tissues on their medial-lateral (ML) axis, such as following sagittal and parasagittal amputations (Figure 1C). Partial amputations can result in duplicated structures (Johnson, 1825). A sagittal incision between the eyes, for instance, can result in the regeneration of two heads (Figure 1D). Removal of irregularly shaped tissue fragments can result in regenerative intercalation of missing anatomy (intercalary regeneration) between fused wound faces. Similarly, one region of the body can be transplanted next to another and regenerative responses can be triggered, including the formation of outgrowths that appear to intercalate missing coordinates between juxtaposed tissues (Figure 1E) (Santos, 1931; Okada and Sugino, 1937; Saló and Baguñà, 1985; Kobayashi et al., 1999; Kato et al., 2001; Witchley et al., 2013; Oderberg et al., 2017). The highly plastic, dramatic body plans that result from these various experiments are the source of endless fascination.
I. The cellular basis for new tissue production in planarian regeneration
Clonogenic neoblasts: pluripotent stem cells produce missing cell types
Neoblasts are dividing cells that are widespread in the planarian body (Figure 2A). Neoblasts were recognized as simple, embryonic-like cells possessing a large nucleus and little cytoplasm, but present in the context of adult tissues (Keller, 1894; Baguna, 2012). Neoblasts are specifically eliminated by irradiation and irradiated animals cannot generate any new tissues (Bardeen and Baetjer, 1904; Dubois, 1949; Reddien et al., 2005b). A host of other histological, BrdU, transplantation, and transcript/protein-labeling experiments demonstrated that all new somatic cells in tissue turnover and regeneration come from neoblasts (Baguñà et al., 1989; Newmark and Sánchez Alvarado, 2000; Reddien et al., 2005b; Eisenhoffer et al., 2008; Baguna, 2012). The neoblasts are therefore collectively pluripotent.
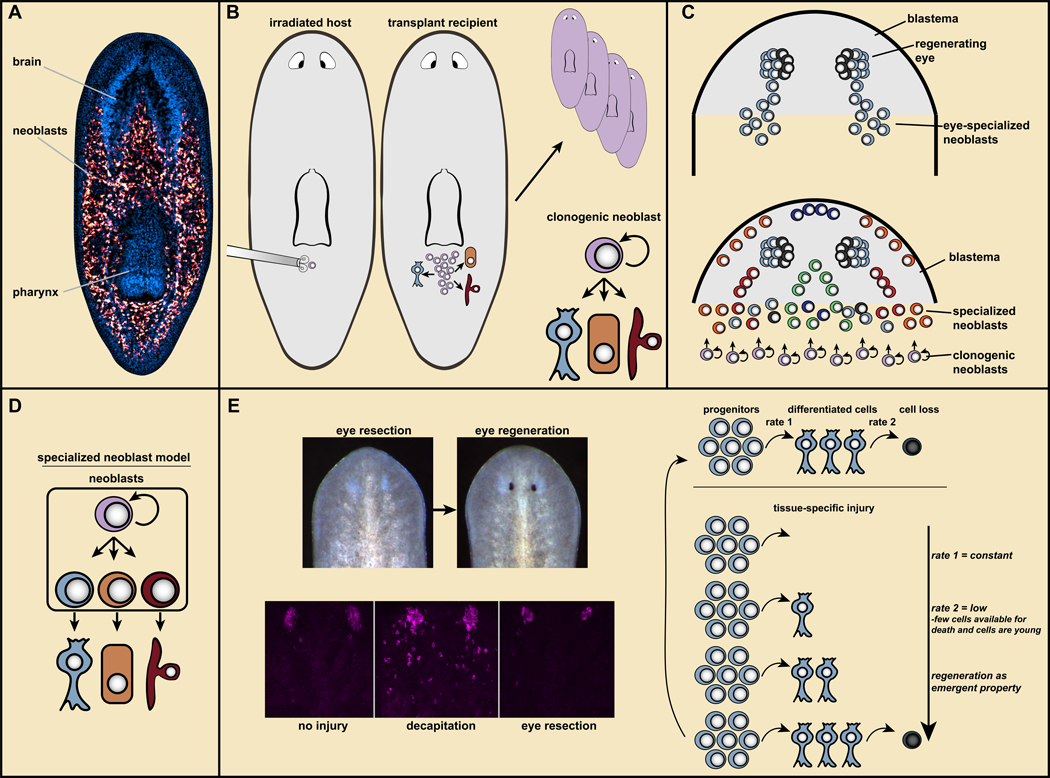
Figure 2. Neoblasts provide the cellular basis for new tissue production in planarian regeneration.
A. Neoblasts (red) are mesenchymal cells distributed broadly. Blue, DAPI; the brain and pharynx are readily visible. From (Wagner et al., 2012). B. Transplantation of single neoblasts can generate colonies with broad differentiation potential and capacity to restore regeneration to lethally irradiated hosts. Resultant animals are genetic clones of the donor (top, right). Bottom right, clonogenic neoblasts (cNeoblasts) are pluripotent stem cells that provide the cellular basis for planarian regeneration. C. Top, specialized neoblasts for the eye produce progenitors that migrate in two trails from the wound into the head blastema, where they coalesce into eyes. Bottom, many specialized neoblast classes produce the differentiated cells of the blastema. The fate of blastema cells is pre-determined based upon which specialized neoblasts they came from. D. Specialized neoblast model. Activation of distinct transcription factors in neoblasts specifies their fate in tissue turnover and regeneration. E. Left: Eyes can be regenerated following resection, but this does not trigger amplification of eye progenitors (magenta; data from (LoCascio et al., 2017)). Right, “target-blind” regeneration: regeneration occurs as an emergent property of constant progenitor production. During regeneration, with no progenitor production-rate change, there are fewer cells in the forming eye accessible to undergo cell death and they are young. Consequently, fewer cell death events per eye passively enable regeneration.
Are neoblasts comprised of multiple different populations of renewing stem cells, each with restricted differentiation potential, or do they include cells pluripotent at the single cell level? The key to addressing this problem involved assessing the potential of individual neoblasts. Irradiation and transplantation experiments demonstrated that some individual neoblasts can produce many more neoblasts (Wagner et al., 2011) (Figure 2B). Upon transplantation, individual neoblasts restored regenerative capacity to lethally irradiated host animals that entirely lacked their own neoblasts - slowly converting a transplant recipient into a genetic clone of the donor (Wagner et al., 2011) (Figure 2B). Furthermore, in animals irradiated with doses that leave sparse surviving neoblasts, neoblast clones are generated that produce neurons, epidermis, and intestine (Wagner et al., 2011). Therefore, at least some neoblasts, the “cNeoblasts”, are pluripotent stem cells that provide the cellular basis for all new tissue production in planarian regeneration (Wagner et al., 2011). An antibody to the protein Tetraspanin, which has enriched cell-surface expression in a subset of neoblasts, allows FACs-based enrichment of cells that can be transplanted and have the functional hallmarks of cNeoblasts (Zeng et al., 2018). The ability to prospectively isolate cells with cNeoblast properties should enable substantial molecular investigation of these pluripotent cells.
The missing-tissue response: proliferative and cell-death responses to tissue loss
Neoblasts respond to injuries by increasing their rate of proliferation. An initial peak in the proliferative response is widespread, occurs approximately six hours after injury, and is followed by a second phase of sustained proliferation near the wound occurring by approximately 48 hours after injury (Baguñà, 1976; Saló and Baguñà, 1984; Wenemoser and Reddien, 2010). The initial proliferative response occurs after any injury type, including injuries such as incisions, that do not require substantial production of new tissues (Wenemoser and Reddien, 2010). By contrast, the second phase of proliferation only occurs at wounds that result in substantial tissue loss, and is referred to as a “missing-tissue” or “regenerative” response (Wenemoser and Reddien, 2010). This response is associated with neoblast accumulation at the wound. Additional responses also distinguish wounds that remove substantial tissue from those that do not. A burst in cell death (TUNEL+ cells) occurs proximal to essentially any wound within four hours of injury(Pellettieri et al., 2010), whereas a second, sustained phase of elevated cell death (involved in morphallaxis) is associated only with injuries that remove substantial tissue (Pellettieri et al., 2010). These proliferative and cell-death responses, detectable by 48–72 h post-amputation and unique to missing tissue injury contexts, are prominent features of planarian regeneration.
Specialized neoblasts: specification of regenerative cell fate occurs in neoblasts
How do neoblasts produce progeny cells that ultimately acquire the correct identity to replace missing tissues? Naive- and specialized-neoblast models were considered to address this question (Reddien, 2013). The naive model posits that neoblasts are a largely homogenous, naive population of cells. Fate specification would occur in their non-dividing, but not yet differentiated, progeny cells, for instance by the position of such cells in a blastema or by neighboring cells. The specialized-neoblast model posits that neoblasts are heterogeneous, with subsets having different fates. Blastema cells would thus have fates predetermined by which specialized-neoblast classes generated them (Figure 2C, D). Substantial evidence now supports the specialized-neoblast model (Reddien, 2013).
Studies of planarian eye and protonephridia regeneration first uncovered specialized-neoblast classes, demonstrating that fate specification for planarian regenerative lineages occurs in neoblasts (Lapan and Reddien, 2011; Scimone et al., 2011; Lapan and Reddien, 2012). Because neoblasts are the only dividing somatic cell population, FACS with DNA labeling can isolate neoblasts in S/G2/M phases, called “X1” cells to reflect their X-ray sensitivity (Hayashi et al., 2006). Some genes typically associated with differentiated cell function (such as the muscle marker DjMHC-A) were expressed in individual X1 cells, which indicated potential functional heterogeneity would exist (Hayashi et al., 2010). Eye specialized neoblasts express eye-associated transcription factors (TFs), such sp6–9, dlx, otxA, six1/2/−1, eya, and ovo, and are required for eye regeneration (Lapan and Reddien, 2011, 2012). Eye-specialized neoblasts have the appearance of all other neoblasts, except for the expression of these eye TF genes.
The planarian protonephridia is a waste and osmoregulatory system that is distributed broadly in the planarian body, and is comprised of multiple cell types (Hyman, 1951; Rink et al., 2011; Scimone et al., 2011; Vu et al., 2015). Similar to the case of the eye, specialized neoblasts expressing TF-encoding genes associated with protonephridia formation (POU2/3 and six1/2–2) were found (Scimone et al., 2011). RNAi studies demonstrated that POU2/3 and six1/2–2 were required for protonephridia-specialized neoblast formation and protonephridia regeneration (Scimone et al., 2011). Subsequent work identified candidate specialized neoblasts for serotonergic (Currie and Pearson, 2013; Marz et al., 2013) and other neurons (Wenemoser et al., 2012; Cowles et al., 2013; Cowles et al., 2014), the pharynx (Adler et al., 2014; Scimone et al., 2014a), the anterior pole (Scimone et al., 2014b; Vásquez-Doorman and Petersen, 2014; Vogg et al., 2014), muscle (Scimone et al., 2017), pigment cells (He et al., 2017), and candidates for numerous additional cell classes (Scimone et al., 2014a).
Single-cell molecular analyses also identified functional heterogeneity in the neoblast population. From systematic single-cell multiplexed qPCR analyses, neoblasts were separated into two major categories with distinct gene expression called sigma and zeta (van Wolfswinkel et al., 2014). zeta-neoblasts are specialized for the planarian epidermis and are the major specialized-neoblast class. The sigma class is heterogenous, but capable of generating zeta-neoblasts (van Wolfswinkel et al., 2014). Gamma-neoblasts represent another abundant neoblast class identified in the multiplexed qPCR data, and are progenitors for the intestine (Wagner et al., 2011; van Wolfswinkel et al., 2014; Flores et al., 2016; González-Sastre et al., 2017). Thousands of neoblasts have now been subjected to SCS (Fincher et al., 2018; Plass et al., 2018; Zeng et al., 2018). From this work the transcriptomes of many classes of specialized neoblasts were revealed, including specialized neoblasts for protonephridia, muscle, intestine, skin, nervous system, parenchymal cells, and other cell types.
The specialized neoblast classes examined in depth so far also all exist in uninjured animals. The emerging picture from this now large set of observations is that the neoblasts include pluripotent stem cells and numerous distinct specialized neoblasts – perhaps one for every cell type of the body (Figure 2C, D).
Specialized neoblasts are produced in a spatially coarse pattern
Whereas neoblast specialization can occur regionally, it is spatially coarse. i.e., specialized neoblasts are distributed much more broadly than the final location of their target differentiated tissue. This attribute was first discovered in uninjured animals for eye-specialized neoblasts and has important implications for the logic of regeneration (described further below) (Lapan and Reddien, 2012). Eye-specialized neoblasts are specified between the centrally located pharynx and the eyes (the prepharyngeal region). These broadly distributed progenitors ultimately incorporate into uninjured eyes to replace cells lost to natural tissue turnover (Lapan and Reddien, 2011, 2012; LoCascio et al., 2017). During regeneration, two migratory eye progenitor trails emerge from the wound area: eye-specialized neoblasts produce non-dividing progeny cells that progressively migrate to specific target locations in the head blastema where they differentiate (Lapan and Reddien, 2011) (Figure 2C). foxA+−pharynx progenitors have similar attributes, being present broadly in the trunk of animals where they maintain the pharynx (Adler et al., 2014; Scimone et al., 2014a). Therefore, regulation of the migration of specialized neoblasts from a coarse pattern to their target destination appears to be an important aspect of specialized-neoblast behavior.
Target-blind regeneration
There are limitless, varied injuries that can be inflicted on planarians, each removing different combinations and amounts of cell types. How is it determined exactly which cell types are missing and how many of each? The capacity of regeneration to detect and replace precisely what is missing has been defined as “regeneration specificity” (LoCascio et al., 2017).
One potential explanation for regeneration specificity would involve a system of feedback cues (Adler and Sanchez Alvarado, 2015). In this view, a given tissue signals its presence to neoblasts, inhibiting its own production. Recent work on planarian eye regeneration, however, demonstrated intriguingly that regeneration specificity is accomplished for this organ without detection of eye absence (LoCascio et al., 2017). As described above, eye-specialized neoblasts produce progenitors that incorporate into existing eyes during homeostatic tissue turnover (Lapan and Reddien, 2011, 2012; LoCascio et al., 2017). Following decapitation, eye progenitors are amplified in large numbers and bring about rapid eye regeneration in a new head. Would absence of only the eye, following eye-specific resection, lead to similar eye-progenitor amplification? What about removal of only a few eye cells – would amplification of only a few progenitors occur? The surprising answer is that specific removal of the eye leads to no change in the number of eye progenitors, but the eye will regenerate nonetheless (Figure 2E) (LoCascio et al., 2017). In animals with one uninjured eye and one regenerating eye (following resection), the two eyes incorporate progenitors at the same low rate. And yet one eye will grow and the other will stay a constant size. How?
Eye size is determined not only by the rate of new cells entering an eye but also by the rate of cells leaving the eye through cell death. Thus, if the rate of new cell incorporation into a regenerating eye is not changing, then the rate of cell loss in the regenerating eye (per eye) should be less than the loss rate for an uninjured eye. Indeed, this was the case (LoCascio et al., 2017). Less cell loss per eye is anticipated to be a passive consequence of regeneration: a regenerating eye has fewer cells accessible for death than an uninjured eye and these cells are all young. Therefore, without need for regulation, less cell death in the regenerating eye than in the uninjured eye should facilitate growth (Figure 2E). By way of analogy, imagine visitors to a museum exhibit arrive one per minute and spend 10 minutes each viewing the exhibit. A fire alarm makes all ten current viewers leave. After the alarm is over, visitors return 1 per minute. After 1 minute there is one viewer, after two minutes two viewers, and after 10 minutes the crowd has been regenerated without need to change viewer arrival rate. Similarly, eyes can regenerate as an emergent property of equilibrium dynamics. Pharynx absence was associated with increased pharynx progenitors, raising the possibility of feedback for this organ, with pharynx absence itself causing increased pharynx progenitor production (Adler et al., 2014). Furthermore, inhibition of the NPHP gene caused nephridia dysfunction and increased nephridia progenitor formation (Vu et al., 2015). However, pharynx removal and NPHP RNAi led to general neoblast proliferation (a missing-tissue response in the case of the pharynx; NPHP RNAi did not eliminate nephridia). As will be discussed below, a broad proliferation response complicates distinction between feedback regulation and generic amplification of progenitors, such as near wounds (LoCascio et al., 2017).
The model based on findings from the eye is called “target-blind” progenitor specification, because the eye-progenitor specification rate is “blind” to the presence or absence of the target organ those progenitors make (Figure 2E) (LoCascio et al., 2017). The simplicity of this target-blind model suggests that it might have broad explanatory power.
II. The instructions for tissue turnover and regeneration
Constitutive positional information
What molecular mechanisms allow animals to regenerate the correct missing body parts? In this section, I will describe positional information, which resides outside of neoblasts and guides tissue maintenance and regeneration. Target-blind regeneration of the eye, described above, involved eye progenitors remaining present after eye resection because of the coarse eye-progenitor-specification zone (LoCascio et al., 2017). By contrast, large injuries that remove eyes and their progenitors require production of eye progenitors from scratch. Following such large injuries, eye progenitors are not only de novo synthesized, but they are generated in much greater numbers than occurs in homeostasis (Lapan and Reddien, 2011, 2012). How does the animal regenerate such large missing regions, involving de novo production of progenitor classes? Major molecular insight into this question emerged from a large series of experiments involving striking RNAi phenotypes in which animals regenerate, but regenerate the incorrect tissues (Figure 3A).
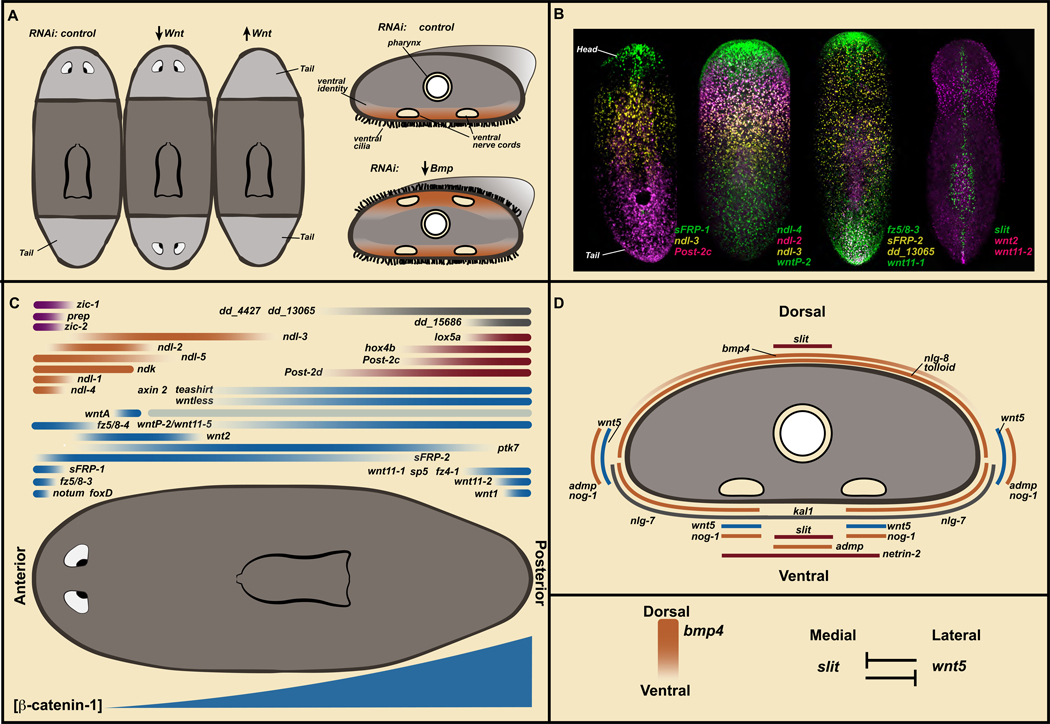
Figure 3. Position-control genes are constitutively and regionally expressed in planarian muscle to control adult body plan maintenance and regeneration.
A. Regeneration-patterning phenotypes. Left, trunk fragments regenerating heads and tails. Inhibition of Wnt pathway genes results in regeneration of posterior-facing heads. Inhibition of negative Wnt pathway regulators leads to regeneration of anterior-facing tails. Right, Bmp pathway inhibition leads to dorsal appearance of ventral attributes. B. PCG expression domains. Anterior, up. Data from (Scimone et al., 2016). C. Cartoon map of PCG transcription domains on the AP axis. Blue, Wnt-signaling related; Orange, FGFRL genes; Red, Hox genes; Purple, anterior transcription factors; Grey, unknown molecular function. Bottom: posterior-to-anterior gradient of β-catenin-1 protein level. D. Cartoon of PCG expression domains on the DV/ML axes. Bmp signaling is active dorsally and wnt5 - slit mutual inhibition mediates ML pattern.
β-catenin-1 RNAi led to dramatic regeneration outcomes: heads were regenerated in place of tails, resulting in two-headed animals, with both heads fighting to pull the same body in different directions (Figure 3A) (Gurley et al., 2008; Iglesias et al., 2008; Petersen and Reddien, 2008). This observation suggested a role for Wnt signaling in the decision-making process at transverse amputation planes to regenerate a head or a tail. Following β-catenin-1 RNAi in uninjured animals, a slow and steady transformation of the body plan occurs during tissue turnover: many heads appear around the animal periphery (Gurley et al., 2008; Iglesias et al., 2008; Petersen and Reddien, 2008). Similarly, inhibition of Wnt signaling components encoding Wnt1 (Adell et al., 2009; Petersen and Reddien, 2009b), Wntless (Adell et al., 2009), Dishevelled (Almuedo-Castillo et al., 2011), or Teashirt (Owen et al., 2015; Reuter et al., 2015) can cause the regeneration of posterior heads in place of tails, and/or homeostatic formation of ectopic heads. Conversely, Wnt signaling upregulation, through APC (Gurley et al., 2008) or notum (Petersen and Reddien, 2011) RNAi can result in the regeneration of tails in place of heads. Numerous genes encoding Wnt-family proteins display regionalized, constitutive transcription along the adult planarian anterior-posterior (AP) axis (Figure 3B, C). For instance, wntP-2 (also known as wnt11–5), wnt11–1, wnt11–2, and wnt1 are all expressed in the posterior (Petersen and Reddien, 2008). By contrast, genes encoding predicted Wnt inhibitors, sFRP-1, sFRP-2, and notum, are expressed in the anterior (Petersen and Reddien, 2008; Gurley et al., 2010; Petersen and Reddien, 2011). β-catenin-1 protein levels also demonstrate a posterior-to-anterior gradient of Wnt activity (Figure 3C) (Sureda-Gómez et al., 2016; Stückemann et al., 2017). These findings indicate that constitutive regional expression of Wnt ligands (posteriorly) and Wnt inhibitors (anteriorly) control regionalization of the planarian AP axis. The discovery of the role of Wnt signaling in planarian AP axis regulation helped reveal a broadly utilized role of Wnt signaling in the polarization of tissues along the AP axis of animals throughout the animal kingdom (Petersen and Reddien, 2009a).
Another prominent model of planarian adult positional information involves Bmp signaling, which regulates the dorsal-ventral (DV) axis (Figure 3A). RNAi of bmp4, which encodes a Bmp signaling ligand, or of pathway components smad1, smad4, or tolloid, leads to striking regeneration and homeostatic phenotypes affecting the DV axis (Molina et al., 2007; Orii and Watanabe, 2007; Reddien et al., 2007). Bmp inhibition results in progressive ventralization, with ventral tissue types, such as nerve cords and ciliated epidermis, appearing dorsally. Eventually, for instance, bmp4 RNAi animals can flip over and glide on their backs! Further study of this pathway revealed roles for other pathway regulators, including admp (Gavino and Reddien, 2011; Molina et al., 2011) and a variety of noggin (nog) and noggin-like (nlg) genes (Molina et al., 2009; Molina et al., 2011). bmp4 is expressed dorsally, in a medial-to-lateral gradient (Orii et al., 1998; Molina et al., 2007; Orii and Watanabe, 2007; Reddien et al., 2007). Other genes encoding Bmp pathway components are also expressed in constitutive and regionalized ways, with a variety of DV- and medial-lateral (ML) -restricted expression domains (Figure 3D). Similar to the case for Wnt and the AP axis, these findings indicate that constitutive regional expression of Bmp signaling pathway genes promotes maintenance and regeneration of pattern of the adult planarian DV axis.
Inhibition of several other constitutively expressed genes causes local organ duplication and expansion of body regions, such as in the head and trunk. nou-darake encodes a defining member of the FGFRL-protein family and its inhibition leads to posterior expansion of the brain and ectopic eyes (Cebria et al., 2002). wntA (Kobayashi et al., 2007; Adell et al., 2009) and fz5/8–4 (Scimone et al., 2016) inhibition can cause a similar brain/eye expansion phenotype. ptk7 RNAi (Lander and Petersen, 2016) as well as ndl-3 and wntP-2 RNAi (Lander and Petersen, 2016; Scimone et al., 2016) causes trunk duplications, with animals developing two pharynges rather than one. On the ML axis, wnt5 is expressed laterally and inhibits the medially expressed slit (Figure 3D). wnt5 RNAi and slit RNAi result in medial-lateral patterning abnormalities (Cebria et al., 2007; Adell et al., 2009; Gurley et al., 2010; Oderberg et al., 2017; Atabay et al., 2018).
Genes, with constitutive and regional expression and association with a patterning RNAi phenotype or that are predicted by homology to be part of a planarian patterning pathway, such as those described above, are called “position-control genes” (PCGs) (Witchley et al., 2013). For multiple PCGs, phenotypes have not yet been revealed, so their role in patterning or other biology awaits elucidation. The strength of the argument for a patterning role for many of these genes comes from numerous RNAi patterning phenotypes (Table 1). PCG expression domains are also striking and highly suggestive of a role in patterning: head-to-tail transcriptional gradients, medial-to-lateral transcriptional gradients, expression at the very head or tail tip, or expression around the periphery of the animal at the DV median plane, among other patterns (Figure 3B–D). These experiments together revealed the existence of constitutively expressed adult positional information for planarian tissue turnover and regeneration (Reddien, 2011) (Figure 3B–D).
Table 1.
PCGs with RNAi phenotypes
| gene | RNAi phenotype | reference(s) |
|---|---|---|
| notum | anterior-facing tail | (Petersen and Reddien, 2011) |
| wntl | posterior-facing head; tailless | (Adell et al., 2009; Petersen and Reddien, 2009) |
| wnt11-2 | medial fusion of posterior anatomy | (Adell et al., 2009; Gurley et al., 2010) |
| wntP-2 / wnt11-5 | posterior-facing head; two pharynges | (Petersen and Reddien, 2009; Sureda-Gómez et al., 2015; Lander and Petersen, 2016; Scimone et al., 2016) |
| wnt11-1 | shorter tail; two pharynges | (Sureda-Gómez et al., 2015) |
| wnt5 | lateral duplication of medial anatomy | (Adell et al., 2009; Gurley et al., 2010; Almuedo-Castillo et al., 2011; Oderberg et al., 2017; Atabay et al., 2018) |
| wntA | posterior expansion of eyes and brain | (Kobayashi et al., 2007; Scimone et al., 2016) |
| frizzled5/8-4 | posterior expansion of eyes and brain | (Scimone et al., 2016) |
| wntless | posterior-facing head | (Adell et al., 2009) (Almuedo-Castillo et al., 2011) |
| teashirt | posterior-facing head | (Owen et al., 2015; Reuter et al., 2015) |
| Djislet | tailless | (Hayashi et al., 2011) |
| prep | failed anterior identity regeneration | (Felix and Aboobaker, 2010) |
| foxD | failed anterior and medial identity regeneration | (Roberts-Galbraith and Newmark, 2013; Scimone et al., 2014; Vogg et al., 2014) |
| zic-1 | failed anterior identity regeneration | (Vasquez-Doorman and Petersen, 2014; Vogg et al., 2014) |
| zic-2 | defective anterior identity regeneration | (Vasquez-Doorman and Petersen, 2014; Vogg et al., 2014) |
| bmp | ventralization | (Molina et al., 2007; Orii and Watanabe, 2007; Reddien et al., 2007) |
| admp | ventralization | (Gavino and Reddien, 2011; Molina et al., 2011) |
| nlg-8 | ventralization | (Molina et al., 2011) |
| tolloid | dorsal ruffling, ML-regeneration defective | (Reddien et al., 2007) |
| ndl-3 | two pharynges | (Lander and Petersen, 2016; Scimone et al., 2016) |
| nog-1 | dorsalization | (Molina et al., 2011) |
| glypican-1 | indented heads; midline defect | (Wenemoser et al., 2012) |
| netrin-2 | CNS defects | (Cebria and Newmark, 2005) |
| ptk7 | two pharynges | (Lander and Petersen, 2016) |
| ndk | posterior expansion of eyes and brain | (Cebria et al., 2002) |
| slit | medialization | (Cebria et al., 2007; Gurley et al., 2010; Oderberg et al., 2017) |
Adult positional information is harbored in planarian muscle
The patterns of PCG expression are reminiscent of patterning gene expression in animal embryos (such as Drosophila), but present in adults. Which cells express these genes in the context of adult tissues? Most PCG expression is restricted to a single layer of subepidermal cells that are peripheral to the neoblasts (Witchley et al., 2013). This raised the possibility that there existed a particular cell population with a prominent role in harboring positional information. Indeed, where any two PCGs were expressed in overlapping spatial domains, the genes were expressed together in the same cells to a substantial degree. The cells expressing PCGs turn out to be, unexpectedly, muscle (Witchley et al., 2013) (Figure 4A). Subsequent single-cell RNA sequencing of muscle from multiple regions along the planarian AP axis revealed that many additional genes display regional expression in muscle (Scimone et al., 2016; Fincher et al., 2018). Continued identification and study of candidate PCGs should help in understanding the regulatory logic of regeneration.
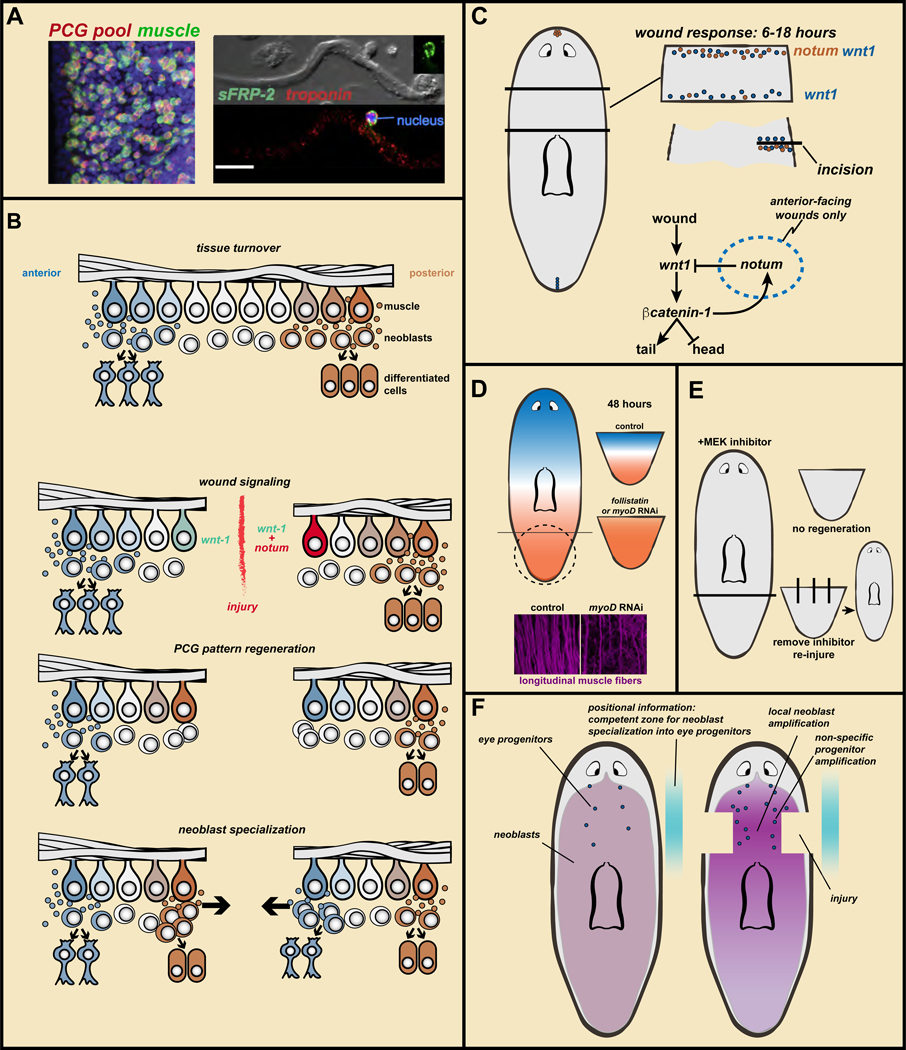
Figure 4. Wound signaling mediates the regeneration of positional information to enable regeneration of missing parts.
A. PCGs are expressed in muscle. Data from (Witchley et al., 2013). Left, a PCG RNA probe pool (red) labels almost all body-wall muscle cells (green, collagen probe). Right, sFRP-2 transcripts around a muscle cell nucleus. Bar, 20 microns. B. Regeneration model: muscle ßPCG expression, specialized neoblasts, and wound-induced re-establishment of PCG expression domains. See text for details. C. notum is preferentially wound-induced at anterior-facing wounds and wnt1 is wound-induced at all wounds. notum inhibits Wnt signaling to promote head regeneration. How notum is selectivity activated at anterior-facing wounds is unknown. D. Without PCG pattern regeneration in follistatin or myoD RNAi animals, regeneration fails to occur. This requires longitudinal muscle fibers (data from (Scimone et al., 2017)). E. MEK inhibitor treatment blocks regeneration. Removal of the inhibitor does not lead to regeneration unless new injuries are inflicted. F. Coincidence of increased neoblast proliferation with positional information leads to amplification of regional progenitor classes, even if the target tissue of those progenitors remains uninjured.
Body-wall muscle includes longitudinal, circular, and diagonal fibers (Cebria, 2016). myoD is required for the regeneration and homeostatic maintenance of longitudinal muscle fibers (Scimone et al., 2017). By contrast, nkx1 is required for the regeneration and homeostatic maintenance of circular fibers (Scimone et al., 2017). Inhibition of both genes together led to a substantial loss of body-wall fibers and loss of PCG expression before animal death. These animals displayed a patterning phenotype, generating extra eyes, further supporting the model that muscle has an instructive role in regulating the adult planarian body plan (Scimone et al., 2017).
Muscle rapidly regenerates pattern of positional information
How positional information regenerates its pattern is a central problem in regeneration. The pattern of PCG expression in muscle rapidly regenerates after injury (Molina et al., 2007; Reddien et al., 2007; Petersen and Reddien, 2008, 2009b; Gurley et al., 2010; Hayashi et al., 2011; Chen et al., 2013; Witchley et al., 2013; Vásquez-Doorman and Petersen, 2014; Owen et al., 2015; Reuter et al., 2015; Wurtzel et al., 2015; Lander and Petersen, 2016; Scimone et al., 2017; Stückemann et al., 2017). For example, beginning between 24–48 hours after amputation, a tail fragment expresses numerous anterior PCGs at the anterior-facing wound and posterior PCGs restrict their expression more posteriorly, reflecting an initial reconstitution of the normal PCG expression relationships (Figure 4B). A converse process happens in an amputated head fragment. Similarly, ML patterns of PCG expression are regenerated after sagittal amputation (Reddien et al., 2007). Some, but not all, of these PCG expression changes during regeneration occur in irradiated animals lacking neoblasts, indicating initial phases of re-establishing PCG expression domains after injury can occur in existing muscle cells without the production of missing differentiated cell types (Reddien et al., 2007; Petersen and Reddien, 2009b; Gurley et al., 2010). This conclusion is also supported by observed transient co-expression of anteriorly and posteriorly expressed PCGs in amputated tail fragments as expression domain changes initiate (Witchley et al., 2013). The rapid regeneration of PCG expression domains as an upstream regulatory step is a central pillar of planarian regeneration.
Wound signaling promotes regeneration of positional information in muscle
In principle, positional information could regenerate its pattern autonomously, as an emergent property of the system (Meinhardt, 1982). By contrast, data suggests that resetting of positional information in planarian muscle is regulated by external input: wound signaling (Figure 4C). Two key molecules, wnt1 and notum, are activated by wound signaling and contribute to the resetting of positional information. wnt1 is activated generically at all wounds by 6 hours after injury (Petersen and Reddien, 2009b). wnt1 RNAi can cause posterior-facing wounds to regenerate ectopic heads or to fail to regenerate, indicating wnt1 promotes posterior and inhibits anterior identity (Adell et al., 2009; Petersen and Reddien, 2009b). wnt1 is also expressed in the planarian posterior pole in a second phase of expression in regeneration (Petersen and Reddien, 2008; Adell et al., 2009; Petersen and Reddien, 2009b). Irradiated, amputated head fragments, lacking neoblasts, cannot make the posterior pole and yet are capable of initiating expression of some posterior PCGs, such as wntP-2 (Petersen and Reddien, 2009b). Irradiated, wnt1 RNAi head fragments however, could not activate wntP-2 at posterior-facing wounds, demonstrating the role of wound-induced wnt1 in the process of re-retting posterior positional information (Petersen and Reddien, 2009b). This finding revealed a pattern-initiating process specific to the context of regeneration, and explains how missing positional information can return by leveraging wound signaling.
If wnt1 is generically activated by wounding and promotes tail regeneration, how do anterior-facing wounds regenerate heads? The answer, at least in part, involves notum. Whereas wnt1 is expressed in the posterior pole in uninjured animals, notum is expressed oppositely, in the anterior pole at the head tip (Petersen and Reddien, 2011). Following amputation, notum is also expressed generically at wounds (many wound types activate it and by ~6 hours post-injury), but unlike wnt1, notum displays preferential expression at anterior-facing wounds (Petersen and Reddien, 2011) (Figure 4C). This result is strikingly shown in body incision injuries, which heal by sealing together without requiring substantial regenerative repair: notum is activated, but only on the posterior (anterior-facing) side of the incision (Petersen and Reddien, 2011) (Figure 4C). Determining how notum selectivity for anterior-facing wounds is accomplished is an important direction. notum RNAi animals regenerate tails in place of heads, indicating that notum promotes head and inhibits tail identity (Petersen and Reddien, 2011). Accordingly, notum RNAi animals displayed ectopic wntP-2 (posterior PCG) expression at anterior-facing wounds (Petersen and Reddien, 2011).
These findings suggest that induction of wnt1 and notum results in a high Wnt signaling environment at posterior-facing wounds and a Wnt inhibitory environment at anterior-facing wounds and that this initiates the changes in PCG expression that ensue (Figure 4C). Because PCG-expression-domain maintenance requires PCG pathways themselves, a reasonable possibility is that interactions among some components of the PCG system result in re-establishment of correct relative patterns in muscle thereafter (Petersen and Reddien, 2009b, 2011; Stückemann et al., 2017). For instance, β-catenin is required for maintaining posterior PCG patterns and preventing anterior PCG patterns from being expressed in the tail (Owen et al., 2015; Reuter et al., 2015; Lander and Petersen, 2016; Scimone et al., 2016; Stückemann et al., 2017).
wnt1 and notum are part of a generic wound response
wnt1 and notum are part of what has been termed a “generic wound response” (Wenemoser et al., 2012; Wurtzel et al., 2015). Approximately 240 genes are transcriptionally activated within the first 12 hours following injury in planarians, regardless of the wound type (Wenemoser et al., 2012; Wurtzel et al., 2015). In other words, wounds that will regenerate completely different anatomy such as a head versus a side start life with essentially the same program. Many of these wound-induced genes are broadly conserved, with functions awaiting elucidation.
Changes in PCG expression and the expression of transcription factor-encoding genes associated with neoblast specialization will be more tailored to the distinct regeneration outcomes at different wounds and occur after this generic wound response (~24–36 hours following injury) (Petersen and Reddien, 2009b; Gurley et al., 2010; Wurtzel et al., 2015; Stückemann et al., 2017). This timing corresponds roughly to a detectable increase in β-catenin levels at posterior-facing wounds (Stückemann et al., 2017). Subsequently, regeneration-context specific, new differentiated cells emerge ~72 hours following injury (Wurtzel et al., 2015).
Wound-induced genes fall into several categories. Those activated within an hour of injury include stress-response genes and are expressed in most tissue types (Wenemoser et al., 2012; Wurtzel et al., 2015). Genes in a second category are preferentially upregulated in a specific tissue: muscle, epidermis, or neoblasts (Wenemoser et al., 2012; Wurtzel et al., 2015). wnt1 and notum are within the muscle wound-induced gene class (Witchley et al., 2013). Systematic efforts to find wound-induced genes (within 12h post-amputation) with asymmetric activation at anterior- versus posterior-facing wounds yielded only one gene: notum (Wurtzel et al., 2015).
Wound-induced positional information re-establishment is required for regeneration
One muscle wound-induced gene, follistatin, is required for the missing-tissue response (Gavino et al., 2013). follistatin RNAi can result in failed regeneration (Gavino et al., 2013; Roberts-Galbraith and Newmark, 2013). follistatin RNAi tail fragments fail to re-establish AP pattern of PCG expression and, accordingly, never regenerate missing cell types (Figure 4D). Instead, amputated fragments endure, maintaining with turnover the tissues they do have (Gavino et al., 2013).
ERK phosphorylation is triggered within minutes of injury, and pharmacologic inhibition of ERK activation blocks regeneration and multiple aspects of the missing-tissue response. ERK signaling is required for wound-induced expression of many genes (such as runt1, wnt1, and notum), indicating that ERK acts upstream of wound-induced gene expression essential for regeneration (Owlarn et al., 2017). After failed regeneration in the presence of a phospho-ERK inhibitor, alleviation of ERK inhibition does not allow regeneration to occur. However, re-injury of such animals, inducing the generic wound response, induces regeneration supporting a view that the generic wound response is important in initiating regeneration (Owlarn et al., 2017) (Figure 4E).
Wound-induced notum and follistatin expression occurs specifically in longitudinal fibers. As noted above, myoD RNAi led to progressive, specific loss of longitudinal fibers (Scimone et al., 2017) (Figure 4D); consequently, notum and follistatin were not induced at wounds in myoD RNAi animals and these animals completely failed to regenerate (Scimone et al., 2017). myoD RNAi animals (like follistatin RNAi animals) were incapable of PCG re-establishment after amputation and it was therefore proposed that these amputated animals could maintain whatever tissues they did have, but failed to regenerate any missing tissues because they didn’t “know” anything was missing (Scimone et al., 2017) (Figure 4D). These findings support a model that wound-induced PCG re-establishment is required for regeneration (Figure 4B).
III. Positional information and stem cells combine to bring about regeneration
PCGs can influence neoblast specification
Because PCG RNAi can impact regeneration and tissue-turnover outcomes, the cellular output of neoblasts is at some level dependent upon PCGs (Figure 4B). The connections between PCGs and neoblast behavior are only beginning to be studied, but several results indicate the type of regulation that might be present. For example, wnt1 RNAi caused ovo+/smedwi-1+ cells (eye specialized neoblasts) to be ectopically present during posterior regeneration four days following amputation (Witchley et al., 2013). Second, β-catenin-1 RNAi followed by RNA sequencing of tails identified targets of β-catenin-1 and some of these targets (such as teashirt and abdBa) displayed β-catenin-1-dependent expression in neoblasts (Reuter et al., 2015). This likely reflects direct Wnt signaling activity in neoblasts, because substantial β-catenin-1 protein accumulation in the planarian posterior occurs in neoblasts (Stückemann et al., 2017). Finally, the planarian epidermis was utilized to study how cells within a lineage determine their position (Wurtzel et al., 2017). zeta-neoblasts produce progeny cells throughout the body that transit through transcriptionally distinct stages over 6+ days as they migrate and enter the epidermis (Eisenhoffer et al., 2008; van Wolfswinkel et al., 2014; Tu et al., 2015; Zhu et al., 2015; Cheng et al., 2018). Whereas these lineage stages are present uniformly, the epidermis itself displays complex pattern (Wurtzel et al., 2017). From single-cell RNA sequencing data it was determined that dorsal and ventral zeta-neoblasts express different regional genes (PRDM1–1 dorsally and kal1 ventrally). These ventral and dorsal expression states propagate from the zeta-neoblast to their non-dividing progeny cells, and further patterning occurs within these migratory progeny thereafter. The dorsal regional identity of zeta-neoblasts required the dorsal PCG bmp4 (Wurtzel et al., 2017). Therefore, PCG expression from muscle impacts the spatial identity of neoblasts, influencing their fate.
Positional information combines with neoblast specialization to initiate new tissue production
The findings described above allow return to the problem posed at the beginning of the section on positional information: how does a small body fragment lacking a progenitor class (specialized neoblast) generate such progenitors de novo and in large numbers? It was proposed, from work on the eye, that de novo progenitor production can be explained simply by the other components (PCGs and neoblast missing-tissue response) of the regeneration model described above (LoCascio et al., 2017) (Figure 4B, F). A tail fragment, for instance, will have regenerated major PCG-expression regions by 48 hours post-injury. Therefore, in this proposal, the environment of tail neoblasts after amputation rapidly obtains “head-like” positional information, and accordingly tail neoblasts make head progenitors, including for the eye (Figure 4B). This simple model could explain the de novo formation of eye progenitors, but what about their production in large numbers? As described above, neoblasts will divide rapidly and accumulate at the wound as part of the missing-tissue response (Baguñà, 1976; Saló and Baguñà, 1984; Wenemoser and Reddien, 2010). Therefore there will be a large number of neoblasts in a “head”-signaling environment and, therefore, a large number of eye progenitors (Figure 4B) (LoCascio et al., 2017). This model is attractive for its simplicity in explaining regeneration: positional information regeneration and neoblast accumulation at the wound occur rapidly and result in generation of whatever progenitors are appropriate for the missing region (LoCascio et al., 2017).
This simple proposal suggests that it is not the absence of eyes themselves that influences the number of eye progenitors in the case of large injury, such as decapitation. Predictions of this model were tested by inflicting large injuries near the location of eye progenitor specification without removing eyes. This injury should amplify neoblasts, and consequently, amplify eye progenitors (Figure 4F) (LoCascio et al., 2017). Indeed this was the case (LoCascio et al., 2017). Coupling a flank injury with eye resection resulted in greater progenitor incorporation and faster eye regeneration. There are indications that this non-specific “bystander” impact on nearby specialized neoblasts will be generalizable to other tissues. Pharynx removal leads to an increase in pharynx progenitor production (Adler et al., 2014); pharynx removal is a large injury that amplifies neoblast proliferation (Adler et al., 2014; LoCascio et al., 2017) and, accordingly, also increases the rate of incorporation of differentiating neoblasts into neighboring ventral nerve cords (LoCascio et al., 2017). Similarly, large injuries flanking the pharynx, but not including the pharynx, resulted in increased progenitor incorporation into the pharynx (LoCascio et al., 2017). These findings show that large wounds can amplify many progenitor types, even when their target tissues remain and suggest a simple view of how positional information combines with fate specification of stem cells to promote regeneration.
Blastema pattern is established involving organizer-like activity of anterior and posterior poles
How is the pattern of tissues within a blastema established? Pattern formation in animals is best studied in the context of embryogenesis. There, embryo-specific cues (such as the point of sperm entry or asymmetric maternal determinants) can be utilized for initial pattern establishment. An intriguing hypothesis is that tissue landmarks at wounds act as regeneration-specific cues (analogous to embryo-specific cues for development) to orchestrate blastema pattern formation (Oderberg et al., 2017).
Numerous lines of evidence indicate clusters of cells at the planarian head tip (anterior pole) and tail tip (posterior pole) have roles in organizing blastema pattern (Figure 5A). These poles are muscle cells expressing patterning factors and are generated from neoblasts during regeneration (Petersen and Reddien, 2008; Adell et al., 2009; Petersen and Reddien, 2009b; Gurley et al., 2010; Hayashi et al., 2011; Petersen and Reddien, 2011; Witchley et al., 2013; Scimone et al., 2014b). Pole formation follows the initial wound-induced PCG pattern re-establishment phase, occurring ~2–3 days after injury. Pole formation requires specific transcription factors; for instance, foxD, zic, prep, pbx, and pitx are required for regeneration of the anterior pole (Felix and Aboobaker, 2010; Blassberg et al., 2013; Chen et al., 2013; Currie and Pearson, 2013; Marz et al., 2013; Roberts-Galbraith and Newmark, 2013; Scimone et al., 2014b; Vásquez-Doorman and Petersen, 2014; Vogg et al., 2014), with foxD being specifically expressed in the pole (Scimone et al., 2014b; Vogg et al., 2014). Djislet is required for regeneration of the posterior pole, and RNAi animals are tailless (Hayashi et al., 2011). Blocking pole formation causes head blastemas to be small, to lack a regenerated midline, to show medial collapse of tissues, and to lack distal-most (anterior-most) PCG expression (Scimone et al., 2014b; Vogg et al., 2014).
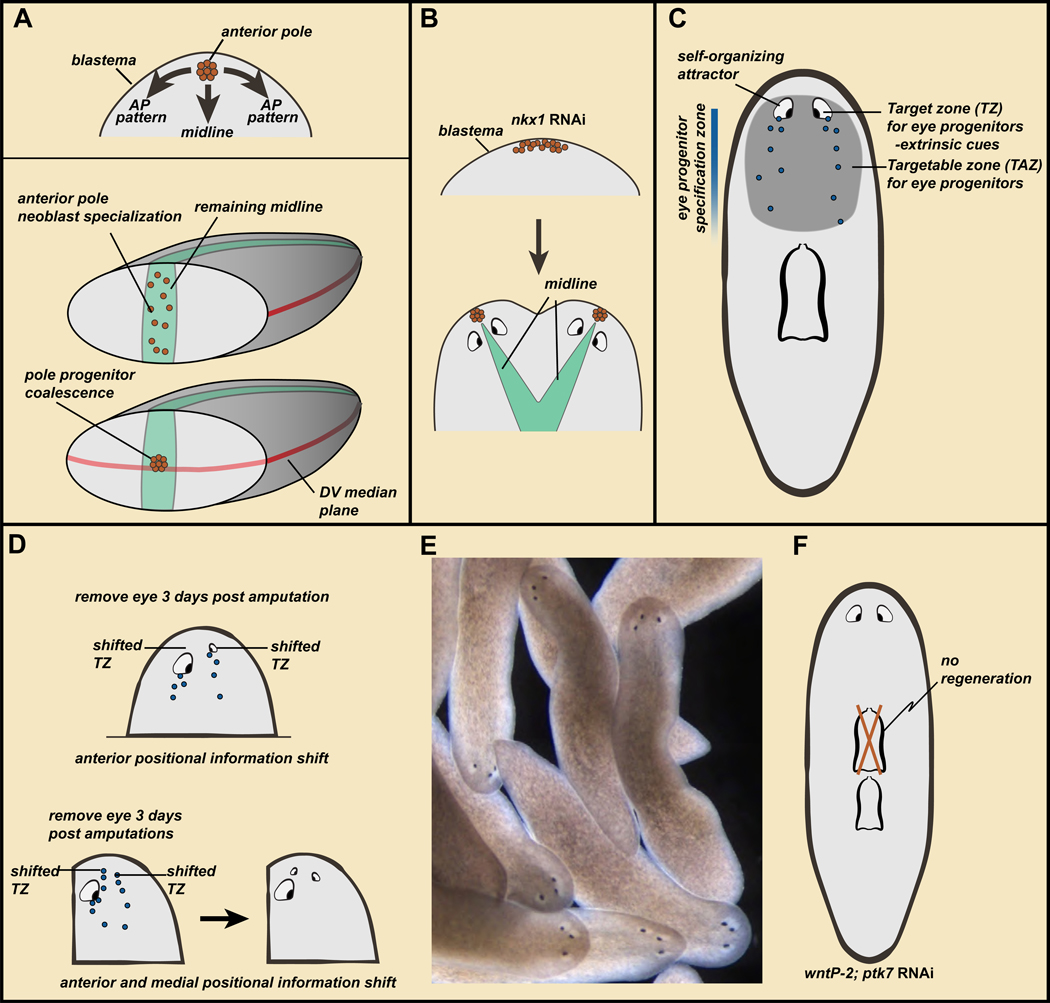
Figure 5. Wound architecture, migratory cues extrinsic to progenitors, and self-organization produce blastema pattern.
A. Top, the anterior pole promotes ML and AP head-blastema pattern. Bottom, wound architectural cues determine the point of anterior pole formation, connecting blastema pattern to the pattern of pre-existing tissue. B. nkx1 RNAi can result in wider initial anterior pole formation in the blastema, with subsequent pole splitting and two heads. C. Model: self-organization and extrinsic targeting cues govern the migratory behavior of progenitors in regeneration. See text for details. D. Amputations lead to PCG shifting, but the self-organizing nature of the eye prevents progenitors from reaching their target zone unless the eye is removed (top) or progenitors are targeted enough medially to miss the eye on their migration path (bottom). E. Wild-type animals with three eyes stably maintain ectopic eyes. Data from (Atabay et al., 2018). F. wntP-2; ptk7 RNAi animals develop a second pharynx; the original anterior pharynx is maintained but not regenerated.
Transplantation of the head tip containing the pole can trigger patterned head-like outgrowths in other parts of the body, consistent with a pattern-organizing role for cells of the head tip (Oderberg et al., 2017). nkx1 RNAi animals fail to maintain circular muscle fibers in tissue turnover, as described above. These animals fail to constrain pole formation normally during regeneration, with two poles occasionally forming next to one another (Figure 5B). In these cases, two heads instead of one can emerge in anterior-facing blastemas (Scimone et al., 2017). These observations regarding the pole suggests where it is placed during anterior regeneration is critical for the pattern that will emerge in that blastema.
Pole progenitors are specified from neoblasts, and three key architectural cues existing in pre-existing tissues at wounds help dictate anterior pole placement (Figure 5A) (Oderberg et al., 2017). First, anterior-pole progenitors only form at anterior-facing wounds involved in head regeneration. Second, pole-progenitor specification initiates at the preexisting slit+ midline at the wound, even if this is asymmetric in the fragment because of the nature of the injury. Because the new pole promotes midline formation in the blastema, this location of pole-progenitor specification links the midline in new tissue to the midline in pre-existing tissue. Finally, pole progenitors coalesce to the pre-existing DV median plane, which is a plane of growth and pattern organization orthogonal to the AP axis. These observations suggest that architectural features of wounds promote propagation of pattern from pre-existing tissue to the blastema through pole placement, facilitating integration of new tissues with prior ones (Figure 5A) (Oderberg et al., 2017).
Self-organization and extrinsic cues guide progenitor migratory targeting
Because progenitors are specified in a broad, spatially coarse region, migratory targeting of progenitors is a central aspect of planarian regeneration. For instance, as described above, eye progenitors are specified in the pre-pharyngeal region and migrate to precise, predictable locations in the head. The position where regenerative progenitors migrate in de novo organ formation has been defined as the progenitor “target zone (TZ)” (Figure 5C) (Atabay et al., 2018). RNAi of several PCGs can influences this; for instance wnt5 RNAi can cause ectopically lateral eye progenitor targeting (Atabay et al., 2018).
The targeting of progenitors is influenced by extrinsic cues, but is also influenced by their target organ itself (Atabay et al., 2018; Hill and Petersen, 2018). Following amputation, PCG patterns in a head fragment will start re-setting within 24–36 hours. This creates a period of discordance between the pattern of PCG expression domains and the location and size of underlying anatomy (Figure 5D). This discordance is resolved by the slow (weeks) rescaling and relative repositioning of remaining tissues, such as the eyes and brain in a head fragment. The existing anatomy of the head will be maintained during this time with progenitor targeting. But where should the progenitors be targeted to - the correct anatomical location (such as existing eyes) or the theoretically correct position with respect to the PCG expression map (Figure 5D) (Atabay et al., 2018)? How progenitors resolve this dilemma was determined by simple experiments in which a head was amputated, and following a three-day delay (allowing PCG resetting) one eye was removed. When this eye regenerated, it was anterior-shifted (Atabay et al., 2018; Hill and Petersen, 2018) (Figure 5D). Progenitors targeted two different locations on the two sides of these head fragments. The remaining eye on one side acted as a self-organizing attractor that trapped incoming progenitors from reaching their TZ.
Shifting the eye TZ medially and anteriorly by combining a parasagittal amputation (to the side of one eye) with decapitation allowed progenitors to bypass the self-organizing influence of a remaining eye resulting in formation of a third eye (Atabay et al., 2018) (Figure 5D). Further similar manipulations on three-eyed animals resulted in 4- and 5-eyed animals, and extra eyes were stably maintained and targeted by eye progenitors, producing stable alternative anatomies in wild-type animals (Atabay et al., 2018) (Figure 5E). Why are ectopic eyes maintained? It was proposed that ectopic eyes were maintained simply because of their self-organizing properties combined with having access to eye progenitors. Eyes are maintained in a specific region –approximating the eye progenitor specification zone – called the Targetable zone (TAZ) and defined as the region progenitors are capable of going to for maintaining an organ (Atabay et al., 2018) (Figure 5C). However, only eyes at the target zone are capable of being regenerated following resection (Atabay et al., 2018; Hill and Petersen, 2018). Spatially coarse progenitor specification is thus a central aspect of regeneration, allowing existing anatomy to have access to progenitors for their maintenance while positional coordinates are shifting. Similar experiments on PCG RNAi animals with a patterning phenotype resulting in two pharynges showed that only one pharynx regenerated (Figure 5F) (Hill and Petersen, 2018). Therefore, extra organs in incorrect locations, such as eyes and pharynges, can be maintained but not regenerated upon their removal. These observations indicate that self-organization and extrinsic targeting cues, combine with a broad progenitor specification zone (the TAZ), to determine the destination of migrating regenerative progenitors (Figure 5C).
IV. A model for planarian regeneration
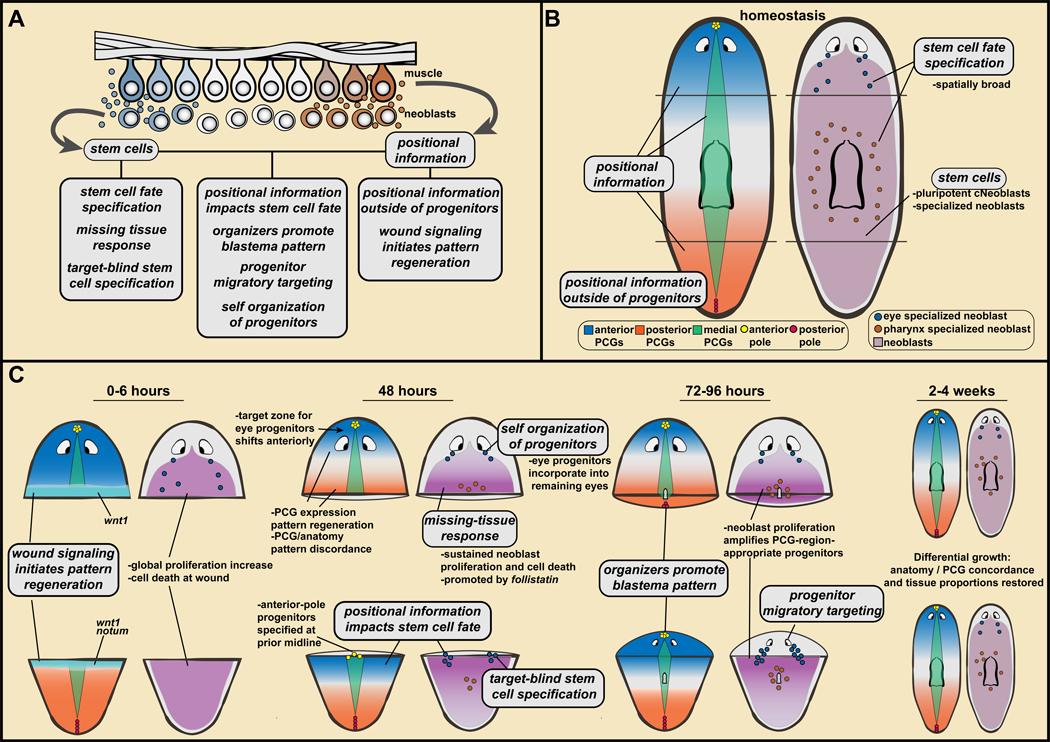
Regeneration is one of the great feats of biology and how it occurs has long been a mystery. Together the findings summarized above lead to a draft model for planarian regeneration. Aspects of this model are only partially understood or might in principle apply to only certain tissues. Thus, much more exploration is needed of the topics presented here and the many questions remaining unaddressed. The pillars of the proposed model are summarized below and in Figure 6:
Figure 6. A model for planarian regeneration involving positional information and stem cells.
A. Pillars of a model for planarian regeneration are described in text section IV. These pillars refer to stem cell and positional information properties, and how these properties together lead to key steps of regeneration. B, C. Application of pillars to explain head and tail regeneration (also, see text).
1. Stem cells.
The proliferative cell population of adult planarians called neoblasts has pluripotent stem cells called cNeoblasts responsible for generating all adult somatic cell types.
2. Stem cell fate specification.
Cell fate specification in regeneration and tissue turnover occurs in the neoblast population, with numerous specialized neoblasts - perhaps a class for every tissue of the adult - existing in uninjured animals. After amputation, specialized neoblasts can be synthesized de novo and appear in amplified numbers.
3. The missing-tissue response.
At wounds with substantial missing tissue there will be extensive neoblast proliferation and accumulation at the wound, as well as a broad increase in cell death and a longer phase of wound-induced gene expression.
4. Target-blind stem cell specification.
Small injuries removing a particular cell type or organ can result in regeneration without changing the rate of progenitor production for that tissue. This has been demonstrated at present for the eye. Regeneration from small regional injuries can be an emergent property resulting from a low homeostatic rate of progenitor production and not requiring surveillance of the presence or absence of target tissues.
5. Positional information.
Patterning molecules are expressed in constitutive and regional patterns in adult planarians, spanning the three body axes. These molecules, called position-control genes (PCGs), are associated with regional tissue identity during tissue turnover and in regeneration. Prominently, a Wnt activity gradient from the posterior regulates the AP axis, dorsally active Bmp regulates the DV axis, and Slit and Wnt5 regulate the ML axis.
6. Positional information outside of progenitors.
Muscle harbors planarian positional information. Instructions stored in a widespread differentiated cell type that is not the source of new cells enables regeneration by allowing positional information to be restored in remaining cells.
7. Wound signaling initiates pattern regeneration.
Wound signaling promotes the re-establishment of proper PCG patterns, with robust PCG-pattern regeneration occurring by 48 hours post-amputation. This process involves wound induction of wnt1 and notum in muscle, which are part of a generic wound response involving >200 genes. wnt1 promotes tail regeneration. notum, a Wnt inhibitor, is preferentially activated at anterior-facing wounds and promotes head regeneration.
8. Positional information impacts stem cell fate.
Simple coincidence of regenerated positional information at a wound and dividing neoblasts can lead to de novo progenitor production for a missing tissue, and in large numbers. It is not the absence of the tissue itself that results in de novo progenitor production (shown most compellingly at present for the eye).
9. Organizers promote blastema pattern.
Pre-existing architectural attributes at anterior-facing wounds initiate blastema pattern. First, tissue is polarized such that wound signaling preferentially activates notum at anterior-facing wounds. Second, anterior-pole-progenitor specification from neoblasts initiates at the pre-existing midline. Third, pole progenitors accumulate at the DV-median plane. The anterior pole promotes blastema pattern in a manner that integrates it with pre-existing tissue pattern.
10. Progenitor migratory targeting.
Specialized neoblasts exist in coarse spatial patterns and replace differentiated cells during natural tissue turnover. Small injuries therefore often do not remove progenitors. Progenitor targeting can occur in the absence of its target tissue, for de novo organ formation, indicating cues from outside the differentiated target tissue influence targeting.
11. Self-organization of progenitors.
If a progenitor encounters its differentiated target on its migratory path it can be incorporated into that structure in a self-organizing process. This enables progenitor targeting to remaining or partially injured body anatomy during regeneration, even though rapid positional information re-setting produces discordance of positional information and anatomy pattern for a substantial period of time during regeneration.
These pillars together can explain many key features of regeneration. Consider application of these concepts to an amputated body fragment, such as the tail (Figure 6): After injury, wound-induced notum in muscle initiates PCG domain reestablishment in muscle, with anterior becoming Wnt-low and posterior being Wnt-high. Previously tail neoblasts near the wound are consequently in an anterior PCG environment, promoting specialization into progenitors for head cell types. Neoblasts proliferate and accumulate at the wound, as part of the missing-tissue response, making a large number of neoblasts accessible for specialization into progenitors for the missing head cell types – without requiring detection of the absence of particular target tissues. Specialized neoblast patterns also change in pre-existing tissue regions because of PCG domain re-scaling in muscle. Therefore, overabundant pre-existing tissue will slowly and steadily shrink (morphallaxis). This process is accelerated by the increased cell death that occurs as part of the missing-tissue response. During this process, progenitors will continue to target their differentiated anatomy, even if it is in the wrong location with respect to positional information because of the self-organizing properties of differentiated structures. Some specialized neoblasts produced at the wound, in particular where the pre-existing midline coincides with an anterior-facing wound plane, are progenitors for the anterior pole. The anterior pole subsequently helps consolidate PCG changes and mediates continued PCG pattern re-establishment in the blastema. Consequently, the fragment makes a new head in a blastema with midline aligned with the prior midline. Cell turnover continues, but with positional information scaled to the new body proportions impacting the location and amount of regional neoblast specialization. This steadily results in a small animal with a complete complement of cell types arranged in their correct spatial relationships and proportions (Figure 6).
Animal growth and shrinking can be seen as simple variations on these themes. After feeding, proliferation is triggered, resulting in more neoblasts in any given region accessible to become specialized into progenitors for target tissues – those tissues will therefore grow. As tissue growth makes the animal bigger, PCG expression domains in muscle would scale to accommodate the new larger body dimensions. The increased neoblasts from feeding are consequently in larger PCG domains and will make more specialized neoblasts for each region, which can maintain the larger body proportionally. As neoblast proliferation rate declines, growth or shrinking would be determined by whether neoblasts are specializing into progenitors at a rate that is above or below that needed to maintain equilibrium for target tissues.
The concepts described above are all important subjects for further inquiry, testing, and elaboration. Further mysteries not yet explored and other significant findings beyond the scope of topics highlighted here are abundant and important for explaining planarian regeneration. For instance, neoblast biology is primed for studying molecular mechanisms of pluripotency, renewal, longevity, and fate determination in stem cells. Planarian wound signaling and the functions of wound-induced genes are also important directions. Patterning problems, such as scaling, the formation of morphogen gradients, and the molecular responses to patterning molecule gradients are also significant. The regulation of organ pattern and scale has just begun to be studied (Hill and Petersen, 2015). Other molecular processes and signaling pathways have vital but understudied roles in regeneration. For instance, Hedgehog signaling impacts the AP axis polarity in regeneration (Rink et al., 2009; Yazawa et al., 2009) and has roles in glia (Wang et al., 2016) and neuron regulation (Currie et al., 2016). Yorkie signaling has important roles in stem cell proliferation control, AP-axis patterning, and the wound response (Lin and Pearson, 2014; Hwang et al., 2015; Lin and Pearson, 2017; de Sousa et al., 2018). Erk signaling has been implicated in anterior regeneration (Umesono et al., 2013). Perturbation of innexin genes or modulation of Ca2+ influx can also impact AP regeneration polarity (Nogi et al., 2009; Oviedo et al., 2010). Studying how regeneration ability varies in planarian evolution is an intriguing direction; β-catenin RNAi ‘awakened’ the ability to regenerate heads in planarian species that normally do not regenerate heads from posterior amputation planes (Liu et al., 2013; Sikes and Newmark, 2013; Umesono et al., 2013). Stem-cell-migratory responses to wounds and organization of cells in regeneration are additional important directions with insights emerging (Abnave et al., 2017; Bonar and Petersen, 2017; Seebeck et al., 2017). Comparison of planarian embryogenesis to regeneration is also an attractive goal (Martín-Durán et al.; Davies et al., 2017). Finally, planarians are powerful experimental organisms for studying a myriad of problems outside of regeneration, such as cell type and organ function and evolution (Roberts-Galbraith and Newmark, 2015).
Uncovering the core modules that underlie regeneration in a diversity of organisms will be critical to understanding the biology that can enable regeneration. This knowledge will facilitate identifying what is different between what happens at wounds in highly regenerative organisms and in those less adept. Regenerative medicine strategies might be informed by understanding the cellular and molecular principles that naturally facilitate regeneration in planarians and other highly regenerative organisms. The findings from planarians identify many central modules that enable their regeneration – including a system of adult positional information, capacity for positional information regeneration involving wound signaling, broad specification and migratory targeting of regenerative stem cells and their progeny, capacity to regenerate without surveillance of target tissue presence, and the self-organization of progenitors into regenerating organs. An important direction is to determine whether these and other key concepts regarding planarian regeneration mechanisms can apply to aspects of regeneration in other organisms. For instance, recent work in Acoels indicates that PCGs are constitutively expressed in adult muscle, despite >550 million years of independent evolution from planarians, raising the possibility that muscle controlled regenerative patterning in the last common ancestor of the Bilateria (Raz et al., 2017). Muscle cells in Drosophila also have regulatory roles for intestinal stem cells (Lin et al., 2008). The rich concepts poised for mechanistic investigation and remaining mysteries make this an exciting era for research into regeneration using planarians as a model. The investigator Brondsted once suggested that “Planarian regeneration displays such an array of astounding phenomena that they could very well force one to believe that planarian regeneration might be performed by witchcraft” (Brøndsted, 1969). The findings on stem cells and positional information described in this article provide a framework for explaining how planarians might actually accomplish their astounding regenerative feats.
Acknowledgements
Thank you to Deniz Atabay, Lauren Cote, Nicholas Polizzi, Amelie Raz, Carice Reddien, Aneesha Tewari, and all other members of the Reddien lab for comments and discussion, Sam LoCascio for suggesting the exhibit analogy, and David Bartel for manuscript comments. P.W.R. is an HHMI Investigator and an associate member of the Broad Institute of Harvard and MIT.
References
- Abnave P, Aboukhatwa E, Kosaka N, Thompson J, Hill MA, and Aboobaker AA (2017). Epithelial-mesenchymal transition transcription factors control pluripotent adult stem cell migration in vivo in planarians. Development 144, 3440–3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adell T, Saló E, Boutros M, and Bartscherer K. (2009). Smed-Evi/Wntless is required for beta-catenin-dependent and -independent processes during planarian regeneration. Development 136, 905–910. [DOI] [PubMed] [Google Scholar]
- Adler CE, and Sánchez Alvarado A. (2015). Types or States? Cellular Dynamics and Regenerative Potential. Trends Cell Biol 25, 687–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler CE, Seidel CW, McKinney SA, and Sánchez Alvarado A. (2014). Selective amputation of the pharynx identifies a FoxA-dependent regeneration program in planaria. Elife 3, e02238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agata K, Tanaka T, Kobayashi C, Kato K, and Saitoh Y. (2003). Intercalary regeneration in planarians. Dev Dyn 226, 308–316. [DOI] [PubMed] [Google Scholar]
- Almuedo-Castillo M, Saló E, and Adell T. (2011). Dishevelled is essential for neural connectivity and planar cell polarity in planarians. Proceedings of the National Academy of Sciences of the United States of America 108, 2813–2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An Y, Kawaguchi A, Zhao C, Toyoda A, Sharifi-Zarchi A, Mousavi SA, Bagherzadeh R, Inoue T, Ogino H, Fujiyama A, et al. (2018). Draft genome of Dugesia japonica provides insights into conserved regulatory elements of the brain restriction gene nou-darake in planarians. Zoological letters 4, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atabay KD, LoCascio SA, de Hoog T, and Reddien PW (2018). Self-organization and progenitor targeting generate stable patterns in planarian regeneration. Science 360, 404–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baguna J. (2012). The planarian neoblast: the rambling history of its origin and some current black boxes. The International journal of developmental biology 56, 19–37. [DOI] [PubMed] [Google Scholar]
- Baguñà J. (1976). Mitosis in the intact and regenerating planarian Dugesia mediterranea n.sp. I. Mitotic studies during growth, feeding and starvation. J Exp Zool 195, 53–64. [Google Scholar]
- Baguñà J, Saló E, and Auladell C. (1989). Regeneration and pattern formation in planarians. III. Evidence that neoblasts are totipotent stem cells and the source of blastema cells. Development 107, 77–86. [Google Scholar]
- Bardeen CR, and Baetjer FH (1904). The inhibitive action of the Roentgen rays on regeneration in planarians. J Exp Zool 1, 191–195. [Google Scholar]
- Blassberg RA, Felix DA, Tejada-Romero B, and Aboobaker AA (2013). PBX/extradenticle is required to re-establish axial structures and polarity during planarian regeneration. Development 140, 730–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonar NA, and Petersen CP (2017). Integrin suppresses neurogenesis and regulates brain tissue assembly in planarian regeneration. Development 144, 784–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brøndsted HV (1969). Planarian Regeneration, 1st edn (London: Pergamon Press; ). [Google Scholar]
- Cebria F. (2016). Planarian Body-Wall Muscle: Regeneration and Function beyond a Simple Skeletal Support. Frontiers in cell and developmental biology 4, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebria F, Guo T, Jopek J, and Newmark PA (2007). Regeneration and maintenance of the planarian midline is regulated by a slit orthologue. Dev Biol 307, 394–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebria F, Kobayashi C, Umesono Y, Nakazawa M, Mineta K, Ikeo K, Gojobori T, Itoh M, Taira M, Sánchez Alvarado A, et al. (2002). FGFR-related gene nou-darake restricts brain tissues to the head region of planarians. Nature 419, 620–624. [DOI] [PubMed] [Google Scholar]
- Chen CC, Wang IE, and Reddien PW (2013). pbx is required for pole and eye regeneration in planarians. Development 140, 719–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng LC, Tu KC, Seidel CW, Robb SMC, Guo F, and Sánchez Alvarado A. (2018). Cellular, ultrastructural and molecular analyses of epidermal cell development in the planarian Schmidtea mediterranea. Dev Biol 433, 357–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowles MW, Brown DD, Nisperos SV, Stanley BN, Pearson BJ, and Zayas RM (2013). Genome-wide analysis of the bHLH gene family in planarians identifies factors required for adult neurogenesis and neuronal regeneration. Development 140, 4691–4702. [DOI] [PubMed] [Google Scholar]
- Cowles MW, Omuro KC, Stanley BN, Quintanilla CG, and Zayas RM (2014). COE loss-of-function analysis reveals a genetic program underlying maintenance and regeneration of the nervous system in planarians. PLoS Genet 10, e1004746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie KW, Molinaro AM, and Pearson BJ (2016). Neuronal sources of hedgehog modulate neurogenesis in the adult planarian brain. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie KW, and Pearson BJ (2013). Transcription factors lhx1/5–1 and pitx are required for the maintenance and regeneration of serotonergic neurons in planarians. Development 140, 3577–3588. [DOI] [PubMed] [Google Scholar]
- Dalyell JG (1814). Observations on Some Interesting Phenomena in Animal Physiology, Exhibited by Several Species of Planariae. Illustrated by Coloured Figures of Living Animals (Edinburgh). [Google Scholar]
- Davies EL, Lei K, Seidel CW, Kroesen AE, McKinney SA, Guo L, Robb SM, Ross EJ, Gotting K, and Alvarado AS (2017). Embryonic origin of adult stem cells required for tissue homeostasis and regeneration. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Sousa N, Rodriguez-Esteban G, Rojo-Laguna JI, Salo E, and Adell T. (2018). Hippo signaling controls cell cycle and restricts cell plasticity in planarians. PLoS Biol 16, e2002399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois F. (1949). Contribution á l ‘ètude de la migration des cellules de règènèration chez les Planaires dulcicoles. Bull Biol Fr Belg 83, 213–283. [Google Scholar]
- Eisenhoffer GT, Kang H, and Sánchez Alvarado A. (2008). Molecular analysis of stem cells and their descendants during cell turnover and regeneration in the planarian Schmidtea mediterranea. Cell Stem Cell 3, 327–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faraday M. (1833). On the Planariae. The Edinburgh new philosophical journal XIV, 183–185. [Google Scholar]
- Felix DA, and Aboobaker AA (2010). The TALE class homeobox gene Smed-prep defines the anterior compartment for head regeneration. PLoS genetics 6, e1000915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fincher CT, Wurtzel O, de Hoog T, Kravarik KM, and Reddien PW (2018). Cell type transcriptome atlas for the planarian Schmidtea mediterranea. Science 360, 874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores NM, Oviedo NJ, and Sage J. (2016). Essential role for the planarian intestinal GATA transcription factor in stem cells and regeneration. Dev Biol 418, 179–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsthoefel DJ, Park AE, and Newmark PA (2011). Stem cell-based growth, regeneration, and remodeling of the planarian intestine. Dev Biol 356, 445–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavino MA, and Reddien PW (2011). A Bmp/Admp regulatory circuit controls maintenance and regeneration of dorsal-ventral polarity in planarians. Curr Biol 21, 294–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavino MA, Wenemoser D, Wang IE, and Reddien PW (2013). Tissue absence initiates regeneration through Follistatin-mediated inhibition of Activin signaling. Elife 2, e00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Sastre A, De Sousa N, Adell T, and Saló E. (2017). The pioneer factor Smed-gata456–1 is required for gut cell differentiation and maintenance in planarians. Int J Dev Biol 61, 53–63. [DOI] [PubMed] [Google Scholar]
- Grohme MA, Schloissnig S, Rozanski A, Pippel M, Young GR, Winkler S, Brandl H, Henry I, Dahl A, Powell S, et al. (2018). The genome of Schmidtea mediterranea and the evolution of core cellular mechanisms. Nature 554, 56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurley KA, Elliott SA, Simakov O, Schmidt HA, Holstein TW, and Sánchez Alvarado A. (2010). Expression of secreted Wnt pathway components reveals unexpected complexity of the planarian amputation response. Dev Biol 347, 24–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurley KA, Rink JC, and Sánchez Alvarado A. (2008). Beta-catenin defines head versus tail identity during planarian regeneration and homeostasis. Science 319, 323–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Asami M, Higuchi S, Shibata N, and Agata K. (2006). Isolation of planarian X-ray-sensitive stem cells by fluorescence-activated cell sorting. Dev Growth Differ 48, 371–380. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Motoishi M, Yazawa S, Itomi K, Tanegashima C, Nishimura O, Agata K, and Tarui H. (2011). A LIM-homeobox gene is required for differentiation of Wnt-expressing cells at the posterior end of the planarian body. Development 138, 3679–3688. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Shibata N, Okumura R, Kudome T, Nishimura O, Tarui H, and Agata K. (2010). Single-cell gene profiling of planarian stem cells using fluorescent activated cell sorting and its “index sorting” function for stem cell research. Dev Growth Differ 52, 131–144. [DOI] [PubMed] [Google Scholar]
- He X, Lindsay-Mosher N, Li Y, Molinaro AM, Pellettieri J, and Pearson BJ (2017). FOX and ETS family transcription factors regulate the pigment cell lineage in planarians. Development 144, 4540–4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill EM, and Petersen CP (2015). Wnt/Notum spatial feedback inhibition controls neoblast differentiation to regulate reversible growth of the planarian brain. Development 142, 4217–4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill EM, and Petersen CP (2018). Positional information specifies the site of organ regeneration and not tissue maintenance in planarians. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang B, An Y, Agata K, and Umesono Y. (2015). Two distinct roles of the yorkie/yap gene during homeostasis in the planarian Dugesia japonica. Dev Growth Differ 57, 209–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman LH (1951). The Invertebrates: Platyhelminthes and Rhynchocoela The acoelomate bilateria, Vol II (New York: McGraw-Hill Book Company Inc.). [Google Scholar]
- Iglesias M, Gomez-Skarmeta JL, Salo E, and Adell T. (2008). Silencing of Smed-betacatenin1 generates radial-like hypercephalized planarians. Development 135, 1215–1221. [DOI] [PubMed] [Google Scholar]
- Johnson JR (1825). Further observations on Planariae. Phil Trans Royal Soc London Part II, 247–256. [Google Scholar]
- Kato K, Orii H, Watanabe K, and Agata K. (2001). Dorsal and ventral positional cues required for the onset of planarian regeneration may reside in differentiated cells. Dev Biol 233, 109–121. [DOI] [PubMed] [Google Scholar]
- Keller J. (1894). Die ungeschlechtliche Fortpflanzungder Süsswasser-Turbellarien. Jen Zeit Naturw 28, 370–407. [Google Scholar]
- Kobayashi C, Nogi T, Watanbe K, and Agata K. (1999). Ectopic Pharynxes arise by regional reorganization after anterior/posterior chimera in planarians. Mech of Dev 89, 25–34. [DOI] [PubMed] [Google Scholar]
- Kobayashi C, Saito Y, Ogawa K, and Agata K. (2007). Wnt signaling is required for anteroposterior patterning of the planarian brain. Dev Biol 306, 714–724. [DOI] [PubMed] [Google Scholar]
- Lander R, and Petersen CP (2016). Wnt, Ptk7, and FGFRL expression gradients control trunk positional identity in planarian regeneration. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapan SW, and Reddien PW (2011). dlx and sp6–9 control optic cup regeneration in a prototypic eye. PLoS Genet 7, e1002226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapan SW, and Reddien PW (2012). Transcriptome Analysis of the Planarian Eye Identifies ovo as a Specific Regulator of Eye Regeneration. Cell Reports 2, 294–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin AY, and Pearson BJ (2014). Planarian yorkie/YAP functions to integrate adult stem cell proliferation, organ homeostasis and maintenance of axial patterning. Development 141, 1197–1208. [DOI] [PubMed] [Google Scholar]
- Lin AYT, and Pearson BJ (2017). Yorkie is required to restrict the injury responses in planarians. PLoS Genet 13, e1006874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin G, Xu N, and Xi R. (2008). Paracrine Wingless signalling controls self-renewal of Drosophila intestinal stem cells. Nature 455, 1119–1123. [DOI] [PubMed] [Google Scholar]
- Liu SY, Selck C, Friedrich B, Lutz R, Vila-Farre M, Dahl A, Brandl H, Lakshmanaperumal N, Henry I, and Rink JC (2013). Reactivating head regrowth in a regeneration-deficient planarian species. Nature 500, 81–84. [DOI] [PubMed] [Google Scholar]
- LoCascio SA, Lapan SW, and Reddien PW (2017). Eye Absence Does Not Regulate Planarian Stem Cells during Eye Regeneration. Dev Cell 40, 381–391 e383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Durán JM, Amaya E, and Romero R. (2010). Germ layer specification and axial patterning in the embryonic development of the freshwater planarian Schmidtea polychroa. Dev Biol 340, 145–158. [DOI] [PubMed] [Google Scholar]
- Marz M, Seebeck F, and Bartscherer K. (2013). A Pitx transcription factor controls the establishment and maintenance of the serotonergic lineage in planarians. Development 140, 4499–4509. [DOI] [PubMed] [Google Scholar]
- Meinhardt H. (1982). Models of Biological Pattern Formation (Academic, London: ). [Google Scholar]
- Molina MD, Neto A, Maeso I, Gomez-Skarmeta JL, Saló E, and Cebrià F. (2011). Noggin and Noggin-like genes control dorsoventral axis regeneration in planarians. Curr Biol 21, 300–305. [DOI] [PubMed] [Google Scholar]
- Molina MD, Saló E, and Cebria F. (2007). The BMP pathway is essential for re-specification and maintenance of the dorsoventral axis in regenerating and intact planarians. Dev Biol 311, 79–94. [DOI] [PubMed] [Google Scholar]
- Molina MD, Saló E, and Cebrià F. (2009). Expression pattern of the expanded noggin gene family in the planarian Schmidtea mediterranea. Gene Expr Patterns 9, 246–253. [DOI] [PubMed] [Google Scholar]
- Morgan TH (1898). Experimental studies of the regeneration of Planaria maculata. Arch Entw Mech Org 7, 364–397. [Google Scholar]
- Newmark P, and Sánchez Alvarado A. (2000). Bromodeoxyuridine specifically labels the regenerative stem cells of planarians. Dev Biol 220, 142–153. [DOI] [PubMed] [Google Scholar]
- Nogi T, Zhang D, Chan JD, and Marchant JS (2009). A novel biological activity of praziquantel requiring voltage-operated Ca2+ channel beta subunits: subversion of flatworm regenerative polarity. PLoS Negl Trop Dis 3, e464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oderberg IM, Li DJ, Scimone ML, Gavino MA, and Reddien PW (2017). Landmarks in Existing Tissue at Wounds Are Utilized to Generate Pattern in Regenerating Tissue. Curr Biol 27, 733–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada YK, and Sugino H. (1937). Transplantation experiments in planaria gonocephala Duges. Zoological Institute, Kyoto Imperial University 7, 373–439. [Google Scholar]
- Orii H, Kato K, K. A, and Watanabe K. (1998). Molecular cloning of bone morphogenetic protein (BMP) gene from the planarian Dugesia japonica. Zool Science 15, 871–877. [Google Scholar]
- Orii H, and Watanabe K. (2007). Bone morphogenetic protein is required for dorso-ventral patterning in the planarian Dugesia japonica. Dev Growth Differ 49, 345–349. [DOI] [PubMed] [Google Scholar]
- Oviedo NJ, Morokuma J, Walentek P, Kema IP, Gu MB, Ahn JM, Hwang JS, Gojobori T, and Levin M. (2010). Long-range neural and gap junction protein-mediated cues control polarity during planarian regeneration. Dev Biol 339, 188–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oviedo NJ, Newmark PA, and Sánchez Alvarado A. (2003). Allometric scaling and proportion regulation in the freshwater planarian Schmidtea mediterranea. Dev Dyn 226, 326–333. [DOI] [PubMed] [Google Scholar]
- Owen JH, Wagner DE, Chen CC, Petersen CP, and Reddien PW (2015). teashirt is required for head-versus-tail regeneration polarity in planarians. Development 142, 1062–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owlarn S, Klenner F, Schmidt D, Rabert F, Tomasso A, Reuter H, Mulaw MA, Moritz S, Gentile L, Weidinger G, et al. (2017). Generic wound signals initiate regeneration in missing-tissue contexts. Nature communications 8, 2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallas PS (1766). Miscellanea zoologica, quibus novae imprimis atque obscurae animalium species describuntur et observationibus iconibusque illustrantur... (Hagae Comitum, apud Pterum van Cleef). [Google Scholar]
- Pellettieri J, Fitzgerald P, Watanabe S, Mancuso J, Green DR, and Sánchez Alvarado A. (2010). Cell death and tissue remodeling in planarian regeneration. Dev Biol 338, 76–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen CP, and Reddien PW (2008). Smed-betacatenin-1 is required for anteroposterior blastema polarity in planarian regeneration. Science 319, 327–330. [DOI] [PubMed] [Google Scholar]
- Petersen CP, and Reddien PW (2009a). Wnt signaling and the polarity of the primary body axis. Cell 139, 1056–1068. [DOI] [PubMed] [Google Scholar]
- Petersen CP, and Reddien PW (2009b). A wound-induced Wnt expression program controls planarian regeneration polarity. Proc Natl Acad Sci U S A 106, 17061–17066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen CP, and Reddien PW (2011). Polarized notum activation at wounds inhibits Wnt function to promote planarian head regeneration. Science 332, 852–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plass M, Solana J, Wolf FA, Ayoub S, Misios A, Glazar P, Obermayer B, Theis FJ, Kocks C, and Rajewsky N. (2018). Cell type atlas and lineage tree of a whole complex animal by single-cell transcriptomics. Science 360, 875. [DOI] [PubMed] [Google Scholar]
- Raz AA, Srivastava M, Salvamoser R, and Reddien PW (2017). Acoel regeneration mechanisms indicate an ancient role for muscle in regenerative patterning. Nature communications 8, 1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien PW (2011). Constitutive gene expression and the specification of tissue identity in adult planarian biology. Trends Genet 27, 277–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien PW (2013). Specialized progenitors and regeneration. Development 140, 951–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien PW, Bermange AL, Kicza AM, and Sánchez Alvarado A. (2007). BMP signaling regulates the dorsal planarian midline and is needed for asymmetric regeneration. Development 134, 4043–4051. [DOI] [PubMed] [Google Scholar]
- Reddien PW, Bermange AL, Murfitt KJ, Jennings JR, and Sánchez Alvarado A. (2005a). Identification of genes needed for regeneration, stem cell function, and tissue homeostasis by systematic gene perturbation in planaria. Dev Cell 8, 635–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien PW, Oviedo NJ, Jennings JR, Jenkin JC, and Sánchez Alvarado A. (2005b). SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science 310, 1327–1330. [DOI] [PubMed] [Google Scholar]
- Reddien PW, and Sánchez Alvarado A. (2004). Fundamentals of planarian regeneration. Ann Rev Cell Dev Bio 20, 725–757. [DOI] [PubMed] [Google Scholar]
- Reuter H, Marz M, Vogg MC, Eccles D, Grifol-Boldu L, Wehner D, Owlarn S, Adell T, Weidinger G, and Bartscherer K. (2015). Beta-catenin-dependent control of positional information along the AP body axis in planarians involves a teashirt family member. Cell Rep 10, 253–265. [DOI] [PubMed] [Google Scholar]
- Rink JC, Gurley KA, Elliott SA, and Sánchez Alvarado A. (2009). Planarian Hh signaling regulates regeneration polarity and links Hh pathway evolution to cilia. Science 326, 1406–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rink JC, Vu HT, and Sánchez Alvarado A. (2011). The maintenance and regeneration of the planarian excretory system are regulated by EGFR signaling. Development 138, 3769–3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts-Galbraith RH, and Newmark PA (2013). Follistatin antagonizes activin signaling and acts with notum to direct planarian head regeneration. Proc Natl Acad Sci U S A 110, 1363–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts-Galbraith RH, and Newmark PA (2015). On the organ trail: insights into organ regeneration in the planarian. Curr Opin Genet Dev 32, 37–46. [DOI] [PubMed] [Google Scholar]
- Saló E, and Baguñà J. (1984). Regeneration and pattern formation in planarians. I. The pattern of mitosis in anterior and posterior regeneration in Dugesia (G) tigrina, and a new proposal for blastema formation. J Embryol Exp Morphol 83, 63–80. [PubMed] [Google Scholar]
- Saló E, and Baguñà J. (1985). Proximal and distal transformation during intercalary regeneration in the planarian Dugesia (S) mediterranea. Roux’s Arch Dev Biol 194, 364–368. [Google Scholar]
- Sánchez Alvarado A, and Newmark PA (1999). Double-stranded RNA specifically disrupts gene expression during planarian regeneration. Proc Natl Acad Sci 96, 5049–5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos FV (1931). Studies on transplantation in planaria. Phys Zool 4, 111–164. [Google Scholar]
- Scimone ML, Cote LE, and Reddien PW (2017). Orthogonal muscle fibres have different instructive roles in planarian regeneration. Nature 551, 623–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scimone ML, Cote LE, Rogers T, and Reddien PW (2016). Two FGFRL-Wnt circuits organize the planarian anteroposterior axis. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scimone ML, Kravarik KM, Lapan SW, and Reddien PW (2014a). Neoblast Specialization in Regeneration of the Planarian Schmidtea mediterranea. Stem Cell Reports 3, 339–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scimone ML, Lapan SW, and Reddien PW (2014b). A forkhead transcription factor is wound-induced at the planarian midline and required for anterior pole regeneration. PLoS genetics 10, e1003999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scimone ML, Srivastava M, Bell GW, and Reddien PW (2011). A regulatory program for excretory system regeneration in planarians. Development 138, 4387–4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seebeck F, Marz M, Meyer AW, Reuter H, Vogg MC, Stehling M, Mildner K, Zeuschner D, Rabert F, and Bartscherer K. (2017). Integrins are required for tissue organization and restriction of neurogenesis in regenerating planarians. Development 144, 795–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikes JM, and Newmark PA (2013). Restoration of anterior regeneration in a planarian with limited regenerative ability. Nature 500, 77–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stückemann T, Cleland JP, Werner S, Thi-Kim Vu H, Bayersdorf R, Liu SY, Friedrich B, Julicher F, and Rink JC (2017). Antagonistic Self-Organizing Patterning Systems Control Maintenance and Regeneration of the Anteroposterior Axis in Planarians. Dev Cell 40, 248–263 e244. [DOI] [PubMed] [Google Scholar]
- Sureda-Gómez M, Martín-Durán JM, and Adell T. (2016). Localization of planarian beta-CATENIN-1 reveals multiple roles during anterior-posterior regeneration and organogenesis. Development 143, 4149–4160. [DOI] [PubMed] [Google Scholar]
- Tu KC, Cheng LC, H TKV, Lange JJ, McKinney SA, Seidel CW, and Sánchez Alvarado A. (2015). Egr-5 is a post-mitotic regulator of planarian epidermal differentiation. Elife 4, e10501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umesono Y, Tasaki J, Nishimura Y, Hrouda M, Kawaguchi E, Yazawa S, Nishimura O, Hosoda K, Inoue T, and Agata K. (2013). The molecular logic for planarian regeneration along the anterior-posterior axis. Nature 500, 73–76. [DOI] [PubMed] [Google Scholar]
- Umesono Y, Watanabe K, and Agata K. (1997). A planarian orthopedia homolog is specifically expressed in the branch region of both the mature and regenerating brain. Develop Growth Differ 39, 723–727. [DOI] [PubMed] [Google Scholar]
- van Wolfswinkel JC, Wagner DE, and Reddien PW (2014). Single-Cell Analysis Reveals Functionally Distinct Classes within the Planarian Stem Cell Compartment. Cell Stem Cell 15, 326–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vásquez-Doorman C, and Petersen CP (2014). zic-1 Expression in planarian neoblasts after injury controls anterior pole regeneration. PLoS genetics 10, e1004452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogg MC, Owlarn S, Perez Rico YA, Xie J, Suzuki Y, Gentile L, Wu W, and Bartscherer K. (2014). Stem cell-dependent formation of a functional anterior regeneration pole in planarians requires Zic and Forkhead transcription factors. Developmental Biology 390, 136–148. [DOI] [PubMed] [Google Scholar]
- Vu HT, Rink JC, McKinney SA, McClain M, Lakshmanaperumal N, Alexander R, and Sánchez Alvarado A. (2015). Stem cells and fluid flow drive cyst formation in an invertebrate excretory organ. Elife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner DE, Ho JJ, and Reddien PW (2012). Genetic regulators of a pluripotent adult stem cell system in planarians identified by RNAi and clonal analysis. Cell Stem Cell 10, 299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner DE, Wang IE, and Reddien PW (2011). Clonogenic neoblasts are pluripotent adult stem cells that underlie planarian regeneration. Science 332, 811–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang IE, Lapan SW, Scimone ML, Clandinin TR, and Reddien PW (2016). Hedgehog signaling regulates gene expression in planarian glia. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenemoser D, Lapan SW, Wilkinson AW, Bell GW, and Reddien PW (2012). A molecular wound response program associated with regeneration initiation in planarians. Genes & Development 26, 988–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenemoser D, and Reddien PW (2010). Planarian regeneration involves distinct stem cell responses to wounds and tissue absence. Dev Biol 344, 979–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witchley JN, Mayer M, Wagner DE, Owen JH, and Reddien PW (2013). Muscle cells provide instructions for planarian regeneration. Cell Reports 4, 633–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzel O, Cote LE, Poirier A, Satija R, Regev A, and Reddien PW (2015). A Generic and Cell-Type-Specific Wound Response Precedes Regeneration in Planarians. Dev Cell 35, 632–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzel O, Oderberg IM, and Reddien PW (2017). Planarian Epidermal Stem Cells Respond to Positional Cues to Promote Cell-Type Diversity. Dev Cell 40, 491–504 e495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazawa S, Umesono Y, Hayashi T, Tarui H, and Agata K. (2009). Planarian Hedgehog/Patched establishes anterior-posterior polarity by regulating Wnt signaling. Proc Natl Acad Sci U S A 106, 22329–22334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng A, Li H, Guo L, Gao X, McKinney S, Wang Y, Yu Z, Park J, Semerad C, Ross E, et al. (2018). Prospectively Isolated Tetraspanin(+) Neoblasts Are Adult Pluripotent Stem Cells Underlying Planaria Regeneration. Cell 173, 1593–1608 e1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu SJ, Hallows SE, Currie KW, Xu C, and Pearson BJ (2015). A mex3 homolog is required for differentiation during planarian stem cell lineage development. Elife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]