Abstract
Gracilaria spinulosa is an economical species of marine red algae. The length of its plastid genome sequence is 179,082 bp; a total of 236 genes were determined, including 203 protein-encoding genes, 3 rRNA genes, 29 tRNA genes, 1 ribonuclease gene, and 1 intron inserted into the trnM gene. The gene content and structure of Gracilariaceae species were relatively well conserved. Phylogenetic analysis showed that G. spinulosa had a closer relationship with Gracilaria salicornia in Gracilaria. The complete plastid genome sequence provided will help the understanding of Gracilaria evolution.
Keywords: Gracilaria spinulosa, plastid genome, phylogenetic analysis, Gracilariaceae
Gracilaria spinulosa (Okamura) Chang & B.M.Xia is a marine red alga belonging to the family Gracilariaceae. Rhodymenia spinulosa (Okamura) is the basionym of Gracilaria spinulosa. It is an agar-producing seaweed (Lin 2006). Previous studies focused on the vegetative and reproductive morphology and taxonomic status analyses of G. spinulosa (Withell et al. 1994; Lin 2006). It was originally described from Taiwan, with bushy and erect Thalli, irregularly dichotomous branches, flattened blades, a discoid holdfast, and occasionally with a short stipe (1–2.5 mm long; Lin 2006). However, no previous genomic studies on G. spinulosa have been reported.
Herein, we report the determination of the G. spinulosa plastid genome sequence. The specimen was collected from Yinggehai, Hainan Province (18°30′36′′ N, 108°42′15′′ E) was sequenced, and was deposited at the Culture Collection of Seaweed at the Ocean University of China (accession number: 2017060066). Total DNA was extracted via the modified CTAB method (Doyle and Doyle 1990). Paired-end reads were sequenced by using Illumina HiSeq system (Illumina, San Diego, CA, USA). Approximately 27 Gb of paired-end (150 bp) sequence data were randomly extracted from the total sequencing output and used as input for NOVOPlasty (Dierckxsens et al. 2017) to assemble the plastid genome. The complete plastid genome, using Gracilaria tenuistipitata var. liui (AY673996) as the seed sequence, was annotated with Geneious R10.1.3. The tRNA genes were identified using tRNAscan-SE Search Server (Schattner et al. 2005).
The complete G. spinulosa plastid genome is a circular DNA molecule measuring 179,082 bp in length, and the overall A + T content of the complete plastid genome was 71.3% (GenBank accession number MN053319). The plastid genome contained 236 genes, including 203 protein-coding, 3 rRNA, and 29 tRNA genes, 1 ribonuclease gene (rnpB), and 1 intron interrupting the trnM gene. The length of the coding region was 145,035 bp, corresponding to 81.0% of the total length. The plastid genome of G. spinulosa was compact, with 10 pairs of overlapping genes found with overlap lengths of 3–95 bp (rpl23–rpl4, rpl14–rps17, trnT–ilvB, ycf40–rps1, ycf29–trnH, psbD–psbC, carA–ycf53, chlI–trnR, atpF–atpD, and rps18–rpl33). The gene numbers and structures were largely similar among Gracilariaceae species published in the NCBI sequence database; their plastid genomes were relatively well conserved, with no gene rearrangement phenomena.
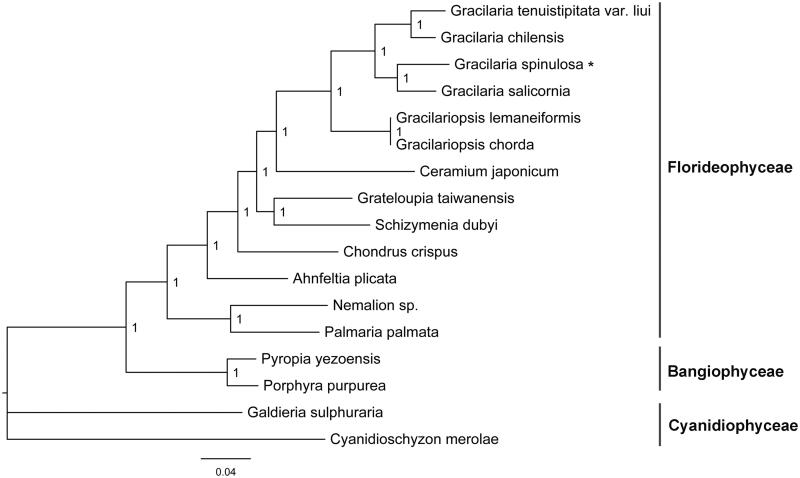
Phylogenetic analysis was conducted using 82 shared plastid protein sequences from 17 red algal plastid genomes, using Cyanidioschyzon merolae as an outgroup. The nucleotide sequences were aligned by using MAFFT (Katoh et al. 2002). Concatenated alignments were generated and poorly aligned regions were removed by using the Gblocks server (http://phylogeny.lirmm.fr/phylo_cgi/one_task.cgi?task_type=gblocks) (Castresana 2000). MrBayes 3.1.2 software was used to construct the amino acids phylogenetic tree (Ronquist and Huelsenbeck 2003). The results showed that all red algal taxa were clearly separated according to their original class (Figure 1). The Gracilaria species formed a branch, in which G. spinulosa showed a closer relationship with Gracilaria salicornia. The determination of the complete plastid genome sequence will help the understanding of Gracilaria evolution.
Figure 1.
Phylogenetic tree (Bayesian method) based on the complete plastid genome sequence of red algae as shown below: Gracilaria spinulosa (MN053319), Gracilaria salicornia (NC_023785), Gracilaria tenuistipitata var. liui (AY673996), Gracilaria chilensis (NC_029860), Gracilariopsis chorda (NC_031149), Gracilariopsis lemaneiformis (KP330491), Grateloupia taiwanensis (KC894740), Schizymenia dubyi (NC_031169), Chondrus crispus (NC_020795), Ceramium japonicum (NC_031174), Nemalion sp. (LT622871), Ahnfeltia plicata (NC_031145), Palmaria palmata (NC_031147), Pyropia yezoensis (KC517072), Porphyra purpurea (U38804), Galdieria sulphuraria (KJ700459), and Cyanidioschyzon merolae (NC_004799). The asterisks after species names indicate newly determined plastid genomes.
Disclosure statement
No conflict of interest for all the authors including the implementation of research experiments and writing this article was reported.
References
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17:540–552. [DOI] [PubMed] [Google Scholar]
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucl Acids Res. 45:e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. 1990. Isolation of plant DNA from fresh tissue. Focus. 12:13–15. [Google Scholar]
- Katoh K, Misawa K, Kuma K-i, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucl Acids Res. 30:3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SM. 2006. Observations on flattened species of Gracilaria (Gracilariaceae, Rhodophyta) from Taiwan. J Appl Phycol. 18:671–678. [Google Scholar]
- Ronquist F, Huelsenbeck JP. 2003. Mrbayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19:1572–1574. [DOI] [PubMed] [Google Scholar]
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucl Acids Res. 33:W686–W689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Withell AF, Millar AJK, Kraft GT. 1994. Taxonomic studies of the genus Gracilaria (Gracilariales, Rhodophyta) from Australia. Australian System Bot. 7:281–352. [Google Scholar]