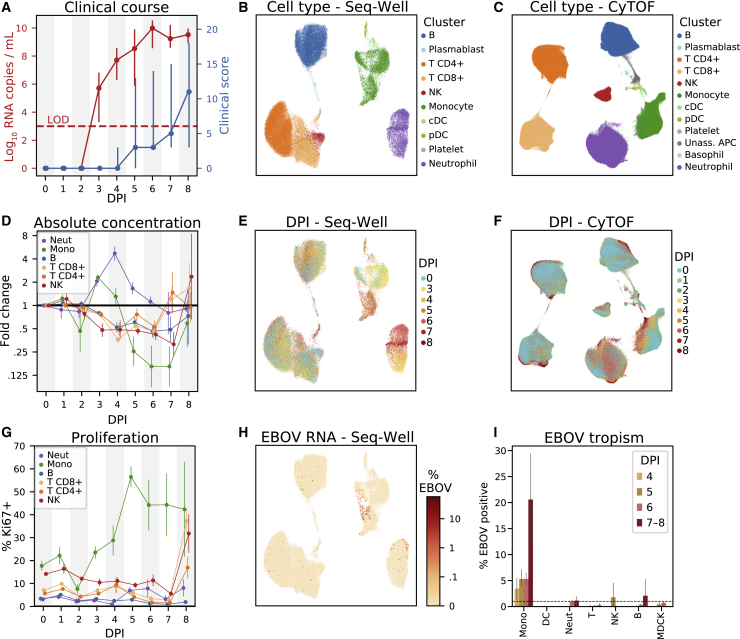
Figure 2.
Changing Cell-Type Abundance, Proliferation Rate, and Infection Status during EVD
(A) Time course of viral load (red, left y axis, log10 scale) and clinical score (blue, right y axis). Markers: mean; error bars: minimum and maximum; LOD, limit of detection by reverse transciption quantitative PCR.
(B and C) Uniform Manifold Approximation and Projection (UMAP) embedding of Seq-Well (B) and CyTOF (C) data, colored by annotated cluster assignment. See also Figures S1 and S2, and Data S1.
(D) Fold change (log2 scale) in the absolute abundance (cells/μL of whole blood) of each cell type relative to baseline based on CyTOF clusters. Error bars: mean ± 1 SE. See also Figures S3A and S3B.
(E and F) UMAP embedding of Seq-Well (E) and CyTOF (F) data, colored by the day post-infection (DPI) on which each cell was sampled.
(G) Percentage of Ki67-positive cells (CyTOF intensity >1.8) of each cell type. Error bars: mean ± 1 SE. See also Figures S3C and S3D.
(H) UMAP embedding of Seq-Well data, colored by the percentage of cellular transcripts mapping to EBOV.
(I) Percentage of infected cells by cell type based on Seq-Well. Dashed line: 1% false positive rate threshold for calling infected cells. Error bars: 95% CI on the mean based on 1,000 bootstraps. See also Figures S1H.