Abstract
HMG-CoA reductase (Hmgcr) is the rate-limiting enzyme in the mevalonate pathway and is inhibited by statins. In addition to cholesterol, Hmgcr activity is also required for synthesizing nonsterol isoprenoids, such as dolichol, ubiquinone, and farnesylated and geranylgeranylated proteins. Here, we investigated the effects of Hmgcr inhibition on nonsterol isoprenoids in the liver. We have generated new genetic models to acutely delete genes in the mevalonate pathway in the liver using AAV-mediated delivery of Cre-recombinase (AAV-Cre) or CRISPR/Cas9 (AAV-CRISPR). The genetic deletion of Hmgcr by AAV-Cre resulted in extensive hepatocyte apoptosis and compensatory liver regeneration. At the biochemical level, we observed decreased levels of sterols and depletion of the nonsterol isoprenoids, dolichol and ubiquinone. At the cellular level, Hmgcr-null hepatocytes showed ER stress and impaired N-glycosylation. We further hypothesized that the depletion of dolichol, essential for N-glycosylation, could be responsible for ER stress. Using AAV-CRISPR, we somatically disrupted dehydrodolichyl diphosphate synthase subunit (Dhdds), encoding a branch point enzyme required for dolichol biosynthesis. Dhdds-null livers showed ER stress and impaired N-glycosylation, along with apoptosis and regeneration. Finally, the combined deletion of Hmgcr and Dhdds synergistically exacerbated hepatocyte ER stress. Our data show a critical role for mevalonate-derived dolichol in the liver and suggest that dolichol depletion is at least partially responsible for ER stress and apoptosis upon potent Hmgcr inhibition.
Keywords: cholesterol synthesis and regulation, liver, endoplasmic reticulum, animal models, 3-hydroxy-3-methylglutaryl-coenzyme A, adeno-associated virus, dolichol, dehydrodolichyl diphosphate synthase subunit, clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9
HMG-CoA reductase (HMGCR) is the rate-limiting enzyme in the mevalonate pathway, which leads to the production of cholesterol (1). HMGCR is an integral glycoprotein localized in the ER and is the target of statins, the most widely used cholesterol-lowering drugs in the world (2). Cholesterol biosynthesis is tightly regulated through multiple mechanisms. First, the expression of HMGCR and all other enzymes in the mevalonate pathway is transcriptionally activated by SREBP-2, which is processed when ER sterol levels are low (3). Second, the translation of HMGCR protein is subject to feedback inhibition by cholesterol (4, 5) or other nonsterol isoprenoids (6). Third, HMGCR undergoes insulin-induced gene (INSIG)-dependent ubiquitination and proteasomal degradation in conditions of high levels of sterols and the branch product geranylgeranyl pyrophosphate in the ER (7, 8), which has a substantial effect on the levels of the enzyme in the liver (9, 10). Lastly, HMGCR activity is regulated by AMP-activated protein kinase (AMPK)-mediated phosphorylation, which can decrease cholesterol biosynthesis in low energy conditions (11). Collectively, these and other regulatory mechanisms finely tune HMGCR activity to maintain cellular cholesterol levels within a narrow physiological window.
Germline deletion of Hmgcr is embryonic lethal in mice around embryonic day 9.5 (E9.5), demonstrating the importance of the mevalonate pathway for normal growth and development. Interestingly, heterozygous mice appeared normal and showed unaffected cholesterol biosynthesis, due to compensatory upregulation of wild-type allele (12). Since this initial study, Hmgcr has been conditionally deleted in a number of tissues including the adipose (13), T-cells (14), skeletal muscle (15), myeloid cells (16), and the liver (17). The liver-specific KO (LSKO) model is of particular interest, as this is the major site for whole-body cholesterol synthesis in rodents (18), and is the target of the statin drugs. Hmgcr LSKO mice were generated by crossing floxed animals with the Albumin-Cre transgenic line. These animals developed hepatic steatosis with apoptosis, hypoglycemia, and eventually liver failure and death. Viability could be rescued by supplementation with high doses of mevalonate in the drinking water. The mechanism of liver injury was at least partially attributed to impairment of H-Ras and Rac1 isoprenylation (17). This work demonstrates the necessity for Hmgcr in hepatocyte viability. However, it is important to note that deletion with Albumin-Cre is not instantaneous but occurs gradually over a period of many weeks, i.e., 40% at birth, 75% at weaning, and complete at 6 weeks of age in one example (19). The gradual nature of Cre excision in this model allows ample time for physiological compensation, making it difficult to study direct effects.
In addition to cholesterol, the mevalonate pathway produces other important nonsterol isoprenoids, such as dolichol, ubiquinone, and farnesyl and geranylgeranyl anchors for protein prenylation. Depletion of these metabolites has been suggested to be responsible for some beneficial effects of statins (20), as well as rare adverse events (21). However, the degree to which these metabolites are reduced upon Hmgcr inhibition, and their relative importance for liver physiology is not well understood. Insight into the role of these metabolites could be gained through genetic deletion of enzymes that occupy important branch points from the direct route to cholesterol. Dehydrodolichyl diphosphate synthase subunit (DHDDS) is a branch point enzyme, which in complex with Nogo-B receptor constitutes a cis-prenyltransferase that catalyzes the first committed step in the synthesis of dolichol, a glycosyl carrier essential for the biosynthesis of N-linked glycoproteins (22–25). Prenyl diphosphate synthase (PDSS)1 and PDSS2 are branch point enzymes that elongate the prenyl side-chain of ubiquinone, which acts as electron carrier in mitochondrial respiration (26). Likewise, farnesyltransferase and geranylgeranyltransferase-I are branch point transferases that catalyze the covalent attachment of either the C-15 isoprene farnesyl or the C-20 isoprene geranylgeranyl group to the C-terminal end of multiple proteins (27). These include the proteins of the Ras superfamily, which are involved in cell growth, differentiation, signaling, and mobility (28).
Recombinant AAVs are small nonintegrating nonenveloped single-stranded DNA viruses that can deliver transgenes to the liver with high efficiency (29). AAV delivery of Cre recombinase is rapidly gaining popularity for inducible deletion of genes in the adult liver (30, 31). AAV can also be used to deliver the CRISPR/Cas9 genome editing system, which can directly disrupt genes, thus bypassing the need to generate new floxed mouse models (32, 33). Here, we used these tools to study the effects of mevalonate pathway inhibition on nonsterol isoprenoids and hepatocyte physiology. A new Hmgcr mouse model was produced through introduction of a floxed allele into the germline, which was used to generate LSKO mice with AAV-Cre delivery to adult mice. In Hmgcr KO livers, we found depletion of certain nonsterol isoprenoids, including dolichol and ubiquinone, along with ER stress-induced apoptosis and impairment of N-linked glycosylation. We demonstrate that inhibiting the dolichol biosynthetic pathway with AAV-CRISPR-mediated deletion of Dhdds partially phenocopied the Hmgcr LSKO mice. Finally, the combined deletion of Hmgcr and Dhdds resulted in exacerbation of hepatocyte ER stress and liver injury. Altogether, our work identifies dolichol as a nonsterol isoprenoid essential for hepatocyte viability, which is sensitive to potent inhibition of the mevalonate pathway.
MATERIAL AND METHODS
Animals
Male C57BL/6J mice were obtained from Jackson Laboratories. Mice harboring a “knockout first” allele for Hmgcr, Hmgcrtm1a(KOMP)Wtsi (referred as Hmgcr FRT-FLOX), were generated by the Knockout Mouse Project at the University of California Davis. These mice were used for generating Hmgcr conditional mice harboring the exon 5 flanked by loxP sites (Hmgcrfl/fl). Details are included in the supplemental data. Liver-specific deletion of Hmgcr was accomplished by delivery of Cre recombinase with AAV8 vectors and expression of Cre under the control of the hybrid liver-specific promoter (HLP) (34). Liver-specific deletion of Dhdds was performed by delivery of the CRISPR/Cas9 genome editing system with AAV8 vectors and expression of Cas9 under the control of the HLP promoter. AAVs were diluted in 300 μl of sterile saline and intraperitoneally injected to 6-week-old mice [2 or 5 × 1011 or 1 × 1012 genome copies (GCs) per mouse]. All treatment conditions were randomly allocated within each cage of mice at the time of injection. Mice were fasted for 5 h prior to injection and again before subsequent blood collections. Blood was collected via retro-orbital bleeding using heparinized Natelson collection tubes, and plasma was isolated by centrifugation at 10,000 g for 20 min at 4°C. Mice were allowed free access to food and water and maintained on a standard chow diet. All experiments were approved by the Baylor College of Medicine IACUC and performed in accordance with institutional guidelines under protocol number AN-6243. Experimental schemes in the figures were created with BioRender.
Plasmid design and cloning
1326-pAAV-HLP-CRE-2A-EGFP-WPRE-BGHpA and 1162-pAAV-HLP-EmGFP-SpA plasmids were produced by standard molecular biology techniques. A guide RNA (gRNA) targeting the exon 5 of Dhdds was designed by manual inspection based on the presence of a canonical NNGRRT protospacer adjacent motif for Staphylococcus aureus Cas9 (SaCas9). Cloning of gRNA (GCAGATGCAGATCACCCAGGAC) was accomplished by annealing oligonucleotides (Sigma-Aldrich), and ligating into the BbsI site of 1313-pAAV-U6-SA-BbsI-MluI-gRNA-HLP-SaCas9-HA-OLLAS-spA (Addgene 109314) (35), to obtain 1560-pAAV-U6-SA-mDhdds-gRNA2-HLP-SaCas9-HA-OLLAS-spA. Complete plasmid sequences are included in the supplemental data.
AAV production
Recombinant AAV8 vectors were generated as previously described (36) with several modifications (33). Plasmids required for AAV packaging, adenoviral helper plasmid pAdDeltaF6 (PL-F-PVADF6) and AAV8 packaging plasmid pAAV2/8 (PL-T-PV0007), were obtained from the University of Pennsylvania Vector Core. Each AAV transgene construct was cotransfected with the packaging constructs into 293T cells (ATCC, CRL-3216) using polyethylenimine. Cell pellets were harvested and purified using a single cesium chloride density gradient centrifugation. Fractions containing AAVs were pooled and then dialyzed against PBS using a 100 kDa Spectra-Por® Float-A-Lyzer® G2 dialysis device (Spectrum Labs) to remove the cesium chloride. Purified AAVs were concentrated using a Sartorius™ Vivaspin™ Turbo 4 ultrafiltration unit and stored at −80°C until use. AAV titers were calculated after DNase digestion using qPCR relative to a standard curve of the transgene plasmid. Primers used for titer are included in supplemental Table S1.
DNA analysis
Genomic DNA was extracted from livers and peripheral tissues using the DNeasy Blood and Tissue kit (Qiagen). One hundred nanograms of DNA were PCR amplified for detecting Hmgcr deletion with APEX TaqRed Master Mix (Apex Bioresearch Products). PCR products were separated by agarose gel electrophoresis. For detecting editing at the Dhdds locus, 100 ng of DNA were PCR amplified with Phusion DNA polymerase (Roche) and the 680 bp product was separated by agarose gel electrophoresis and gel extracted using the QIAquick gel extraction kit (Qiagen). The PCR product was Sanger sequenced and the indel percentage was determined by inference of CRISPR edits (ICE) analysis (https://ice.synthego.com/#/) using a control chromatogram for comparison. For detecting AAV-genome integration events at the double-strand break site, 100 ng of DNA were PCR amplified with APEX TaqRed Master Mix (Apex Bioresearch Products) using a forward primer annealing upstream of the cut site in the Dhdds locus and a reverse primer specific for the viral inverted terminal repeat sequence. The AAV-CRISPR genome was PCR amplified using primers specific for the Cas9 sequence. All primers are listed in supplemental Table S1.
RNA analysis
RNA was isolated from liver and peripheral tissues using RNeasy Mini Kit (Qiagen). One microgram of RNA was used for generating cDNA by the iScript cDNA synthesis kit (Bio-Rad). cDNA was diluted 1:25 and used as template for qPCR analysis using iTaq Universal SYBR Green Supermix (Bio-Rad) and ViiA 7 real-time PCR system (Thermo Fisher Scientific). A dissociation curve was carried out at the end of qPCR for assessing the homogeneity of the PCR products. Relative gene expression was calculated using the ΔΔCt method and graphed as fold change relative to TATA-box binding protein (Tbp). All primers are listed in supplemental Table S1.
Western blot
Liver and most of the peripheral tissues were homogenized in ∼10 vol of RIPA buffer [50 mM Tris (pH 8.0), 1 mM EDTA, 1% Triton X-100, 0.1% sodium dodecyl sulfate, 0.5% sodium deoxycholate, 150 mM sodium chloride, and protease inhibitors (Roche)] using a Bead Blaster 24 (Benchmark). Gastrocnemius, diaphragm, and heart tissues were homogenized in ∼10 vol of RIPA buffer using a Polytron PT2100 homogenizer (Kinematica). For membrane fraction isolation, ∼100 mg of liver samples were homogenized in 0.5 ml of lysing buffer (PBS supplemented with protease inhibitors) and centrifuged for discarding nuclei and cell debris. Supernatants were centrifuged at 57,000 g for 1 h at 4°C, using an OPTIMA TLX ultracentrifuge with a Beckman TLA 100.3 rotor. Pellets (membrane fraction) were resuspended in 0.2 ml of lysing buffer. Protein concentrations were determined using BCA assay (Thermo-Pierce). Liver and peripheral tissue lysates (30–50 μg) or plasma (1 μl) were diluted in 4× LDS buffer (Life Technologies) supplemented with 5% β-mercaptoethanol and separated by SDS-PAGE using 4–12% gradient gels (Life Technologies). Proteins were transferred to PVDF membranes (Millipore) followed by blocking for 2 h at room temperature in a 2:1 solution of Odyssey blocking buffer (Li-Cor) and PBS with 0.05% Tween-20 (PBS-T). Primary antibodies were diluted in 1% BSA in PBS-T, and membranes were incubated overnight at 4°C. Secondary antibodies were incubated at room temperature for 1 h and imaged using an Odyssey Classic (Li-Cor). All antibodies are listed in supplemental Table S2.
Immunohistochemistry
Livers were formalin-fixed for 24 h and then gradually dehydrated with ethanol and paraffin-embedded. Immunohistochemistry and hematoxylin and eosin staining were performed by the Texas Digestive Diseases Morphology Core at Baylor College of Medicine, following standard protocols. Briefly, liver sections were deparaffinized and subjected to antigen retrieval with Target Retrieval Solution (DAKO). The sections were then incubated with 3% hydrogen peroxide, followed by incubation in normal serum to block nonspecific protein binding. Sections were incubated for 1 h at room temperature with anti-Ki67 (1:60; CRM325, Biocare). The Ki67 antibody was then detected with a Rabbit-on-Rodent HRP-Polymer (RMR622H, Biocare) and visualized with DAB chromogen (DB801, Biocare). TUNEL stainings were performed and detected using ApopTag Peroxidase In Situ Apoptosis Detection Kit (Millipore, S7100). All slides were counterstained with hematoxylin, dehydrated, and mounted with a permanent mounting medium. A Nikon Ci-L bright field microscope was used for imaging at the Integrated Microscopy Core (Baylor College of Medicine). Ki67- and TUNEL-positive cell quantification was performed by manual count of positive hepatocytes in five 200× magnification images per liver taken across the whole section. Section area was quantified using ImageJ24 (https://imagej.nih.gov/ij/).
Transmission electron microscopy
Livers were fixed in glutaraldehyde and processed for transmission electron microscopy at the Texas Digestive Diseases Morphology Core at Baylor College of Medicine.
Lipidomics
Sterols, isoprenyl phosphates, and isoprenoids were extracted from ∼150 mg of liver samples. Sterols were analyzed as previously described (37) with some modifications. Lipidomics analyses were conducted at the Genome British Columbia Proteomics Centre, University of Victoria, Canada. A detailed method for lipidomics is available in the supplemental data.
Plasma analyses
Alanine aminotransferase (ALT) activity was measured using the Teco ALT (SGPT) kinetic liquid kit. Alkaline phosphatase (ALP) activity was measured using the Alkaline Phosphatase Assay Kit (Colorimetric) (ab83369, Abcam). Total cholesterol was measured using the Wako Cholesterol E kit. Triglycerides were measured using the Infinity Triglycerides Reagent (Thermo Fisher). As control of impaired glycosylation, plasma samples were enzymatically deglycosylated using Protein Deglycosylation Kit II (NEB).
Cell treatments
NIH3T3 cells (CRL-1658, ATCC) were cultured in DMEM (Fisher Scientific) supplemented with 10% fetal bovine serum (Sigma-Aldrich) and 1% penicillin/streptomycin (VWR), at 37°C and 5% CO2. Cells were treated with 10 μM of farnesyltransferase inhibitor FTI-277 trifluoroacetate salt (Sigma-Aldrich) or 30 μM of geranylgeranyltransferase-I inhibitor GGTI 298 TFA salt (Selleckchem) for 48 h, or 30 μM of Lovastatin (Sigma-Aldrich) for 24 h.
Statistics
All data are shown as the mean ± SD. Comparisons involving two groups were evaluated by a two-tailed Student’s t-test. For comparisons involving three or more groups, a one-way ANOVA was applied, with Tukey’s posttest used to test for significant differences among groups. In all cases, significance was assigned at P < 0.05.
RESULTS
Efficient deletion of Hmgcr in the liver by AAV-Cre
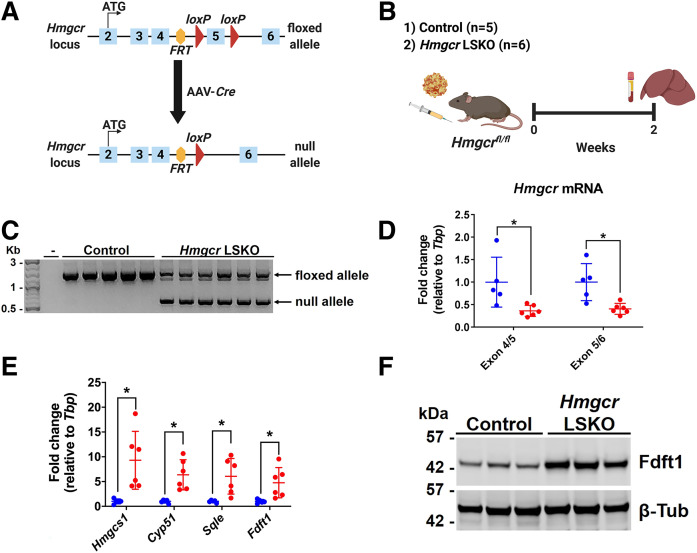
We sought to develop a novel genetic model of acute deletion of Hmgcr in the livers of adult mice. To this end, we first generated Hmgcr+/− mice harboring the nonfunctional Hmgcr FRT-FLOX allele by ES cell-mediated gene transfer (supplemental Fig. S1A). No Hmgcr−/− mice were born from Hmgcr+/− × Hmgcr+/− breeding pairs (supplemental Fig. S1B), confirming early embryonic lethality following complete disruption of Hmgcr (12). Then, this line was crossed with FLP-transgenic mice and backcrossed to the C57BL/6J background, finally resulting in the generation of a mouse line harboring the exon 5 of Hmgcr flanked by loxP sites (Hmgcrfl/fl) (Fig. 1A). To delete Hmgcr in the liver, we injected Hmgcrfl/fl mice with an AAV8 vector encoding the Cre recombinase under the control of the HLP promoter (Fig. 1A, B). At 2 weeks postinjection, Hmgcr LSKO mice showed the Hmgcr-null allele in the liver (Fig. 1C), with no genetic deletion detected in any of the peripheral tissues analyzed (supplemental Fig. S2). The Hmgcr mRNA level was found to be significantly decreased in livers from Hmgcr LSKO mice compared with control mice (Fig. 1D). Moreover, the deletion of Hmgcr resulted in transcriptional upregulation of Srebp-2-targeted genes of the mevalonate pathway (Fig. 1E, F), but not in any of the peripheral tissues analyzed (supplemental Fig. S3).
Fig. 1.
Efficient deletion of Hmgcr in the liver by AAV-Cre. A: Schematic diagram of genetic deletion of Hmgcr (exon 5) by AAV-Cre in the liver of Hmgcrfl/fl mice. B: Six-week-old Hmgcrfl/fl mice were injected with 2 × 1011 GCs of AAV-Cre or AAV-GFP (control). Two weeks postinjection, liver and plasma were collected for analyses. C: Genotyping PCR on liver DNA showing two main bands corresponding to the Hmgcr floxed (1,364 bp) and null (544 bp) alleles. Minus (-) indicates water only control. D: qPCR analysis of Hmgcr mRNA levels in livers from control (blue) and Hmgcr LSKO (red) mice. E: qPCR analysis of representative genes of the mevalonate pathway in the liver of Hmgcr LSKO (red) and control (blue) mice. F: Western blot analysis of Fdft1 in liver lysates from Hmgcr LSKO and control mice, with β-tubulin (β-Tub) used as a loading control. Hmgcs1, HMG-CoA synthase 1; Cyp51, cytochrome P450 family 51; Sqle, squalene epoxidase; Fdft1, farnesyl-diphosphate farnesyltransferase 1. Data are shown as mean ± SD with significance determined by two-tailed Student’s t-test (n = 5 control and 6 Hmgcr LSKO mice). *P < 0.05.
Hmgcr deletion results in depletion of sterol and nonsterol isoprenoids in the liver
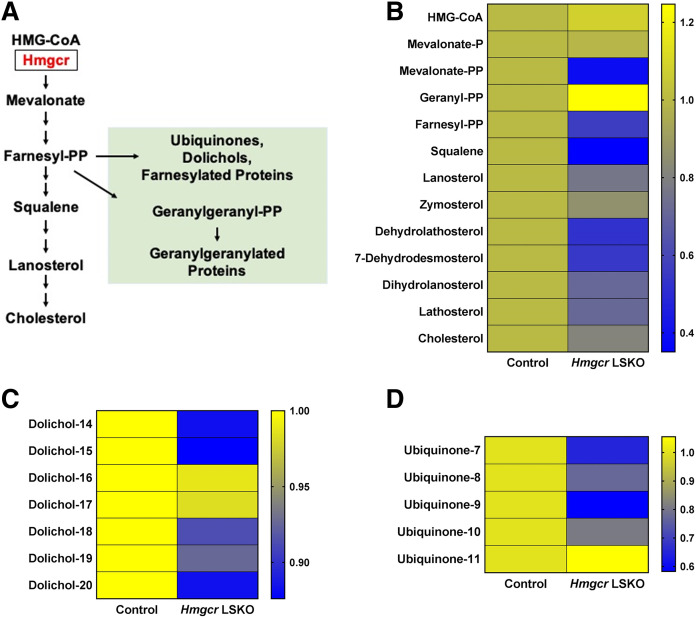
Next, we aimed to investigate the effects of Hmgcr deletion on the metabolites in the mevalonate pathway, with particular interest in the nonsterol isoprenoids (Fig. 2A). We used a targeted lipidomics approach for simultaneously measuring selected sterol and nonsterol isoprenoids. Livers from Hmgcr LSKO mice showed an overall decrease of cholesterol and sterol isoprenoids as compared with control mice (Fig. 2B). In terms of nonsterol isoprenoids, Hmgcr KO livers showed lower levels of dolichols and ubiquinones compared with control mice (Fig. 2C, D), with dolichol-18 and ubiquinone 9 being respectively the most abundant species in mice. No changes were observed in plasma total cholesterol and triglyceride levels (supplemental Fig. S4). Together, these data show that the deletion of Hmgcr results in depletion of both sterols and important nonsterol isoprenoids in the liver.
Fig. 2.
Hmgcr deletion results in depletion of sterol and nonsterol isoprenoids in the liver. A: Schematic diagram of the mevalonate pathway and selected branch point products. Representative metabolites are shown. Nonsterol isoprenoids are shown in green. Heat maps showing sterol isoprenoids (B), dolichols (C), and ubiquinones (D) measured in livers from Hmgcr LSKO and control mice by a lipidomics approach. Data are presented as relative abundance of each metabolite in Hmgcr KO compared with control livers (n = 5 control and 6 Hmgcr LSKO mice).
Hmgcr LSKO mice show transient liver injury and regeneration
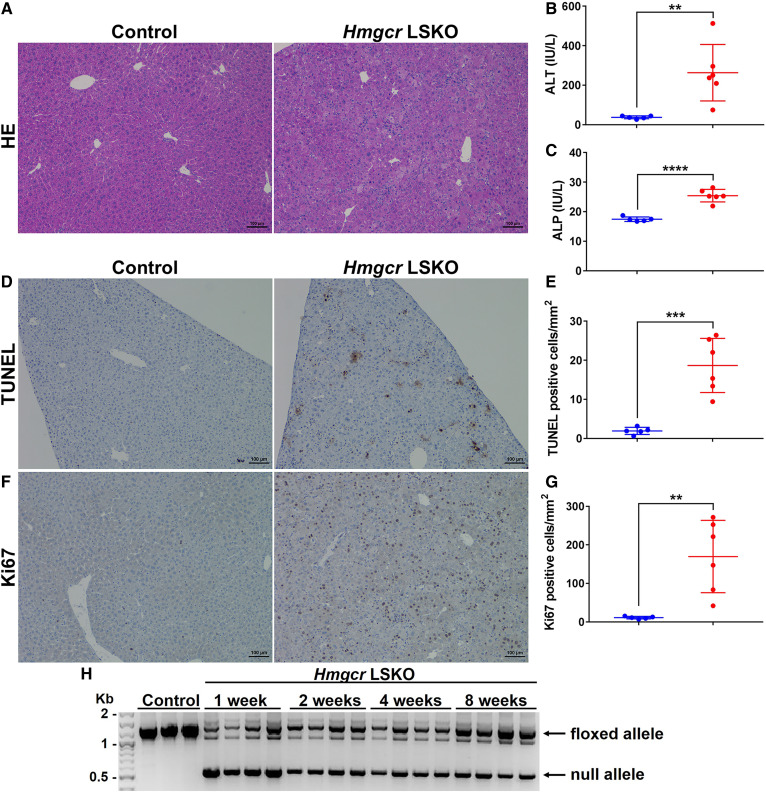
Chronic deletion of Hmgcr in the liver was reported to cause severe liver damage and eventually liver failure (17). We investigated the effects of acute Hmgcr deletion on liver pathophysiology in our model. Hmgcr KO livers showed altered histology, characterized by enlarged and dying hepatocytes (Fig. 3A). This resulted in significant increases in plasma ALT and ALP levels as compared with control mice (Fig. 3B, C). Many hepatocytes died by apoptosis, as shown by positive TUNEL staining in Fig. 3D and E. We also observed a massive burst of proliferating hepatocytes by Ki67 staining at this time point (Fig. 3F, G). Hepatocyte apoptosis gradually decreased at 4 and 8 weeks post-AAV-Cre injection (supplemental Fig. S5). When we looked at our model at longer time points, we noticed a progressive loss of the Hmgcr-null allele as well as enrichment of the Hmgcr-floxed allele over time (Fig. 3H), indicating selective expansion of cells that escaped complete Hmgcr deletion. In contrast to previous work (17), the viability of the mice was not adversely affected by the loss of Hmgcr in the liver. Overall, these data show that the deletion of Hmgcr results in transient hepatocyte apoptosis followed by compensatory liver regeneration.
Fig. 3.
Hmgcr LSKO mice show increased hepatocyte apoptosis and liver regeneration. A: Hematoxylin and eosin (HE) staining of control and Hmgcr KO liver sections. ALT (B) and ALP (C) measurement in plasma from control (blue) and Hmgcr LSKO (red) mice. TUNEL staining of liver sections (D) and quantification of TUNEL-positive hepatocytes per square millimeter in control (blue) and Hmgcr KO (red) livers (E). Ki67 staining of liver sections (F) and quantification of Ki67-positive hepatocytes per square millimeter in control (blue) and Hmgcr KO (red) livers (G). H: Hmgcr genotyping PCR on liver DNA from Hmgcr LSKO mice at 1, 2, 4, and 8 weeks post-AAV-Cre injection. The two main bands correspond to the Hmgcr floxed (1,364 bp) and null (544 bp) alleles. Representative IHC images are shown (scale bar is 100 μm). Data are presented as mean ± SD, with significance determined by two-tailed Student’s t-test (n = 5 control and 6 Hmgcr LSKO mice). **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Hmgcr LSKO mice show hepatocyte ER stress-induced apoptosis and impaired N-linked glycosylation
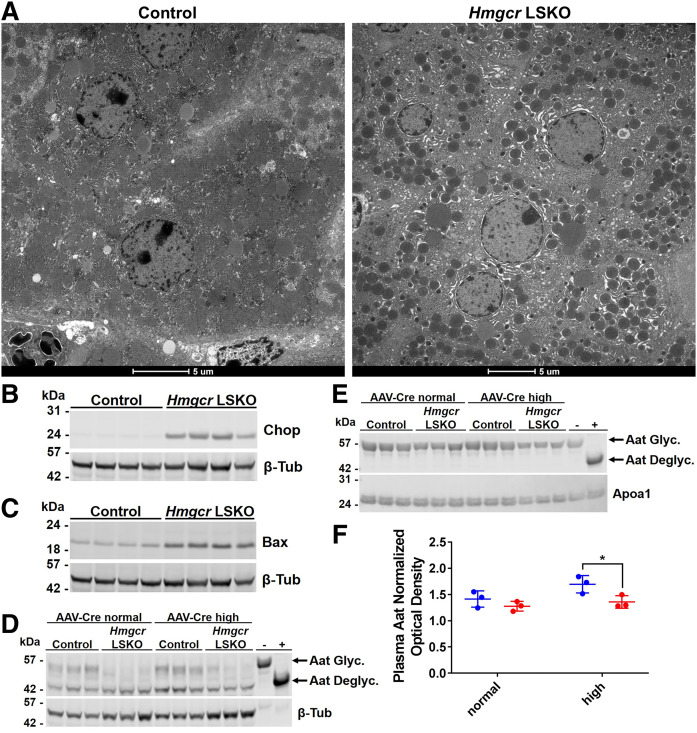
To further investigate the nature of liver injury, we performed a transmission electron microscopy analysis of livers following Hmgcr deletion (Fig. 4A). We observed swollen mitochondria and nucleus fragmentation as common features of apoptosis in Hmgcr KO hepatocytes (supplemental Table S3). Most importantly, we detected evidence of ER swelling (Fig. 4A, supplemental Table S3), which is commonly observed in conditions of ER stress (38). In parallel, we observed induction of the ER stress marker, Chop, and the apoptosis marker, Bax, in liver lysates from Hmgcr LSKO mice (Fig. 4B, C). Given the importance of dolichyl-phosphate for N-linked glycosylation in the ER (22), we hypothesized an ongoing impairment of the N-linked glycosylation process in Hmgcr KO livers. To test this, we used Western blot for analyzing the gel mobility of α-1 antitrypsin (Aat), which harbors three N-glycosylation sites (39). We observed decreased levels of mature (fully glycosylated) Aat in liver lysates as well as plasma from Hmgcr LSKO mice regardless the dose of AAV-Cre used for deleting Hmgcr (Fig. 4D–F). The previously described Hmgcr LSKO model showed impairment of protein prenylation (17). In our model, we found that the farnesylation of prelamin-A was not impaired in Hmgcr KO livers even when a 5-fold higher dose of AAV-Cre was used (supplemental Fig. S6A, B). On the contrary, the geranylgeranylation of RhoA was impaired in Hmgcr KO livers only upon injection of the highest dose of AAV-Cre, resulting in the abnormal accumulation of RhoA in the cytosol (supplemental Fig. S6C, D). Together, these data show that the deletion of Hmgcr results in hepatocyte ER stress-induced apoptosis and impaired N-linked glycosylation.
Fig. 4.
Hmgcr LSKO mice show hepatocyte ER stress-induced apoptosis and impaired N-glycosylation. A: Representative transmission electron microscopy of hepatocytes in liver from control and Hmgcr LSKO mice. Scale bar is 5 μM. Western blot analysis of Chop (B), Bax (C), and Aat (D) in liver lysates from control and Hmgcr LSKO mice, with β-tubulin used as a loading control. E: Western blot analysis of Aat in plasma from control and Hmgcr LSKO mice, with Apoa1 used as a loading control. F: Densitometry analysis of plasma Aat in control (blue) and Hmgcr LSKO (red) mice. Hmgcr LSKO samples in D and E)are from Hmgcrfl/fl mice injected with either 2 × 1011 (AAV-Cre normal) or 1012 (AAV-Cre high) GCs of AAV-Cre. In panels D and E, plasma was treated with an enzymatic deglycosylation kit (+) or sham (−) as control of the effect of glycosylation on protein mobility. Aat Glyc., glycosylated (mature) Aat; Aat Deglyc., deglycosylated Aat. Representative Western blots are shown. Densitometry data are normalized to Apoa1 and shown as mean ± SD with significance determined by two-tailed Student’s t-test (n = 3). *P < 0.05.
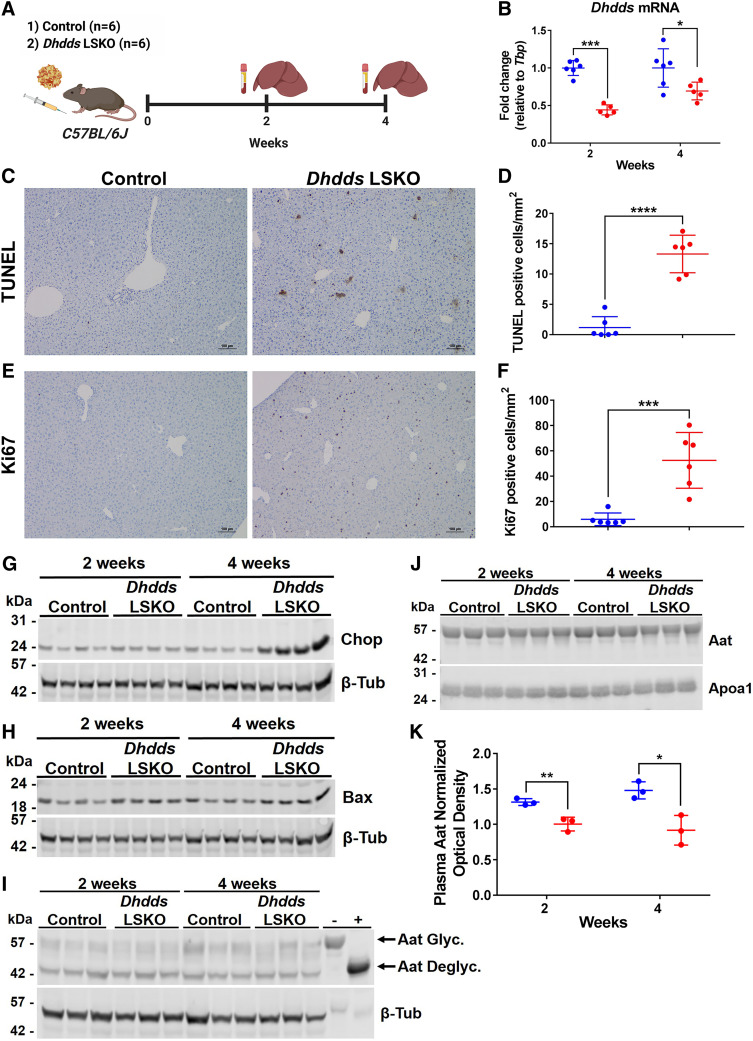
Dhdds LSKO mice show hepatocyte ER stress-induced apoptosis and impaired N-linked glycosylation
Next, we hypothesized that the depletion of newly synthesized dolichyl phosphate could be responsible for the ER stress-induced apoptosis observed in Hmgcr KO livers (Figs. 2, 4). We used the AAV-CRISPR system to somatically delete Dhdds, which encodes a branch point enzyme of the mevalonate pathway that, in complex with Nogo-B receptor, catalyzes the first committed step of dolichol biosynthesis (22). We injected mice with AAV-CRISPR or AAV-GFP (control) and collected liver and plasma samples at 2 and 4 weeks postinjection (Fig. 5A). AAV-CRISPR treatment resulted in the liver-restricted SaCas9 expression by the HLP promoter and efficient indel formation in exon 5 of Dhdds (supplemental Fig. S7), which was accompanied by a significant decrease of hepatic Dhdds mRNA levels (Fig. 5B). No detectable Dhdds editing was observed in any of the peripheral tissues analyzed (supplemental Fig. S8). The livers from Dhdds LSKO mice showed severe hepatocyte apoptosis and regeneration at 4 weeks postinjection (Fig. 5C–F). Most importantly, we observed a large induction of Chop along with increased levels of Bax in livers from Dhdds LSKO mice (Fig. 5G, H). However, the observed liver damage was not reflected by significant elevation of ALT or ALP in plasma (supplemental Fig. S9), contrary to what was observed in Hmgcr LSKO mice (Fig. 3). We next tested whether the deletion of Dhdds would result in impairment of N-linked glycosylation. Although there were not obvious differences in hepatic Aat levels (Fig. 5I), we found decreased levels of mature (fully glycosylated) Aat in plasma from Dhdds LSKO mice, along with a band of lower molecular weight at 4 weeks postinjection (Fig. 5J, K), suggesting impairment of N-glycosylation and processing. When we looked at the effects of Dhdds editing at a longer time point (6 weeks), we found that Dhdds mRNA levels had returned to normal with no evidence of increased ER stress or N-glycosylation impairment (supplemental Fig. S10), suggesting compensatory liver regeneration following the hepatocyte apoptosis observed at 4 weeks (Fig. 5C–F). Overall, these data show that the liver-specific deletion of Dhdds results in transient hepatocyte ER stress-induced apoptosis and impaired N-linked glycosylation.
Fig. 5.
Dhdds LSKO mice show hepatocyte ER stress-induced apoptosis. A: Six-week-old C57BL/6J mice were injected with 5 × 1011 GCs of AAV-CRISPR or AAV-GFP (control) and liver and plasma samples were collected at 2 and 4 weeks postinjection for analysis. B: qPCR analysis of Dhdds mRNA levels in livers from control (blue) and Dhdds LSKO (red) mice. TUNEL staining of liver sections (C) and quantification of TUNEL-positive hepatocytes per square millimeter in control (blue) and Dhdds LSKO (red) livers at 4 weeks postinjection (D). Ki67 staining of liver sections (E) and quantification of Ki67-positive hepatocytes per square millimeter in control (blue) and Dhdds LSKO (red) livers at 4 weeks postinjection (F). Representative images from mice at 4 weeks post-AAV injection are shown (scale bar is 100 μm). Western blot analysis of Chop (G), Bax (H), and Aat (I) in liver lysates from control and Dhdds LSKO mice, with β-tubulin used as a loading control. J: Western blot analysis of Aat in plasma from control and Dhdds LSKO mice, with Apoa1 used as a loading control. Representative Western blots are shown. In panel I, plasma was treated with an enzymatic deglycosylation kit (+) or sham (−) as control of the effect of glycosylation on protein mobility. Aat Glyc., glycosylated (mature) Aat; Aat Deglyc., deglycosylated Aat. K: Densitometry analysis of plasma Aat relative to Apoa1 in control (blue) and Dhdds LSKO (red) mice. All data are presented as mean ± SD with significance determined by two-tailed Student’s t-test (n = 6 mice per group, n = 3 in densitometry analysis). *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.001.
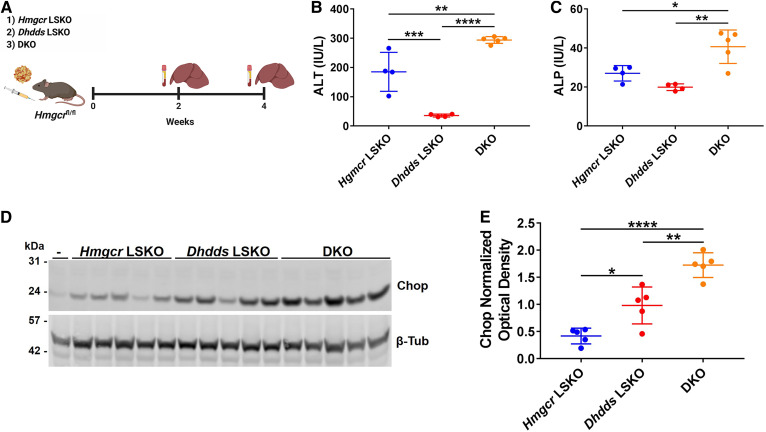
Combined deletion of Hmgcr and Dhdds exacerbates liver injury and hepatocyte ER stress
We next tested to determine whether there is a genetic interaction between Hmgcr and Dhdds inhibition in regard to ER stress. We injected Hmgcrfl/fl mice with AAV-Cre and AAV-CRISPR in order to simultaneously delete Hmgcr and Dhdds in the liver [double KO (DKO) mice; Fig. 6A]. As controls, Hmgcrfl/fl mice were injected with either AAV-Cre or AAV-CRISPR alone. DKO mice showed efficient deletion of Hmgcr along with CRISPR editing of Dhdds, which was readily detected by PCR for integration of the AAV genome at the cut site. Cre deletion and AAV-CRISPR editing resulted in significantly decreased levels of Hmgcr and Dhdds mRNA, respectively (supplemental Figs. S11, S12). We observed higher levels of ALT and ALP in plasma (Fig. 6B, C) as well as Chop in liver lysates from DKO mice as compared with Hmgcr and Dhdds LSKO mice (Fig. 6D, E). These data show that further restriction of isoprenoid flux to dolichol synthesis via Dhdds disruption acts synergistically with Hmgcr deletion to cause ER stress-induced apoptosis.
Fig. 6.
DKO mice show exacerbation of hepatocyte liver injury and ER stress. A: Six-week-old Hmgcrfl/fl mice were injected with 2 × 1011 GCs of AAV-Cre and 5 × 1011 GCs of AAV-CRISPR (DKO mice), and liver and plasma samples were collected at 2 and 4 weeks postinjection for analysis. AAV-CRISPR-empty (no gRNA) (5 × 1011 GCs) or AAV-GFP (2 × 1011 GCs) was included in the AAV dilutions respectively injected in Hmgcr LSKO and Dhdds LSKO mice, to deliver the same total AAV GCs per mouse. ALT (B) and ALP (C) assay on plasma from Hmgcr LSKO, Dhdds LSKO, and DKO mice at 2 weeks postinjection. D: Western blot analysis of Chop in liver lysates from Hmgcr LSKO, Dhdds LSKO, and DKO mice at 4 weeks postinjection, with β-tubulin used as a loading control. -, liver lysate from a Hmgcrfl/fl mouse used as negative control. Representative Western blots are shown. E: Densitometry analysis of Chop relative to β-tubulin loading control. Data are shown as mean ± SD with significance determined by one-way ANOVA followed by Tukey test (n = 4–7 mice per group). *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
DISCUSSION
Here, we report a new Hmgcr LSKO mouse model and its effects on isoprenoid metabolism and liver physiology. In contrast to the previous Albumin-Cre model, acute deletion of Hmgcr in the livers of adult mice is not lethal. Loss of Hmgcr results in transient liver injury characterized by hepatocyte apoptosis and compensatory liver regeneration. Livers from Hmgcr LSKO mice show depletion of important nonsterol isoprenoids, dolichol and ubiquinone, along with ER stress and defective N-linked glycosylation. We show that AAV-CRISPR-mediated deletion of Dhdds phenocopies the loss of Hmgcr in regard to ER stress and impaired N-linked glycosylation. Finally, further restriction of metabolic flux to dolichol, via the combined deletion of Hmgcr and Dhdds, synergistically exacerbates ER stress and liver injury.
The previously described Albumin-Cre Hmgcr deletion model resulted in liver failure and eventually death. Interestingly, cholesterol biosynthesis was only modestly affected, whereas protein isoprenylation was shown to be impaired, suggesting a direct role for prenylation defects in the liver failure (17). However, this phenotype may be a consequence of chronic deficiency of Hmgcr since birth and potentially biased by adaptation mechanisms and compensatory changes. Moreover, it is reasonable to speculate that other nonsterol isoprenoids would also have been depleted at the time point of analysis. Here, we found that the acute deletion of Hmgcr in the liver resulted in depletion of dolichol and ubiquinone, suggesting that these nonsterol isoprenoids are particularly sensitive to potent mevalonate pathway inhibition. In contrast to the findings of Nagashima et al. (17), we did not observe major effects on protein prenylation. In fact, we observed accumulation of RhoA in liver cytosolic fractions as marker of geranylgeranylation impairment (40) only after delivery of a 5-fold higher AAV-Cre dose, whereas prelamin-A farnesylation was never affected. Therefore, our data suggest that protein prenylation is not the branch product that is most sensitive to Hmgcr inhibition in the liver and unlikely to account for hepatocyte apoptosis and liver injury. T-cells and myeloid cells have been reported to be more sensitive to the effects of Hmgcr deletion or high-dose statins on prenylation of the Ras superfamily, possibly reflecting cell-specific differences in enzymatic activities or availability of their protein substrates (14, 16, 41).
Dolichol is a branch point product of the mevalonate pathway produced from farnesyl pyrophosphate. DHDDS and Nogo-B receptor form a cis-prenyltransferase that catalyzes the first committed step of dolichol biosynthesis, connecting the mevalonate pathway to N-linked glycosylation (23–25). Newly synthesized dolichyl-phosphates are converted to dolichol-linked oligosaccharides required for N-linked glycosylation (22). Defects in the dolichol oligosaccharide assembly pathway or accumulation of hypoglycosylated misfolded glycoproteins cause a chronic induction of the unfolded protein response pathway and ER stress (42). In Hmgcr KO livers, we noticed hepatocyte ER stress-induced apoptosis along with impairment of N-linked glycosylation. All these data suggested that the depletion of dolichol could be responsible for this phenotype. We confirmed this hypothesis by inhibiting the synthesis of dolichol via genetic deletion of Dhdds. Indeed, we observed hepatocyte ER stress and apoptosis, along with impairment of N-linked glycosylation. These results are consistent with the marked induction of ER stress and impaired N-linked glycosylation observed in Nogo-B receptor mutant fibroblasts (23). Moreover, mutations of DHDDS in humans have been associated with inherited retinitis pigmentosa because of impaired glycosylation of opsin, and type I congenital disorders of glycosylation (43, 44). Finally, the synergistic effect on liver ER stress by the disruption of dolichol biosynthesis combined with the genetically induced inhibition of Hmgcr further demonstrates the importance of mevalonate-derived dolichol synthesis for hepatocytes. Besides N-linked glycosylation, other dolichol-dependent modifications, such as O-mannosylation, C-mannosylation, and glycosylphosphatidylinositol (GPI)-anchor modification, may also be impaired in our models. In this regard, the elevation of ALP activity in plasma of Hmgcr LSKO and DKO mice might be a direct result of liver damage but could also be due to defective attachment of the GPI-anchor, which anchors ALP to the plasma membrane.
The Nogo-B receptor has recently been identified as a negative regulator of hepatic lipogenesis and LXRα activity. Albumin-Cre-mediated deletion of the Nogo-B receptor in the liver resulted in increased levels of plasma triglycerides and hepatic steatosis. In this report, loss of the Nogo-B receptor was accompanied by increased nuclear translocation of LXRα, activation of lipogenic gene expression, and lipid accumulation in hepatocytes (45). Given the known interaction of the Nogo-B receptor with Dhdds (23, 24), it would be interesting to know whether there is a direct connection between cis-prenyltransferase activity of this complex and hepatic lipogenesis. Liver-specific deletion of Dhdds using either floxed mice or AAV-CRISPR deletion could be useful in resolving this issue.
In Hmgcr KO livers, we also observed decreased levels of ubiquinone, a branch product that acts as electron carrier in the electron transport chain. Although we cannot fully exclude an effect of ubiquinone depletion on hepatocyte ER stress and apoptosis, several reports show that ubiquinone deficiency affects mainly brain and kidney. In humans, mutations in PDSS1 and PDSS2 have been respectively associated with neurological disorder and Leigh syndrome with nephropathy (46, 47). Pdss2 mutant mice recapitulated the ubiquinone deficiency and kidney disease phenotype, with no extrarenal disease (26, 48). More importantly, the liver-specific deletion of Pdss2 resulted in depletion of ubiquinone, but no evidence of liver pathology (26, 49). Altogether, these reports suggest that the liver is not primarily affected by ubiquinone deficiency or may compensate for its depletion. For instance, the liver can scavenge the ubiquinone carried by circulating lipoproteins (50), perhaps at levels adequate to support normal mitochondrial respiration.
The acute Hmgcr LSKO model showed extensive hepatocyte death and liver regeneration. When we investigated the effects of Hmgcr deletion over time, we observed a progressive loss of the Hmgcr-null allele and a corresponding increase in the floxed allele. This is clear evidence for expansion of hepatocytes that escaped complete removal of Hmgcr with AAV-Cre. The dose of virus used (2 × 1011 GCs) is sufficient to mediate complete excision in >99% of hepatocytes (30), so this is an excellent model to study acute effects. However, because AAV-Cre is nonintegrating, the viral transgene is lost with cell division, essentially allowing the liver to be repopulated with Hmgcr+ hepatocytes over time. It is also possible that some of the regenerating cells are not hepatocyte-derived, an event that can happen in very severe cases of liver injury (31). Regardless of the cellular source of the Hmgcr+ hepatocytes, there is sufficient metabolic capacity to protect these animals from liver failure. This is in direct contrast to the Albumin-Cre transgenic model, which showed a more severe phenotype and died at 6 weeks of age (17), which is expected for such an integrated transgene. For this reason, our study adds value to the current models, because it is a tractable system to inactivate mevalonate synthesis, while avoiding any confounding developmental compensation.
Here, we also show that AAV-CRISPR is an efficient genetic tool for deleting genes in the mevalonate pathway in the livers of adult mice, with the great advantage of bypassing the need to generate new floxed animals. Similar to the Hmgcr LSKO mice, our Dhdds LSKO model also showed transient hepatocyte apoptosis and liver regeneration, which resolved at a later time point. It is notable that Dhdds disruption did not cause equivalent liver damage to the Hmgcr LSKO and DKO mice, at least based on the lack of significant increases in ALT and ALP. This could be due to the different efficiency and kinetics of gene disruption between the AAV-Cre and the AAV-CRISPR systems. Optimization of gRNA design and vector dose may overcome these limitations. However, we also cannot exclude that different cell death mechanisms other than apoptosis may occur (i.e., necrosis) and differentially impact ALT elevations in the Hmgcr versus Dhdds KO mice. Finally, the depletion of other nonsterol isoprenoids may also contribute to the greater degree of liver damage observed in Hmgcr LSKO compared with Dhdds LSKO mice.
In conclusion, we report a genetic model to acutely inhibit the mevalonate pathway in the liver. We show that the deletion of Hmgcr results in depletion of the nonsterol isoprenoids, dolichol and ubiquinone, and severe ER stress. We demonstrate that the genetic inhibition of dolichol biosynthesis partially mimics the liver phenotype observed in Hmgcr LSKO mice. Moreover, the combined inhibition of the mevalonate pathway and dolichol biosynthesis exacerbates liver injury and hepatocyte ER stress. Altogether, our data show a critical role for mevalonate-derived dolichol in the liver and suggest that this metabolite could play an important role in mediating the effects of statins on liver physiology.
Data availability
All data are contained within the article and supplemental file.
Supplementary Material
Acknowledgments
The authors thank Dr. Stephen G. Young (University of California Los Angeles) for providing anti-prelamin-A antibody.
Footnotes
This article contains supplemental data.
Author contributions—M.D.G. and W.R.L. conceptualization; M.D.G. and W.R.L. design; M.D.G. experiments; M.D.G. analysis; A.H. viral vector production; K.E.J., J.C.B., A.M.D., A.H., A.L., R.H.H., and M.F. technical and intellectual support; K.R.P. transmission electron microscopy data analysis; K.R.P. pathological scores; J.H. and C.H.B. lipidomics; M.D.G. and W.R.L. writing-original draft; M.D.G., K.E.J., J.C.B., A.M.D., A.H., A.L., R.H.H., M.F., K.R.P., J.H., C.H.B, and W.R.L. writing-editing and review.
Author ORCIDs—Christoph H. Borchers http://orcid.org/0000-0003-2394-6512; William R. Lagor http://orcid.org/0000-0002-1703-5125
Funding and additional information—This work was supported by the National Institutes of Health (HL132840 and DK124477 to W.R.L.; T32HL07676 and T32GM08321 to K.E.J.; T32HL07676 to A.M.D.; and R01-HL132840-02S1 to M.F.), the American Heart Association (19POST34430092 to M.D.G. and 19PRE34380467 to A.M.D.). This work was also supported by the Texas Digestive Diseases Morphology Core (P30DK56338) and the Integrated Microscopy Core (DK56338 and CA125123). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
Abbreviations—
- Aat
- α-1 antitrypsin
- ALP
- alkaline phosphatase
- ALT
- alanine aminotransferase
- Dhdds
- dehydrodolichyl diphosphate synthase subunit
- DKO
- double KO
- GC
- genome copy
- gRNA
- guide RNA
- Hmgcr
- HMG-CoA reductase
- HLP
- hybrid liver-specific promoter
- Insig
- insulin-induced gene
- LSKO
- liver-specific KO
- PDSS
- prenyl diphosphate synthase
- SaCas9
- Staphylococcus aureus Cas9
Manuscript received June 26, 2020, and in revised form October 12, 2020. Published, JLR Papers in Press, October 27, 2020, DOI 10.1194/jlr.RA120001006.
REFERENCES
- 1.Goldstein J. L., and Brown M. S.. 1990. Regulation of the mevalonate pathway. Nature. 343: 425–430. [DOI] [PubMed] [Google Scholar]
- 2.Silverman M. G., Ference B. A., Im K., Wiviott S. D., Giugliano R. P., Grundy S. M., Braunwald E., and Sabatine M. S.. 2016. Association between lowering LDL-C and cardiovascular risk reduction among different therapeutic interventions: a systematic review and meta-analysis. JAMA. 316: 1289–1297. [DOI] [PubMed] [Google Scholar]
- 3.Brown M. S., and Goldstein J. L.. 1997. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 89: 331–340. [DOI] [PubMed] [Google Scholar]
- 4.Chambers C. M., and Ness G. C.. 1998. Dietary cholesterol regulates hepatic 3-hydroxy-3-methylglutaryl coenzyme A reductase gene expression in rats primarily at the level of translation. Arch. Biochem. Biophys. 354: 317–322. [DOI] [PubMed] [Google Scholar]
- 5.Ness G. C., and Chambers C. M.. 2000. Feedback and hormonal regulation of hepatic 3-hydroxy-3-methylglutaryl coenzyme A reductase: the concept of cholesterol buffering capacity. Proc. Soc. Exp. Biol. Med. 224: 8–19. [DOI] [PubMed] [Google Scholar]
- 6.Johnson B. M., and DeBose-Boyd R. A.. 2018. Underlying mechanisms for sterol-induced ubiquitination and ER-associated degradation of HMG CoA reductase. Semin. Cell Dev. Biol. 81: 121–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sever N., Yang T., Brown M. S., Goldstein J. L., and DeBose-Boyd R. A.. 2003. Accelerated degradation of HMG CoA reductase mediated by binding of insig-1 to its sterol-sensing domain. Mol. Cell. 11: 25–33. [DOI] [PubMed] [Google Scholar]
- 8.Schumacher M. M., Elsabrouty R., Seemann J., Jo Y., and DeBose-Boyd R. A.. 2015. The prenyltransferase UBIAD1 is the target of geranylgeraniol in degradation of HMG CoA reductase. eLife. 4: e05560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ness G. C., Chambers C. M., and Lopez D.. 1998. Atorvastatin action involves diminished recovery of hepatic HMG-CoA reductase activity. J. Lipid Res. 39: 75–84. [PubMed] [Google Scholar]
- 10.Engelking L. J., Liang G., Hammer R. E., Takaishi K., Kuriyama H., Evers B. M., Li W. P., Horton J. D., Goldstein J. L., and Brown M. S.. 2005. Schoenheimer effect explained–feedback regulation of cholesterol synthesis in mice mediated by Insig proteins. J. Clin. Invest. 115: 2489–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loh K., Tam S., Murray-Segal L., Huynh K., Meikle P. J., Scott J. W., van Denderen B., Chen Z., Steel R., LeBlond N. D., et al. 2018. Inhibition of adenosine monophosphate-activated protein kinase-3-hydroxy-3-methylglutaryl coenzyme A reductase signaling leads to hypercholesterolemia and promotes hepatic steatosis and insulin resistance. Hepatol. Commun. 3: 84–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohashi K., Osuga J., Tozawa R., Kitamine T., Yagyu H., Sekiya M., Tomita S., Okazaki H., Tamura Y., Yahagi N., et al. 2003. Early embryonic lethality caused by targeted disruption of the 3-hydroxy-3-methylglutaryl-CoA reductase gene. J. Biol. Chem. 278: 42936–42941. [DOI] [PubMed] [Google Scholar]
- 13.Yeh Y. S., Jheng H. F., Iwase M., Kim M., Mohri S., Kwon J., Kawarasaki S., Li Y., Takahashi H., Ara T., et al. 2018. The mevalonate pathway is indispensable for adipocyte survival. iScience. 9: 175–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lacher S. M., Bruttger J., Kalt B., Berthelet J., Rajalingam K., Wortge S., and Waisman A.. 2017. HMG-CoA reductase promotes protein prenylation and therefore is indispensible for T-cell survival. Cell Death Dis. 8: e2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Osaki Y., Nakagawa Y., Miyahara S., Iwasaki H., Ishii A., Matsuzaka T., Kobayashi K., Yatoh S., Takahashi A., Yahagi N., et al. 2015. Skeletal muscle-specific HMG-CoA reductase knockout mice exhibit rhabdomyolysis: A model for statin-induced myopathy. Biochem. Biophys. Res. Commun. 466: 536–540. [DOI] [PubMed] [Google Scholar]
- 16.Sakai K., Nagashima S., Wakabayashi T., Tumenbayar B., Hayakawa H., Hayakawa M., Karasawa T., Ohashi K., Yamazaki H., Takei A., et al. 2018. Myeloid HMG-CoA (3-hydroxy-3-methylglutaryl-coenzyme A) reductase determines atherosclerosis by modulating migration of macrophages. Arterioscler. Thromb. Vasc. Biol. 38: 2590–2600. [DOI] [PubMed] [Google Scholar]
- 17.Nagashima S., Yagyu H., Ohashi K., Tazoe F., Takahashi M., Ohshiro T., Bayasgalan T., Okada K., Sekiya M., Osuga J., et al. 2012. Liver-specific deletion of 3-hydroxy-3-methylglutaryl coenzyme A reductase causes hepatic steatosis and death. Arterioscler. Thromb. Vasc. Biol. 32: 1824–1831. [DOI] [PubMed] [Google Scholar]
- 18.Dietschy J. M., Turley S. D., and Spady D. K.. 1993. Role of liver in the maintenance of cholesterol and low density lipoprotein homeostasis in different animal species, including humans. J. Lipid Res. 34: 1637–1659. [PubMed] [Google Scholar]
- 19.Postic C., and Magnuson M. A.. 2000. DNA excision in liver by an albumin-Cre transgene occurs progressively with age. Genesis. 26: 149–150. [DOI] [PubMed] [Google Scholar]
- 20.Oesterle A., Laufs U., and Liao J. K.. 2017. Pleiotropic effects of statins on the cardiovascular system. Circ. Res. 120: 229–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ward N. C., Watts G. F., and Eckel R. H.. 2019. Statin toxicity. Circ. Res. 124: 328–350. [DOI] [PubMed] [Google Scholar]
- 22.Wolfe L. A., Morava E., He M., Vockley J., and Gibson K. M.. 2012. Heritable disorders in the metabolism of the dolichols: A bridge from sterol biosynthesis to molecular glycosylation. Am. J. Med. Genet. C. Semin. Med. Genet. 160C: 322–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park E. J., Grabinska K. A., Guan Z., Stranecky V., Hartmannova H., Hodanova K., Baresova V., Sovova J., Jozsef L., Ondruskova N., et al. 2014. Mutation of Nogo-B receptor, a subunit of cis-prenyltransferase, causes a congenital disorder of glycosylation. Cell Metab. 20: 448–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grabińska K. A., Edani B. H., Park E. J., Kraehling J. R., and Sessa W. C.. 2017. A conserved C-terminal RXG motif in the NgBR subunit of cis-prenyltransferase is critical for prenyltransferase activity. J. Biol. Chem. 292: 17351–17361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grabińska K. A., Park E. J., and Sessa W. C.. 2016. cis-Prenyltransferase: new insights into protein glycosylation, rubber synthesis, and human diseases. J. Biol. Chem. 291: 18582–18590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peng M., Falk M. J., Haase V. H., King R., Polyak E., Selak M., Yudkoff M., Hancock W. W., Meade R., Saiki R., et al. 2008. Primary coenzyme Q deficiency in Pdss2 mutant mice causes isolated renal disease. PLoS Genet. 4: e1000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang S. H., Chang S. Y., Tu Y., Lawson G. W., Bergo M. O., Fong L. G., and Young S. G.. 2012. Severe hepatocellular disease in mice lacking one or both CaaX prenyltransferases. J. Lipid Res. 53: 77–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wennerberg K., Rossman K. L., and Der C. J.. 2005. The Ras superfamily at a glance. J. Cell Sci. 118: 843–846. [DOI] [PubMed] [Google Scholar]
- 29.De Giorgi M., and Lagor W. R.. 2019. Gene delivery in lipid research and therapies. Methodist DeBakey Cardiovasc. J. 15: 62–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bauer R. C., Sasaki M., Cohen D. M., Cui J., Smith M. A., Yenilmez B. O., Steger D. J., and Rader D. J.. 2015. Tribbles-1 regulates hepatic lipogenesis through posttranscriptional regulation of C/EBPalpha. J. Clin. Invest. 125: 3809–3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pastore N., Huynh T., Herz N. J., Calcagni A., Klisch T. J., Brunetti L., Kim K. H., De Giorgi M., Hurley A., Carissimo A., et al. 2020. TFEB regulates murine liver cell fate during development and regeneration. Nat. Commun. 11: 2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ran F. A., Cong L., Yan W. X., Scott D. A., Gootenberg J. S., Kriz A. J., Zetsche B., Shalem O., Wu X., Makarova K. S., et al. 2015. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 520: 186–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jarrett K. E., Lee C., De Giorgi M., Hurley A., Gillard B. K., Doerfler A. M., Li A., Pownall H. J., Bao G., and Lagor W. R.. 2018. Somatic editing of Ldlr with adeno-associated viral-CRISPR is an efficient tool for atherosclerosis research. Arterioscler. Thromb. Vasc. Biol. 38: 1997–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McIntosh J., Lenting P. J., Rosales C., Lee D., Rabbanian S., Raj D., Patel N., Tuddenham E. G., Christophe O. D., McVey J. H., et al. 2013. Therapeutic levels of FVIII following a single peripheral vein administration of rAAV vector encoding a novel human factor VIII variant. Blood. 121: 3335–3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li A., Lee C. M., Hurley A. E., Jarrett K. E., De Giorgi M., Lu W., Balderrama K. S., Doerfler A. M., Deshmukh H., Ray A., et al. 2018. A self-deleting AAV-CRISPR system for in vivo genome editing. Mol. Ther. Methods Clin. Dev. 12: 111–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lagor W. R., Johnston J. C., Lock M., Vandenberghe L. H., and Rader D. J.. 2013. Adeno-associated viruses as liver-directed gene delivery vehicles: focus on lipoprotein metabolism. Methods Mol. Biol. 1027: 273–307. [DOI] [PubMed] [Google Scholar]
- 37.Tang Z., and Guengerich F. P.. 2010. Dansylation of unactivated alcohols for improved mass spectral sensitivity and application to analysis of cytochrome P450 oxidation products in tissue extracts. Anal. Chem. 82: 7706–7712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oslowski C. M., and Urano F.. 2011. Measuring ER stress and the unfolded protein response using mammalian tissue culture system. Methods Enzymol. 490: 71–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCarthy C., Saldova R., Wormald M. R., Rudd P. M., McElvaney N. G., and Reeves E. P.. 2014. The role and importance of glycosylation of acute phase proteins with focus on alpha-1 antitrypsin in acute and chronic inflammatory conditions. J. Proteome Res. 13: 3131–3143. [DOI] [PubMed] [Google Scholar]
- 40.Berndt N., and Sebti S. M.. 2011. Measurement of protein farnesylation and geranylgeranylation in vitro, in cultured cells and in biopsies, and the effects of prenyl transferase inhibitors. Nat. Protoc. 6: 1775–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Healy A., Berus J. M., Christensen J. L., Lee C., Mantsounga C., Dong W., Watts J. P. Jr., Assali M., Ceneri N., Nilson R., et al. 2020. Statins disrupt macrophage Rac1 regulation leading to increased atherosclerotic plaque calcification. Arterioscler. Thromb. Vasc. Biol. 40: 714–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cherepanova N., Shrimal S., and Gilmore R.. 2016. N-linked glycosylation and homeostasis of the endoplasmic reticulum. Curr. Opin. Cell Biol. 41: 57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zelinger L., Banin E., Obolensky A., Mizrahi-Meissonnier L., Beryozkin A., Bandah-Rozenfeld D., Frenkel S., Ben-Yosef T., Merin S., Schwartz S. B., et al. 2011. A missense mutation in DHDDS, encoding dehydrodolichyl diphosphate synthase, is associated with autosomal-recessive retinitis pigmentosa in Ashkenazi Jews. Am. J. Hum. Genet. 88: 207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sabry S., Vuillaumier-Barrot S., Mintet E., Fasseu M., Valayannopoulos V., Heron D., Dorison N., Mignot C., Seta N., Chantret I., et al. 2016. A case of fatal type I congenital disorders of glycosylation (CDG I) associated with low dehydrodolichol diphosphate synthase (DHDDS) activity. Orphanet J. Rare Dis. 11: 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hu W., Zhang W., Chen Y., Rana U., Teng R. J., Duan Y., Liu Z., Zhao B., Foeckler J., Weiler H., et al. 2016. Nogo-B receptor deficiency increases liver X receptor alpha nuclear translocation and hepatic lipogenesis through an adenosine monophosphate-activated protein kinase alpha-dependent pathway. Hepatology. 64: 1559–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mollet J., Giurgea I., Schlemmer D., Dallner G., Chretien D., Delahodde A., Bacq D., de Lonlay P., Munnich A., and Rotig A.. 2007. Prenyldiphosphate synthase, subunit 1 (PDSS1) and OH-benzoate polyprenyltransferase (COQ2) mutations in ubiquinone deficiency and oxidative phosphorylation disorders. J. Clin. Invest. 117: 765–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.López L. C., Schuelke M., Quinzii C. M., Kanki T., Rodenburg R. J., Naini A., Dimauro S., and Hirano M.. 2006. Leigh syndrome with nephropathy and CoQ10 deficiency due to decaprenyl diphosphate synthase subunit 2 (PDSS2) mutations. Am. J. Hum. Genet. 79: 1125–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Quinzii C. M., Garone C., Emmanuele V., Tadesse S., Krishna S., Dorado B., and Hirano M.. 2013. Tissue-specific oxidative stress and loss of mitochondria in CoQ-deficient Pdss2 mutant mice. FASEB J. 27: 612–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Falk M. J., Polyak E., Zhang Z., Peng M., King R., Maltzman J. S., Okwuego E., Horyn O., Nakamaru-Ogiso E., Ostrovsky J., et al. 2011. Probucol ameliorates renal and metabolic sequelae of primary CoQ deficiency in Pdss2 mutant mice. EMBO Mol. Med. 3: 410–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tomasetti M., Alleva R., Solenghi M. D., and Littarru G. P.. 1999. Distribution of antioxidants among blood components and lipoproteins: significance of lipids/CoQ10 ratio as a possible marker of increased risk for atherosclerosis. Biofactors. 9: 231–240. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article and supplemental file.