Abstract
Epidemiology of bat Betacoronavirus, subgenus Sarbecovirus is largely unknown, especially outside China. We detected a sarbecovirus phylogenetically related to severe acute respiratory syndrome coronavirus 2 from Rhinolophus cornutus bats in Japan. The sarbecovirus’ spike protein specifically recognizes angiotensin-converting enzyme 2 of R. cornutus, but not humans, as an entry receptor.
Keywords: betacoronavirus, bat sarbecovirus, Rhinolophus cornutus, respiratory infections, severe acute respiratory syndrome coronavirus 2, SARS-CoV-2, SARS, COVID-19, coronavirus disease, zoonoses, viruses, coronavirus, Japan
During the past 20 years, coronaviruses belonging to the genus Betacoronavirus have caused multiple human epidemic or pandemic diseases, including severe acute respiratory syndrome (SARS), Middle East respiratory syndrome (MERS), and coronavirus disease (COVID-19). Two viruses of the subgenus Sarbecovirus are severe acute respiratory syndrome coronavirus (SARS-CoV), which causes SARS, and SARS-CoV-2, which causes COVID-19. Although Rhinolophus spp. bats in Asia, Europe, and Africa are considered natural reservoirs of sarbecoviruses (1–3), the epidemiology and distribution of these viruses remain largely unknown, especially outside China. Previously, partial RNA-dependent RNA polymerase (RdRp) genes of betacoronaviruses were detected in little Japanese horseshoe bats (Rhinolophus cornutus) (4). However, limited sequence information left the genetic and virological properties unclear. We detected and determined the entire genome sequence of a bat sarbecovirus belonging to a phylogenetic clade that includes SARS-CoV-2 from R. cornutus bats in Japan. Further, we used a pseudotyped virus system to characterize an entry step of this virus into cells.
The Study
R. cornutus is an endemic bat species in Japan and is found nationwide. These bats primarily inhabit caves and abandoned tunnels in the countryside during daytime and capture insects at night outside their roosts. R. cornutus bats often cohabit with other insectivorous bats, such as R. ferrumequinum or Myotis macrodactylus, and occasionally with wild animals, such as the masked palm civet (Paguma larvata), in their daytime roosts.
In 2013, we captured 4 R. cornutus bats in a cave in Iwate prefecture, Japan, and extracted RNA from fresh feces. Then, we used real-time reverse transcription PCR (rRT-PCR) to detect the partial RdRp gene of sarbecovirus from 2 samples by using a pair of primers designed to detect betacoronavirus. In 2020, we performed RNA sequencing and determined the full genome sequence of 1 sample, Rc-o319, which exhibited lower cycle threshold value by rRT-PCR.
We performed a BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) analysis of the full genome of Rc-o319, which showed Rc-o319 had the highest nucleotide homology to SARS-CoV-2 HKG/HKU-904a/2020 strain (GenBank accession no. MT365032) with a query cover of 96% and sequence identity of 81.47%. The maximum-likelihood analysis with sarbecoviruses demonstrated that the full genome and spike protein (S) gene of Rc-o319 were positioned within a specific clade that included SARS-CoV-2 (Figure 1, panels A, B). Amino acid sequences of open reading frame 1ab (ORF1ab) and S of Rc-o319 also were positioned within the SARS-CoV-2 clade (Figure 1 panels C and D). The phylogenetic trees maintained the same topology between ORF1ab and S, indicating that no recombination event occurred in Rc-0319, which was supported by similarity plot analysis (Appendix Figure 1). The nucleotide and amino acid sequences of Rc-o319 were more homologous to those of viruses belonging to SARS-CoV-2 clade than the SARS-CoV clade (Table). These data suggest that Rc-o319 genetically is related to SARS-CoV-2.
Figure 1.
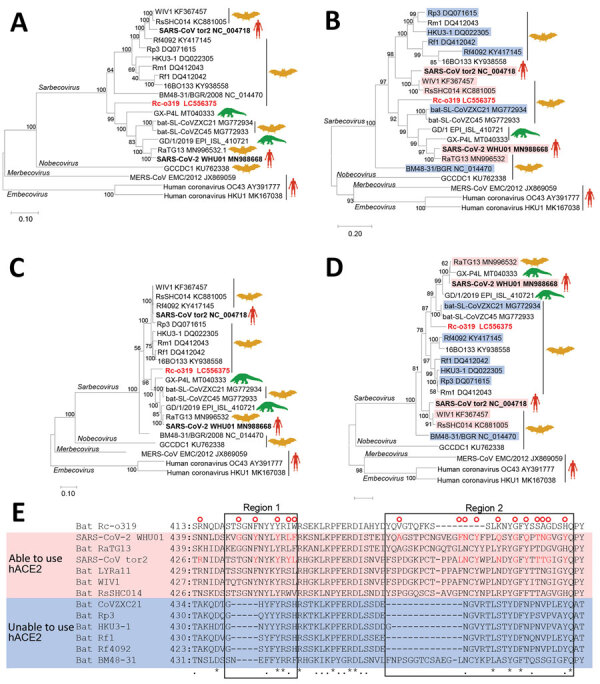
Phylogenetic analysis of sarbecovirus sequenced from little Japanese horseshoe bats (Rhinolophus cornutus) and genetically related to human SARS-CoV-2, Japan. A–D) Phylogenetic trees were generated by using maximum-likelihood analysis combined with 500 bootstrap replicates and show relationships between bat-, human-, and pangolin-derived sarbecoviruses. Phylogenetic trees are shown for nucleotide sequences of the full genome (A), the S protein gene and amino acid sequences (B), the ORF1ab (C), and the S protein (D). Red text indicates positions of Rc-o319, the sarbecovirus sequenced in this study. For panels B and D, magenta bands indicate viruses with S proteins that bind to human ACE2; blue bands indicate viruses with S proteins that do not bind to human ACE2. Bootstrap values are shown above and to the left of the major nodes. Scale bars indicate nucleotide or amino acid substitutions per site. E) Amino acid sequence alignment of the RBM of S proteins that are able or unable to bind to human ACE2. Amino acid residues of the RBM that contact human ACE2 of SARS-CoV-2 and SARS-CoV are indicated in the upper side by red circles. The 2 regions of S protein RBM known to interact with human ACE2 are indicated by boxes labeled region 1 and region 2. ACE2, angiotensin-converting enzyme 2; hACE2, human angiotensin-converting enzyme 2; ORF1ab, open reading frame 1ab; RBM, receptor-binding motif; S, spike protein; SARS-CoV, severe acute respiratory syndrome coronavirus; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Table. Identities of nucleotide and amino acid sequences of genome, genes, and proteins to representative sarbecoviruses used to investigate sarbecovirus Rc-o319 detected in bats, Japan.
| Virus |
Entire genome |
ORF1ab |
S |
ORF3a |
E |
M |
ORF6 |
ORF7a |
ORF7b |
ORF8 |
N |
ORF10 |
|
|
Nucleotide % |
|||||||||||
| SARS-CoV-2 | 81.5 | 80.0 | 73.0 | 83.2 | 97.4 | 86.6 | 86.6 | 78.4 | 77.3 | 53.3 | 88.3 | 94.9 |
| RaTG13 | 81.2 | 79.8 | 73.3 | 83.9 | 96.9 | 85.4 | 87.1 | 77.4 | 78.0 | 53.0 | 87.8 | 94.0 |
| pangolin/P4L | 80.4 | 79.8 | 72.5 | 83.5 | 97.8 | 85.5 | 85.5 | 74.5 | – | 53.2 | 86.8 | 91.5 |
| CoVZXC21 | 80.4 | 80.2 | 72.5 | 79.9 | 96.1 | 86.0 | 83.3 | 75.1 | – | 50.8 | 85.9 | 53.9 |
| SARS-CoV-1 | 81.0 | 78.8 | 73.6 | 76.6 | 93.1 | 87.0 | 77.1 | 73.4 | 74.8 | – | 86.1 | 63.3 |
| Rf1 | 80.6 | 78.7 | 70.9 | 75.1 | 92.6 | 85.8 | 78.8 | 72.2 | 75.6 | 68.6 | 84.9 | 63.3 |
| BM48–31 |
79.6 |
77.4 |
69.3 |
70.4 |
90.5 |
80.6 |
61.3 |
65.1 |
57.9 |
– |
77.0 |
64.1 |
|
|
Amino acid, % |
|||||||||||
| SARS-CoV-2 | NA | 88.2 | 76.7 | 87.0 | 98.7 | 91.0 | 83.6 | 73.8 | 69.8 | 27.5 | 89.5 | 87.2 |
| RaTG13 | NA | 88.1 | 77.6 | 86.6 | 98.7 | 91.0 | 83.6 | 73.0 | 72.1 | 28.2 | 89.7 | 84.6 |
| pangolin/P4L | NA | 88.2 | 76.9 | 86.2 | 98.7 | 91.0 | 80.3 | 68.9 | – | 28.8 | 89.2 | 79.5 |
| CoVZXC21 | NA | 87.9 | 75.5 | 83.3 | 98.7 | 91.4 | 77.1 | 68.9 | – | 20.5 | 88.1 | 28.2 |
| SARS-CoV-1 | NA | 85.9 | 75.2 | 72.5 | 93.4 | 97.7 | 66.7 | 69.7 | 68.2 | – | 88.9 | 59.0 |
| Rf1 | NA | 85.5 | 73.6 | 69.6 | 92.1 | 94.1 | 66.7 | 68.0 | 72.7 | 66.7 | 87.4 | 59.0 |
| BM48–31 |
NA |
83.7 |
71.9 |
64.5 |
93.4 |
88.1 |
48.5 |
52.5 |
58.1 |
– |
85.9 |
61.5 |
| *NA, not available; ORF, open reading frame; –, no ORFs found. | ||||||||||||
To gain insight into the zoonotic potential of Rc-o319, we focused on angiotensin-converting enzyme 2 (ACE2) receptor binding motif (RBM) of the S protein (Figure 1, panel E). RBM has 2 regions (1 and 2); both are essential to human ACE2 (hACE2) recognition (5,6). Several S proteins of bat-origin sarbecoviruses that lack the ability to bind the hACE2 contain amino acid deletions in both regions (Figure 1, panel E) (5). However, the RBM of Rc-o319 S is unique because it has 9 aa deletions in region 2 only, which was not observed in other bat sarbecoviruses. Of note, most residues that contact hACE2, which were detected in the S protein of SARS-CoV-2 and SARS-CoV, were different or missing in the S protein of Rc-o319. Thus, current data do not enable inference for whether the Rc-o319 can use hACE2 as a cell entry receptor.
To evaluate the potential of ACE2 as a receptor for Rc-o319, we adopted a pseudotyped vesicular stomatitis virus (VSV) system, in which VSV glycoprotein envelope (G) gene is replaced by green fluorescent protein (GFP) gene. We generated VSV pseudotyped with S proteins of Rc-o319 (VSV-Rc-o319), SARS-CoV (VSV-SARS), SARS-CoV-2 (VSV-SARS-2), or VSV-G (VSV-VSV-G) in human embryonic kidney 293T (HEK293T) cells. We also constructed ACE2 expression plasmids from hACE2, R. ferrumequinum bats (Rf-ACE2), and R. sinicus bats (Rs-ACE2). R. ferrumequinum is another bat species inhabiting in Japan, and R. sinicus bats are a major host reservoir of bat sarbecoviruses. We also prepared a chimeric bat ACE2 from R. cornutus and R. ferrumequinum bats, Rc/Rf chimera, which has the N terminus of S protein interaction domain of Rc-ACE2 and the remaining region from Rf-ACE2 (Appendix Figure 2). The HEK293T cells expressing Rc-ACE2, Rf-ACE2, Rs-ACE2, Rc/Rf-ACE2, or hACE2 were produced by transfecting each ACE2-expression plasmid (Appendix Figure 3) and inoculating them with pseudotyped VSVs; their infectivity was titrated by counting GFP-positive cells at 20 hours postinfection (Figure 2). Our results showed that VSV-Rc-o319 infected Rc-ACE2– and Rc/Rf-ACE2–expressing cells, but not Rf-ACE2–, Rs-ACE2–, or hACE2–expressing cells or control cells transfected with empty vector plasmid. In contrast, VSV-SARS and VSV-SARS-2 more effectively infected hACE2-expressing cells than bat ACE2–expressing cells and control cells. VSV-VSV-G infected all tested cells to comparable levels, confirming ACE2-independent infectivity of VSV. These results suggest high specificity of ACE2 receptor between sarbecovirus and host cells and possibly a limited zoonotic potential of Rc-o319 in terms of cell receptor usage without adaptation to humans.
Figure 2.
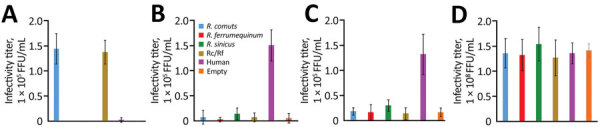
Infectivity titers of sarbecoviruses from bats and humans used to investigate bat sarbecovirus Rc-o319, which is genetically related to human SARS-CoV-2, Japan. Cells expressing each host-origin angiotensin-converting enzyme 2 were inoculated with VSV pseudotyped with spike proteins of Rc-o319 (A), SARS-CoV (B), SARS-CoV-2 (C), or glycoprotein of VSV (D). At 20 hour postinfection, GFP-positive cells were counted and the infectivity titers were calculated. Error bars indicate SDs from 3 independent experiments. CoV, coronavirus; GFP, green fluorescent protein; Rc/Rf, chimera of Rhinolophus cornutus and R. ferrumequinum; SARS, severe acute respiratory syndrome; VSV, vesicular stomatitis virus.
We next analyzed the membrane fusion step of Rc-o319 S. A previous study showed that human sarbecovirus S protein was proteolytically activated by cellular transmembrane serine protease 2 (TMPRSS2), in vitro and in vivo, inducing efficient virus-cell membrane fusion at the cell surface (7). We prepared a fusion assay, in which HEK293T cells were cotransfected with S-expression plasmid of Rc-o319, SARS-CoV, or SARS-CoV-2 and Rc-ACE2–, Rf-ACE2–, Rs-ACE2–, or hACE2-expression plasmid, together with fluorescent reporter Venus-expression plasmid with and without TMPRSS2-expression plasmid, and incubated for 24 h to assess syncytium formation. We observed that the S protein of SARS-CoV and SARS-CoV-2 required both hACE2 and TMPRSS2 for fusion activity (Appendix Figure 4), confirming the previous findings (7,8). In contrast, the S protein of Rc-o319 induced cell fusion only in Rc-ACE2–expressing cells, both in the presence and absence of TMPRSS2. These results suggest that unlike human sarbecoviruses, Rc-o319 uses Rc-ACE2 as a functional receptor, leading to membrane fusion independent of S-cleavage by TMPRSS2.
Conclusions
Among R. cornutus bats in Japan, we detected sarbecovirus Rc-o319, which is phylogenetically positioned in the same clade as SARS-CoV-2. Sarbecoviruses belonging to this clade previously were detected from other Rhinolophus spp. bats and pangolins (family Manidae) in China and could have played a role in the emergence of SARS-CoV-2 (9–11). We provide a hypothesis that a bat sarbecovirus with zoonotic potential might exist even outside China, because Rhinolophus spp. bats inhabit Asia, Europe, and Africa (12).
Receptor usage is one factor for cross-species transmission of viruses. Unlike a previous report that showed that some bat SARS-CoV–related viruses could use human and civet ACE2 and R. sinicus ACE2 as entry receptors (5,13,14), VSV-Rc-o319 was found to use only homologous Rc-ACE2, which arguably suggests that Rc-o319 and its related viruses might not jump the species barrier easily and cause infection. However, Rc-o319 and its related viruses could be transmitted accidentally from R. cornutus bats to cohabitant animals, such as civets, which are potential intermediate hosts for human infection (15). Therefore, further epidemiologic surveillance of bat betacoronaviruses, including evaluation of their zoonotic potential, is essential because betacoronaviruses that caused SARS, MERS, and COVID-19 outbreaks in humans during the past 20 years likely originated from bat betacoronaviruses.
Additional methods used for detection and characterization of bat sarbecovirus phylogenetically related to severe acute respiratory syndrome coronavirus 2, Japan.
Acknowledgments
S.M. is supported by a Kakenhi Grant-in-Aid for Challenging Research (Exploratory) from the Japan Society for the Promotion of Science (grant no. 17K19319).
Biography
Dr. Murakami is an associate professor at the Graduate School of Agricultural and Life Sciences, University of Tokyo, Tokyo, Japan. His research interests include epidemiologic and molecular biological studies of animal viruses, including coronaviruses and influenza viruses.
Footnotes
Suggested citation for this article: Murakami S, Kitamura T, Suzuki J, Sato R, Aoi T, Fujii M, et al. Detection and characterization of bat sarbecovirus phylogenetically related to SARS-CoV-2, Japan. Emerg Infect Dis. 2020 Dec [date cited]. https://doi.org/10.3201/eid2612.203386
These authors contributed equally to this article.
References
- 1.Drexler JF, Gloza-Rausch F, Glende J, Corman VM, Muth D, Goettsche M, et al. Genomic characterization of severe acute respiratory syndrome-related coronavirus in European bats and classification of coronaviruses based on partial RNA-dependent RNA polymerase gene sequences. J Virol. 2010;84:11336–49. 10.1128/JVI.00650-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hu B, Zeng LP, Yang XL, Ge XY, Zhang W, Li B, et al. Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. PLoS Pathog. 2017;13:e1006698. 10.1371/journal.ppat.1006698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tao Y, Tong S. Complete genome sequence of a severe acute respiratory syndrome-related coronavirus from Kenyan bats. Microbiol Resour Announc. 2019. 8(28):e00548–19. [DOI] [PMC free article] [PubMed]
- 4.Suzuki J, Sato R, Kobayashi T, Aoi T, Harasawa R. Group B betacoronavirus in rhinolophid bats, Japan. J Vet Med Sci. 2014;76:1267–9. 10.1292/jvms.14-0012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Letko M, Marzi A, Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol. 2020;5:562–9. 10.1038/s41564-020-0688-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–20. 10.1038/s41586-020-2180-5 [DOI] [PubMed] [Google Scholar]
- 7.Glowacka I, Bertram S, Müller MA, Allen P, Soilleux E, Pfefferle S, et al. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J Virol. 2011;85:4122–34. 10.1128/JVI.02232-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamamoto M, Kiso M, Sakai-Tagawa Y, Iwatsuki-Horimoto K, Imai M, Takeda M, et al. The anticoagulant nafamostat potently inhibits SARS-CoV-2 S protein-mediated fusion in a cell fusion assay system and viral infection in vitro in a cell-type-dependent manner. Viruses. 2020;12:E629. 10.3390/v12060629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–3. 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lau SKP, Luk HKH, Wong ACP, Li KSM, Zhu L, He Z, et al. Possible bat origin of severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis. 2020;26:1542–7. 10.3201/eid2607.200092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang T, Wu Q, Zhang Z. Probable pangolin origin of SARS-CoV-2 associated with the COVID-19 outbreak. Curr Biol. 2020;30:1346–1351.e2. 10.1016/j.cub.2020.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stoffberg S, Jacobs DS, Mackie IJ, Matthee CA. Molecular phylogenetics and historical biogeography of Rhinolophus bats. Mol Phylogenet Evol. 2010;54:1–9. 10.1016/j.ympev.2009.09.021 [DOI] [PubMed] [Google Scholar]
- 13.Ge XY, Li JL, Yang XL, Chmura AA, Zhu G, Epstein JH, et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503:535–8. 10.1038/nature12711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang XL, Hu B, Wang B, Wang MN, Zhang Q, Zhang W, et al. Isolation and characterization of a novel bat coronavirus closely related to the direct progenitor of severe acute respiratory syndrome coronavirus. J Virol. 2015;90:3253–6. 10.1128/JVI.02582-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guan Y, Zheng BJ, He YQ, Liu XL, Zhuang ZX, Cheung CL, et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–8. 10.1126/science.1087139 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional methods used for detection and characterization of bat sarbecovirus phylogenetically related to severe acute respiratory syndrome coronavirus 2, Japan.


