Abstract
NAFLD is an important public health issue closely associated with the pervasive epidemics of diabetes and obesity. Yet, despite NAFLD being among the most common of chronic liver diseases, the biological factors responsible for its transition from benign nonalcoholic fatty liver (NAFL) to NASH remain unclear. This lack of knowledge leads to a decreased ability to find relevant animal models, predict disease progression, or develop clinical treatments. In the current study, we used multiple mouse models of NAFLD, human correlation data, and selective gene overexpression of steroidogenic acute regulatory protein (StarD1) in mice to elucidate a plausible mechanistic pathway for promoting the transition from NAFL to NASH. We show that oxysterol 7α-hydroxylase (CYP7B1) controls the levels of intracellular regulatory oxysterols generated by the “acidic/alternative” pathway of cholesterol metabolism. Specifically, we report data showing that an inability to upregulate CYP7B1, in the setting of insulin resistance, results in the accumulation of toxic intracellular cholesterol metabolites that promote inflammation and hepatocyte injury. This metabolic pathway, initiated and exacerbated by insulin resistance, offers insight into approaches for the treatment of NAFLD.
Keywords: cholesterol toxicity, oxysterol 7α-hydroxylase, inflammation, liver injury, nonalcoholic fatty liver disease, nonalcoholic steatohepatitis, oxysterol, nonalcoholic fatty liver
NAFLD has emerged as the most common chronic liver disease and is soon to become a major cause for liver cancer and transplantation (1). With one-fourth of the population affected, it will have a dramatic global economic impact on healthcare systems and patient’s well-being (2). NAFLD is a disease spectrum ranging from simple steatosis [nonalcoholic fatty liver (NAFL)] to steatohepatitis (NASH) with or without liver fibrosis or cirrhosis. The natural history of disease progression is complex, supported by findings of rapid fibrosis progression on one hand and spontaneous disease regression on the other hand (3, 4). Unfortunately, we do not have a pathophysiology-driven explanation for the observed phenotypic variability of this liver disease.
NAFLD is considered as the hepatic manifestation of the metabolic syndrome, as it is strongly associated with conditions of dietary excess such as obesity, dyslipidemia, type 2 diabetes mellitus (T2DM), and hypertension (5). Insulin resistance and lipid excess within the liver are considered as the earliest markers or evidence of progression of NAFLD followed by inflammation and subsequent fibrosis (6). Interestingly, excess liver lipid is contributed to by increased de novo fatty acid synthesis within the liver, further exacerbating dietary induced liver lipid excess (7). What complicates this relatively straightforward sequence of metabolic events is that only a small percentage of those with fatty liver progress to NASH. The amount of lipid excess does not seem to correlate with disease activity or progression, i.e., the presentation of liver lipid excess that occurs in lean NASH patients (8, 9). Furthermore, the coexistence of NASH in type 1 diabetes mellitus suggests other causes than insulin resistance promoting fatty liver to NASH (10). In the absence of a common thread, NAFLD has become described as a complex metabolic entity of liver lipid excess. It is obvious that only a better understanding of the pathophysiologic derangement promoting progression from fatty liver to NASH will allow us to develop effective treatment approaches other than weight loss.
Our laboratory hypothesized that liver lipid excess in NAFLD and NASH might be linked to dysregulation of cholesterol metabolism through the alternative pathway of bile acid synthesis (Fig. 1A) (11). As previously hypothesized by Javitt and colleagues and our own group, this pathway was the initial pathway of bile acid synthesis; which was likely not originally a pathway to form bile acids, but a pathway that controlled the levels of the regulatory oxysterols regarded as key metabolic regulators of both cholesterol and lipid biosynthesis (12–14). Moreover, the inner mitochondrial membrane has low levels of cholesterol and the alternative pathway may have also evolved to remove excess cholesterol via side-chain oxidation by sterol 27-hydroxylase (CYP27A1) and further metabolism by oxysterol 7α-hydroxylase (CYP7B1). As a vital metabolic pathway, it is pervasive within body tissues. The ultimate ability to form bile acids likely developed within hepatocytes as a way to aid the absorption of dietary lipids. However, an inability to increase rates of bile acids to aid in the absorption of larger amounts of dietary lipids without generating toxic amounts of oxysterols and their metabolic products likely led to the neutral pathway of bile acid synthesis; a pathway where 7α-hydroxylation of cholesterol is the initial metabolic step allowing for the synthesis of nonregulatory less toxic metabolites (14).
Fig. 1.
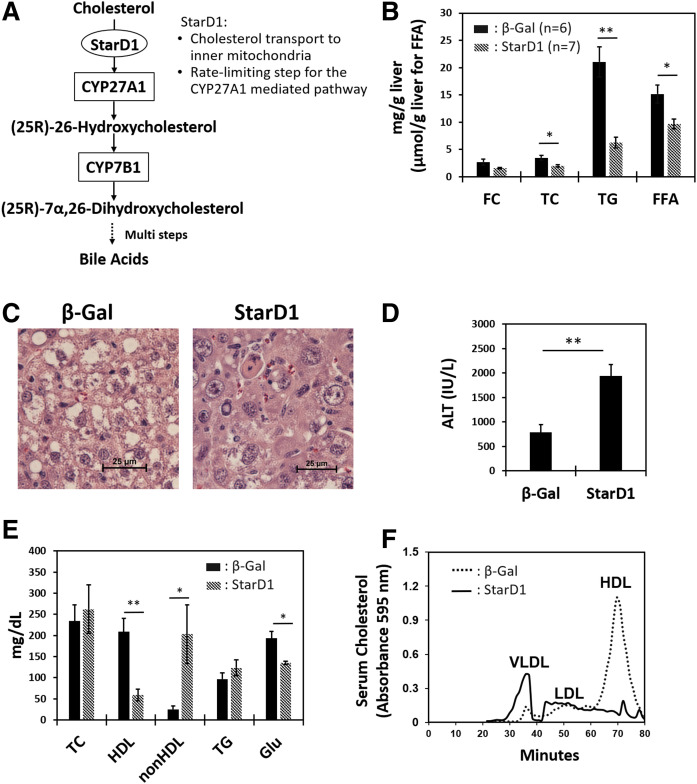
StarD1 overexpression leads to removal of cholesterol/lipids from livers of WD-fed B6/129 mice. StarD1 and β-Gal as control were overexpressed for the last 7 days in male mice fed a WD (Harlan-Teklad TD.88137) for 14 days. A: Key steps of the alternative pathway of bile acid synthesis in hepatocytes. B: StarD1 overexpression decreased liver TC, FC, FFA, and TG levels. C: Liver histology by H&E staining (40×) visually confirmed the measured biochemical reduction in liver lipid levels found with increased StarD1 expression. D: Increased ALT was observed in StarD1-overexpressed mice. E: Serum biochemical analysis revealed an increase in non-HDL (VLDL + LDL) level and a marked decrease in HDL and glucose (Glu). F: FPLC analysis demonstrated a marked increase in serum VLDL coupled to a dramatic decrease in the HDL fraction; findings consistent with serum biochemical analysis shown in panel E. β-Gal control, n = 6; StarD1 overexpression, n = 7. Welch’s t-tests were performed between β-Gal control and StarD1-overexpressed mice, and significance is indicated by *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001.
In this regard, Setchell et al. (15) reported that the absence of CYP7B1 in children led to oxysterol-induced liver inflammation and cirrhosis within the first year of life. Dai et al. (16) subsequently showed that the absence of CYP7B1 led first to the presence of fat within the liver with subsequent liver inflammation in a pattern found in the transition from fatty liver to NASH. Interestingly, Tang, Pettersson, and Norlin (17) showed that upregulation of the insulin signal pathway seemed able to upregulate Cyp7b1. Biddinger et al. (18), in a cholesterol gallstone mouse model, reported that Cyp7b1 mRNA levels were markedly suppressed in liver selective insulin receptor knockout mice. However, the potential importance of regulation of CYP7B1 by insulin has never been truly addressed in the context of NAFLD and NASH.
NAFLD is a rheostat of disease. Within the existing literature, the timing of the transition of fatty liver to NASH and to fibrosis is not clearly delineated, i.e., lacking a clear starting point from which to initiate investigation. As a result, the lines of initiation appear to have become blurred within the multitude of ensuing metabolic consequences of cellar toxicity, inflammatory cell infiltrate, cell to cell interactions, collagen synthesis, liver architectural changes, and increased cell metabolic contents such as bile acids associated with cholestasis. Thus, establishing inciting events appeared unlikely in the absence of coupling several early-event in vivo models. In the current study, we chose to pursue five overlapping mouse models of NAFL, coupled to human data, and describe evidence for a new model explaining how the dysregulation in cellular oxysterol levels initiated by the chronic suppression of CYP7B1 plays a role in the transition from NAFLD to NASH.
METHODS
Materials and chemicals
All chemicals were of the highest purity commercially available. HPLC grade solvents were purchased from Fisher Scientific (New Lawn, NJ), and all other reagents were from Sigma-Aldrich, Inc. (St. Louis, MO), unless indicated otherwise: oxysterol standards, 7α-hydroxycholesterol (7α-HC), 24(S)-hydroxycholesterol (24-HC), 25-hydroxycholesterol (25-HC), (25R)-26-hydroxycholesterol (26-HC), 7α,24(S)-dihydroxycholesterol (7,24-diHC), 7α,25-dihydroxycholesterol (7α,25-diHC), and (25R)-7α,26-dihydroxycholesterol (7α,26-diHC) were purchased from Avanti Polar Lipids, Inc. (Alabaster, AL). d6-25-HC (26,26,26,27,27,27-[2H6]25-HC) was purchased from Cayman Chemical (Ann Arbor, MI). Tauro (T) α-Muricholic acid (MCA), αMCA, TβMCA, βMCA, TωMCA, ωMCA, cholic acid (CA)-3-sulfate (S), chenodeoxycholic acid (CDCA)-3S, ursodeoxycholic acid (UDCA)-3S, deoxycholic acid (DCA)-3S, lithocholic acid (LCA)-3S, glyco (G)CA-3S, GCDCA-3S, GUDCA-3S, GDCA-3S, GLCA-3S, TCA-3S, TCDCA-3S, TUDCA-3S, TDCA-3S, and TLCA-3S were kind gifts from Professor Takashi Iida (Tokyo, Japan). [2,2,4,4-d4]CA [d4-CA, internal standard (IS) for unconjugated bile acids], [2,2,4,4-d4]GCA (d4-GCA, IS for glycine-conjugated bile acids), and [2,2,4,4-d4]TCA (d4-TCA, and IS for taurine-conjugated and double-conjugated bile acids) were obtained from C/D/N Isotopes, Inc. (Pointe-Claire, Quebec, Canada). All other oxysterol standards and bile acids were from Steraloids, Inc. (Newport, RI). SV Total RNA Isolation System was obtained from Promega Co. (Madison, WI). Cyp7b1 antibody was available from Proteintech Group Inc. (Rosemont, IL). Anti-GRP78 (BiP) and anti-GAPDH antibodies were from Abcam (Cambridge, MA). Monoclonal anti-CCAAT-enhancer-binding homologous protein (CHOP) antibody was from Thermo Fisher Scientific (Waltham, MA). Human liver tissues were obtained from the National Institutes of Health (NIH)-sponsored Liver Tissue Distribution Center at the University of Minnesota.
Animal models
An F2 intercross of the mouse strains 129S1/SvlmJ and C57Bl/6J (B6/129) that develop classic fatty liver with progression to NASH on a supplemental WD chow, were obtained from Dr. Sandra Erickson (University of California, San Francisco). The mice were bred at the animal facility of McGuire Veterans Affairs Medical Center with a 12 h light cycle (6:00 AM to 6:00 PM). All mice had free access to water. Unless specified differently, mice were fed an ad libitum WD with 42% of calories from fat and 43% from carbohydrates (Harlan-Teklad TD.88137; Envigo, Frederick, MD) for 2–6 weeks. One group of mice was prolonged on the WD feeding for 32 weeks for reaffirming the B6/129 mouse as previously characterized as a viable NASH model. Controls were age-matched B6/129 mice fed a regular chow (RC; irradiated Teklad LM-485). All mice were 12 weeks old at the time they were euthanized unless otherwise noted. For the study of in vivo steroidogenic acute regulatory protein (StarD1) overexpression (Figs. 1–3), Ad-StarD1 or Ad-β-galactosidase (β-Gal; for control) recombinant virus (1 × 109 pfu) was injected into the tail vein of B6/129 mice on the seventh day from start of WD feeding. These adenovirus constructs were prepared as previously described (19). WD feeding was continued for another week, and the mice were euthanized on the fourteenth day. Blood was centrifuged at 1,620 g for 15 min at 4°C, and serum in the supernatant was collected. All specimens were snap-frozen in liquid nitrogen and stored at −78°C until further analysis. A portion of liver was also fixed in 10% neutral buffered formalin and sent to the Department of Pathology at Virginia Commonwealth University to be processed and stained with H&E. Images were taken with a Nikon Ti-U microscope and NIS Elements software.
Fig. 3.
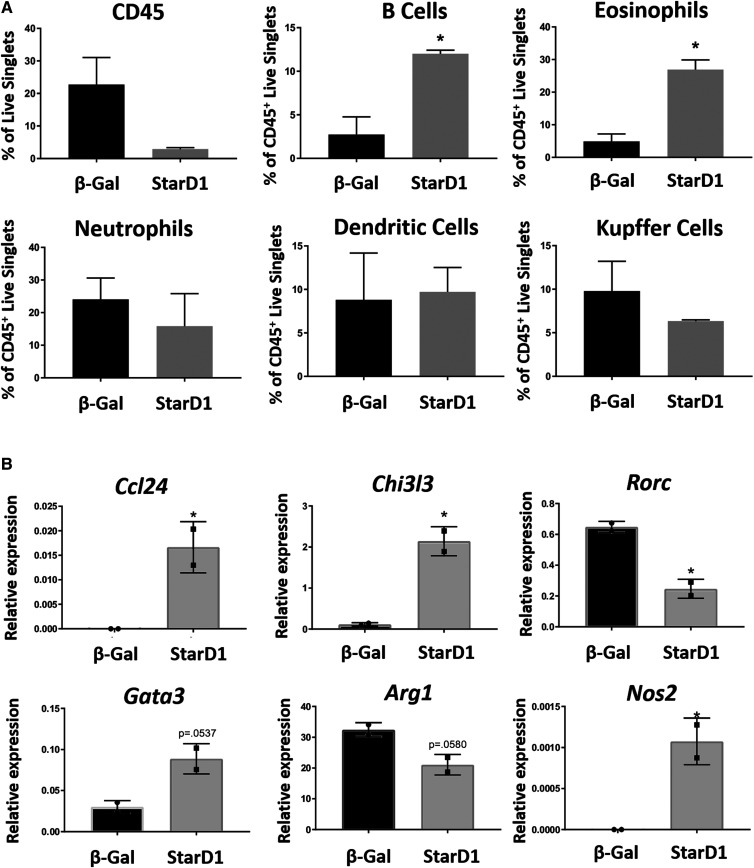
Sterile inflammation is associated with increased oxysterol formation in mice. StarD1 was overexpressed in B6/129 male mice to drive endogenous oxysterol formation as outlined in the Methods. Mouse livers were evaluated at day 14, which was 7 days following StarD1 overexpression, by both flow cytometry and qPCR. All cells were gated off of live singlets. A: Measurement of total percent hematopoietic cells (CD45+), percent B cells (CD45+B220+MHCII+), percent eosinophils (CD45+CD11b+CCR3+), percent neutrophils (CD45+CD11bhiF4/80lowLy6G+Ly6C+), percent dendritic cells (CD45+CD11c+MHCII+), and percent Kupffer cells (CD45+CD11b+F4/80+). B: Normalized relative mRNA expression of inflammation markers related to NAFLD, normalized to the housekeeping gene (Tbp). Statistics were performed as a Student’s t-test between groups in A and a Mann-Whitney nonparametric comparison in B. Error bars depict SEM. *P ≤ 0.05 versus β-Gal control.
The STAM™ mice were prepared and characterized at SMC Laboratories Inc. (Tokyo, Japan) as previously described (20). Briefly, 200 μg of streptozotocin (STZ) were subcutaneously injected into C57Bl/6J male mice at 2 days after birth. The mice were fed a low-fat RC diet (CE-2; CLEA Japan Inc.) until 4 weeks of age, and then fed an ad libitum HFD (HFD32; CLEA Japan Inc) until 8 weeks (NASH stage). STZ injection led to approximately 60% of β cells being impaired when they were evaluated at 9 weeks of age (supplemental Fig. S1). One group of mice was continued on their RC diet for an additional 4 weeks (fed RC for 8 weeks).
All mice were fasted 4 h prior to being euthanized unless otherwise noted. At the time of euthanasia, mice were subjected to isoflurane inhalation. All animal protocols were approved by the IACUC of the McGuire Veterans Affairs Medical Center and the Virginia Commonwealth University; both are accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International and comply with the Guide for Care and Use of Laboratory Animals published by the NIH.
Measurement of serum and liver biochemical parameters
Serum total cholesterol (TC), HDL-C, LDL-C, TGs, glucose, aspartate aminotransferase (AST), alanine aminotransferase (ALT), and alkaline phosphatase levels were measured by the enzymatic procedures run on Siemens Vista 1500 instrumentation. Serum insulin was measured by the sensitive ELISA sandwich assay method using Crystal Chem ultra-sensitive mouse insulin ELISA kit (Elk Grove Village, IL) according to manufacturer’s instructions. For plasma insulin measurement of the STZ-treated mice, Ultra-Sensitive Mouse Insulin ELISA kit (Morinaga Institute of Biological Science, Inc., Yokohama, Japan) was used. Plasma glucagon of C57Bl/6J mice was measured using Glucagon Quantikine ELISA kit (R&D Systems, Inc., Minneapolis, MN). Liver TC and free cholesterol (FC) were measured by enzymatic assay using the Cholesterol E and Free Cholesterol E kits (FUJIFILM Wako Chemicals U.S.A. Co, Richmond, VA). Infinity Triglycerides kit (Thermo Fisher Scientific) was used for liver TG measurement. The serum lipoprotein profile was obtained by the FPLC using an Agilent 1100 series HPLC system as follows: 100 μl of serum was injected onto a Superose 6, 10/300 GL column (GE Healthcare, Chicago, IL). NaCl (154 mM) containing 1 mM EDTA (pH 8.0) was used for the elution buffer at a flow rate of 0.2 ml/min. Effluents were monitored at 280 nm (21). Lipoproteins were identified by coelution with VLDL, LDL and HDL standards (Sigma-Aldrich). Starting at 20 min, fractions were collected every 1.2 min (240 μl/fraction) until 100 min. Cholesterol content was measured by adapting the Cholesterol E kit, adding 20 μl of a 10× reagent buffer to 180 μl of each fraction in a 96-well plate, heating at 37°C, and measuring absorbance at 595 nm.
Measurement of nonparenchymal cells from B6/129 mouse livers
Primary hepatocytes were removed from the livers of the StarD1-overexpressed B6/129 mice using the collagenase-perfusion technique as previously described (22). Briefly, the liver was digested with 0.025% collagenase in William’s E culture medium (40 ml) at 37°C for 10 min. The digest was filtered through a double thickness of sterile gauze. Hepatocytes were removed by centrifugation (60 g at 4°C for 5 min), and nonparenchymal cells (approximately 9.0 × 106 cells) in the supernatant were collected. The live-dead staining was conducted using Zombie Aqua (#423102; BioLegend, San Diego, CA) according to manufacturer’s protocol. Cells were washed with FACS buffer (5% FBS in PBS with 2 mM EDTA). Fc receptors were blocked with 5 μg 2.4G2 for 10 min at 4°C. Antibodies were added for 45 min at 4°C. Cells were washed twice with FACS buffer and fixed in fixation buffer (BioLegend, #420801) for 10 min at room temperature. Flow cytometry data were collected on a BD LSR Fortessa™ X-20 (BD, Franklin Lakes, NJ) and analyzed in FlowJo (BD) software, all as previously described (23). Antibodies used in this study were as follows: APC-conjugated anti-mouse CD45, PE/Cy7-conjugated anti-mouse F4/80, PE-conjugated anti-mouse CCR3, BV421-conjugated anti-mouse MHCII, FITC-conjugated anti-mouse CD11c, PE-conjugated anti-mouse B220, APCFIRE750-conjugated anti-mouse Ly6C, FITC-conjugated anti-mouse Ly6G (all from BioLegend), and BUV395-conjugated anti-mouse CD11b (BD).
Real-time quantitative PCR
Total RNA from the liver tissue was isolated with the Promega Total RNA Isolation system according to manufacturer’s protocol. qPCR was performed with 2 μg of RNA using the Affimetrix (Santa Clara, CA) VeriQuest SYBR Green PCR Master Mix in an Applied Biosystems 7500 PCR system (Foster City, CA). Primers are listed in supplemental Table S2. All data were produced in triplicates for each mRNA. The comparative CT method was used to calculate relative quantification of gene expression. The mRNA expression levels obtained for each gene were normalized to the expression of the Gapdh housekeeping gene by using the following equation: relative mRNA expression = 2−(Ct of each gene−Ct of Gapdh) (where Ct is the threshold cycle). For Fig. 3, total RNA was extracted from the nonhepatocyte-enriched fraction using Invitrogen™ TRIzol™ reagent per the manufacturer’s instructions. RNA was quantified using a NanoDrop ND-100 spectrophotometer. One microgram of total RNA was reverse transcribed using SuperScript IV (Thermo Fisher Scientific) with oligo(dT20). The mRNA expression levels obtained for each gene were normalized to the expression level of the housekeeping gene, tbp: Normalized relative expression = 2−ΔΔCt (24).
Western blot
Protein samples from B6/129 mouse livers were separated on 12% SDS-PAGE gel and then transferred onto a PVDF membrane using a Bio-Rad semidry transfer cell apparatus. The membrane was incubated in TBST (TBS and Polysorbate 20) with 5% nonfat dry milk for 2 h at room temperature. The membrane was then incubated overnight in TBST with 2.5% nonfat dry milk containing a dilution of a primary antibody at 4°C, as indicated. The membrane was then washed three times in TBST for 30 min at room temperature. After washing, the membrane was incubated in a 1:2,000 dilution of the corresponding HRP-conjugated IgG in TBST with 2.5% nonfat dry milk for 1.5 h at room temperature. Finally, the membrane was washed three more times in TBST. Protein bands were visualized using SuperSignal West Pico chemiluminescent substrate and developed on a Bio-Rad ChemiDoc Touch imaging system. Quantification of protein expression levels of Cyp7b1, BiP, and GAPDH were performed using ImageJ software. Relative levels of Cyp7b1 and BiP expression were normalized by GAPDH.
Oxysterol analysis in the liver tissue
Quantification of oxysterols was performed by LC/ESI-MS/MS according to the reported method (25) with modification: Liver tissue (100 mg) was digested in 1.0 mg/ml Proteinase K (0.5 ml in water) at 55°C for 6 h. Chloroform/methanol (1:2, v/v, 2 ml) was added to the digest, and the mixture was ultra-sonicated in a Branson type B-220 ultra-sonic bath (Danbury, CT) for 30 min. After centrifugation at 1,620 g for 5 min, the supernatant was collected in a clean glass tube. This procedure was repeated twice, and the combined extract was evaporated to dryness under a nitrogen stream at 40°C. The residue was redissolved in ethanol (1 ml). Aliquots (100 μl each) of this solution were used for oxysterol and bile acid analysis (see next section). For oxysterol analysis, a 100 μl aliquot of this solution was mixed with 10 μl of d6-25-HC (100 pmol/ml in methanol) and 10 μl of butylated hydroxytoluene (5 mg/ml in ethanol). After adding water (0.5 ml) and n-hexane (1.0 ml), the mixture was thoroughly vortexed (approximately 2 min), centrifuged at 180 g for 5 min, and the organic upper layer was collected in a glass tube. The bottom layer was washed again with n-hexane (1.0 ml) by the same procedure, and the combined extract was evaporated under reduced pressure. The residue was redissolved in n-hexane (0.5 ml) and loaded onto an InertSep NH2 cartridge (100 mg/1 ml; GL Sciences Inc., Tokyo, Japan) to remove cholesterol. Prior to loading the sample, the column was rinsed with 1.0 ml of chloroform/methanol (1:1) and conditioned with 3 ml of n-hexane. After loading the sample, the column was washed with n-hexane (1.0 ml), and the desired oxysterols were eluted with 1.0 ml of chloroform/methanol (20:1). Oxysterols were then derivatized to nicotinyl ester form according to the method by Sidhu et al. (25) with modifications. Derivatization reagent (100 μl), which was a mixture of nicotinic acid (80 mg), N,N′-dimethyl-4-aminopyridine (30 mg), and 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC; 100 mg) in N,N′-dimethylformamide (1.0 ml), was added to the oxysterol sample, and the mixture was incubated at 60°C for 1 h. Water (0.5 ml) and n-hexane (1.0 ml) were added, and the solution was thoroughly vortexed (approximately 2 min). After centrifugation at 180 g for 5 min, the n-hexane layer was collected and evaporated under a reduced pressure. The residue was dissolved in acetonitrile (100 μl), and an aliquot (10 μl) was injected into a Shimadzu LCMS-8050 system equipped with an ESI probe. A Nexera X2 HPLC pump with SunShell C30 column (100 × 2.1 mm inner diameter, 2.6 μm particle size; ChromaNik Tecnology Inc., Osaka, Japan) was used for the chromatographic separation. The column was heated at 35°C. Mobile phase A (5 mM ammonium acetate with 0.1% formic acid) and mobile phase B [isopropanol-acetonitrile (3:2) containing 0.1% formic acid] were used for gradient elution as follows: 0–2 min 70% B, 2–6 min 70–76% B, 6–16 min 78% B, 16–18 min 96% B, 18–20 min 96% B, 20–20.1 min 96–70% B, and 20.1–25 min 70% B. Flow rate was kept constant at 0.25 ml/min. The nebulizer gas flow was set at 3.0 liters per minute, and the heating gas and the drying gas were set at 10 liters per minute. The interface temperature was 300°C; the desolvation line temperature was 250°C; and the heat block temperature was set at 400°C with an interface voltage of 4,000 V. The collision gas (argon) pressure was 270 kPa. Quantification of all analytes was achieved in positive multiple reaction monitoring mode (supplemental Table S1). Oxysterols were quantified according to the calibration curve constructed by IS against known concentration of authentic standards.
Bile acid analysis in liver tissue and gallbladder bile
A 100 μl aliquot of the liver extract suspended in ethanol (see the Oxysterol analysis in the liver tissue section above) was combined with 100 μl of IS solution, which was a mixture of 1.0 nmol/ml of d4-CA, d4-GCA, d4-TCA, d4-GCDCA, d4-TCDCA, d5-CDCA-3S, d5-GCDCA-3S, and d5-TCDCA-3S in 50% ethanol. After dilution with water (800 μl), the mixture was loaded to an InertSep C18-B cartridge (100 mg/1 ml), which was preconditioned with 1 ml of methanol and 3 ml of water. The cartridge was washed with water (1 ml) and the desired bile acids were eluted with 1 ml of 90% ethanol. After evaporation of the solvent, the residue was dissolved in 1 ml of 20% acetonitrile and then 5 μl of this solution was injected into the Shimadzu LCMS-8050 system. An InertSustain C18 column (150 × 2.1 mm inner diameter, 3 μm particle size; GL Sciences) was used at 40°C. Mobile phase A (5 mM ammonium acetate) and mobile phase B (acetonitrile) were used for gradient elution as follows: 0–0.5 min 14% B, 0.5–5 min 14–22% B, 5–36 min 22–60% B, 36–46 min 60–98% B, 46–50 min 98% B, 50–50.1 min 98–14% B, and 50.1–56.0 min 14% B. Flow rate was kept constant at 0.2 ml/min. The nebulizer gas flow was set at 3.0 liters per minute. The heating gas and the drying gas were set at 10 liters per minute. The interface temperature, desolvation line temperature, and heat block temperature were set at 300°C, 250°C, and 400°C, respectively. The interface voltage was −3,000 V. Quantitative analysis was performed using multiple reaction monitoring transition pairs for each analyte in negative ion mode. For unconjugated bile acids that did not yield major fragment, the same mass was monitored for both parent and daughter ions. Bile acids were quantified according to the calibration curve constructed by IS against known concentration of authentic standards. Supplemental Table S1 presents bile acids and their abbreviations quantified in this study.
For gallbladder bile acid analysis (Fig. 2B), a HPLC method was used (26). Bile (2–5 μl) was diluted with sodium acetate buffer [100 mM (pH 5.6), 250 μl] containing 15 units of cholylglycine hydrolase (Clostridium welchii; Sigma-Aldrich, #C4018) and 150 units of sulfatase (Type H-1; Sigma-Aldrich, #S9626), and the mixture was incubated at 37°C for 16 h. After adding isopropanol (250 μl), norDCA (50 nmol, IS), and 0.1 N NaOH (3 ml), the mixture was applied to a Waters Sep Pak tC18 cartridge (500 mg sorbent), which had been primed with methanol (10 ml) and water (10 ml). The cartridge was washed by water (5 ml), 10% acetone (4 ml), and water (5 ml). Retained bile acids were eluted with methanol (6 ml). Methanol was evaporated under a nitrogen stream. Bile acids were derivatized to their 24-phenacyl esters and analyzed by HPLC-UV as described (26). Each bile acid was quantified as total of unconjugated, 24-aminoacyl conjugated (taurine and glycine), 3-sulfated, 3-glucuronidated, and multi-conjugated forms. “Cholic acids (CAs)” is defined as the total of CA, TCA, GCA, CA3S, CA3G, TCA3S, and GCA3S.
Fig. 2.
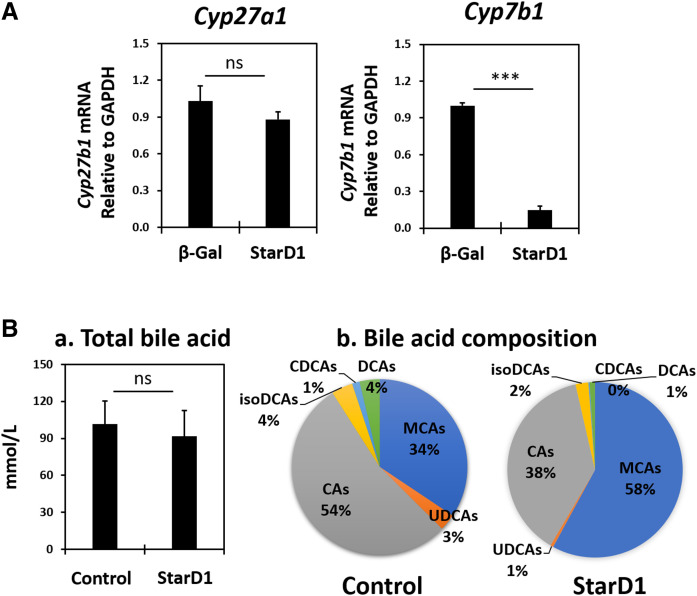
StarD1 overexpression in B6/129 mice led to hepatic Cyp7b1 mRNA downregulation without altering gallbladder total bile acid concentration. A: Effect of Ad-StarD1 overexpression on hepatic Cyp7b1 and Cyp27a1 mRNA expression. B: Gallbladder bile acid concentration (a) and composition (b) of StarD1-overexpressed B6/129 male mice. Bile acids were measured by HPLC after deconjugation (see Methods). Each bile acid was the sum of unconjugated, 24-aminoacyl conjugated, 3-sulfated, 3-glucuronidated, and double-conjugated forms of bile acids. i.e., CAs represents the total of CA, TCA, GCA, CA3S, CA3G, TCA3S, and GCA3S. MCAs include three isomers (α-, β-, and ω-MCA) and their conjugated forms. Abbreviations are shown in supplemental Fig. S1 (n = 4 in each group). Welch’s t-tests were performed between β-Gal control and StarD1-overexpressed mice and significance indicated by ***P ≤ 0.001.
Statistical analysis
All data are reported as mean ± SE. Where indicated, data were subjected to Welch’s t-test and determined to be significantly different at P ≤ 0.05.
RESULTS
StarD1 overexpression in B6/129 male mice reduced liver lipids but increased ALT and serum non-HDL
The data in Fig. 1 presents the effect of adenoviral StarD1 overexpression on lipid levels in WD-fed B6/129 male mice. We have previously reported that increasing hepatic StarD1 gene expression in mice increases bile acid synthesis (27). As liver cholesterol and lipid content are coordinately regulated, we hypothesized that increasing cholesterol metabolism would not only decrease liver cholesterol but also decrease liver lipid. StarD1 and β-Gal (as control) were overexpressed for the last 7 days in male mice fed a WD for 14 days. StarD1 overexpression significantly decreased hepatic TC, TG, and FFA levels (Fig. 1B). Liver histology with H&E staining confirmed the biochemical reduction in the hepatic lipid levels found with increased StarD1 expression (Fig. 1C). Interestingly, despite lowered hepatic lipids, a marked increase in ALT (3.5-fold), a marker of hepatic injury, was observed in the StarD1-overexpressing mice (Fig. 1D). Additionally, serum TG levels were slightly increased (Fig. 1E). More specifically, an increase in non-HDL (VLDL + LDL) coupled to a decrease in HDL in serum was observed. Serum glucose was significantly lowered with StarD1 overexpression. FPLC revealed an increase in VLDL coupled to a decrease in the HDL (Fig. 1F), which was consistent with serum biochemical analysis. These results suggested that the decrease in hepatic lipids with increased expression of StarD1 in B6/129 mice was secondary to facilitated hepatic lipid secretion into the serum.
StarD1 overexpression in B6/129 male mice led to lower level of hepatic Cyp7b1 mRNA expression
The data in Fig. 2A presents the effect of adenoviral StarD1 overexpression on Cyp27a1 and Cyp7b1 mRNA, two key genes involved in oxysterol synthesis in mitochondria and their subsequent metabolism, as previously described (14). With StarD1 overexpression in B6/129 male mice, Cyp7b1 mRNA expression level was only 15% of control, while the Cyp27a1 mRNA level remained unchanged. We have recently shown that decreased Cyp7b1 mRNA expression inversely correlated with increased oxysterol levels following StarD1 overexpression (14). More specifically, increased expression of StarD1 significantly increased 24-HC and 26-HC levels in B6/129 mouse liver. Furthermore, as Setchell et al. (15) reported, the absence of CYP7B1 in humans led to liver inflammation and subsequent cirrhosis within the first year of life in association with accumulation of 24-HC, 25-HC, 26-HC and 3β-hydroxy-5-cholestenoic acid (3β-HCA). These observations prompted us to hypothesize that excess 24-HC and 26-HC, and/or their metabolites, might lead to liver toxicity and be the cause of increased ALT in the StarD1 overexpression mouse model. StarD1 overexpression previously has been shown to increase rates of bile acid synthesis in mice and rats (28). Therefore, bile acid-induced hepatocyte toxicity was also considered. However, the findings were not supportive: gallbladder bile acid analysis (representative of enterohepatic circulating pool) showed that total bile acid concentrations were unchanged with/without StarD1 overexpression (Fig. 2B). Furthermore, elevated composition of muricholic acids (MCAs) with reduced CA derivatives (CAs), in contrast to those in control mice, created a more hydrophilic bile acid pool. Cyp8b1 mRNA levels in WD-fed mice corresponded with reduced CAs, being ∼20% of control RC-fed mice (P ≤ 0.001, n = 4) (data not shown in Fig. 2). Therefore, bile acid composition or concentration seemed unlikely as either a direct or indirect cause of liver inflammation.
StarD1 overexpression led to sterile inflammation
In order to assess sterile inflammation in association with increased oxysterol formation, StarD1-overexpressed B6/129 mouse livers were further evaluated. After hepatocytes were removed, nonparenchymal cells from B6/129 mouse livers were measured by both flow cytometry and qPCR. CD45+ antigen-positive cell analysis (Fig. 3A) showed a significant increase in the percentage of both B cells and eosinophils in StarD1-overexpressed mice compared with β-Gal controls. There was no observable difference between neutrophils, dendritic cells, or Kupffer cells. We additionally analyzed the nonhepatocyte portion of liver by qPCR (Fig. 3B). The gene expression for chemokine (C-C motif) ligand 24 (Ccl24), chitinase-like 3 (Chi3l3), and nitric oxide synthase 2 (Nos2) were all significantly elevated in StarD1-overexpressed mice compared with β-Gal controls. GATA binding protein 3 (Gata3) expression appeared increased, but differences did not reach statistical significance. RAR-related orphan receptor C (Rorc) expression was significantly reduced in StarD1-overexpressed mice compared with β-Gal controls, while arginase 1 (Arg1) expression trended down.
WD feeding of B6/129 male mice led to lower hepatic Cyp7b1 mRNA level of expression and accumulation of intercellular 26-HC and 3β-HCA
Based on findings obtained in the StarD1 overexpression model, we examined the role of Cyp7b1 and hepatic oxysterols with NAFLD progression using a diet-induced NASH mouse model. Male B6/129 mice were fed a WD for 2–6 weeks to induce fatty liver. Table 1 presents body weight, liver lipids, and serum biochemical parameters of these mice. With longer periods of WD feeding, body weight and hepatic TG (lipid) and cholesterol contents became increasingly elevated, evidence for developing fatty liver. Liver histology (H&E staining) (Fig. 4A) showed obvious lipid accumulation, but there was no histologic evidence of inflammatory cell infiltrates in 2- to 6-week WD-fed B6/129 mice. Prolonged WD feeding for 32 weeks led to characteristic inflammatory nodule formation, as seen in NASH, reaffirming the B6/129 mouse as previously characterized as a viable NASH model. Despite the absence of visual histologic evidence of inflammatory cell infiltrate, 2 week WD-fed mice had a more than 2-fold elevation of ALT and AST (Table 1). These data suggested that early biochemical inflammation was already occurring with just 2 weeks of WD feeding.
TABLE 1.
Body weight, liver lipids, and serum parameters of timed WD-fed male B6/129 mice
| RC | WD | ||
| 2 Weeks | 4 Weeks | ||
| Body weight (g) | 36.2 ± 1.19 | 39.6 ± 0.770 | 42.1 ± 0.524a |
| Liver TG (mg/g) | 18.9 ± 2.25 | 270 ± 41.8b | 452 ± 46.7b |
| Liver TC (mg/g) | 10.3 ± 3.05 | 8.90 ± 0.859 | 24.1 ± 1.64a |
| Serum TG (mg/dl) | 101 ± 15.0 | 73.6 ± 5.98c | 137 ± 15.4 |
| Serum TC (mg/dl) | 151 ± 14.9 | 231 ± 20.0a | 227 ± 6.45b |
| ALT (U/ml) | 32.6 ± 2.77 | 74.0 ± 12.3a | 102 ± 11.3b |
| AST (U/ml) | 89.0 ± 27.6 | 84.0 ± 3.29a | 81.0 ± 6.24 |
| Serum glucose (mg/dl) | 284 ± 37.4 | 369 ± 12.9 | 358 ± 10.7 |
| Serum insulin (ng/ml) | 1.15 ± 0.305 | 4.65 ± 0.752b | 3.4 ± 1.2 |
| Serum glucagon (pg/ml) | 52.1 ± 4.19 | 41.2 ± 3.78 | 66.7 ± 19.4 |
| HOMA-IR | 0.875 ± 0.314 | 4.14 ± 0.595b | 3.45 ± 1.26 |
Values are expressed as mean ± SE. B6/129 mice fed WD for 2 weeks (n = 5) and 4 weeks (n = 4). All mice were 11 weeks old when euthanized. Controls were the age-matched B6/129 mice fed a RC diet (n = 5).
P < 0.05 versus control group (RC), by Welch’s t-test.
P < 0.01 versus control group (RC), by Welch’s t-test.
P < 0.001 versus control group (RC), by Welch’s t-test.
Fig. 4.
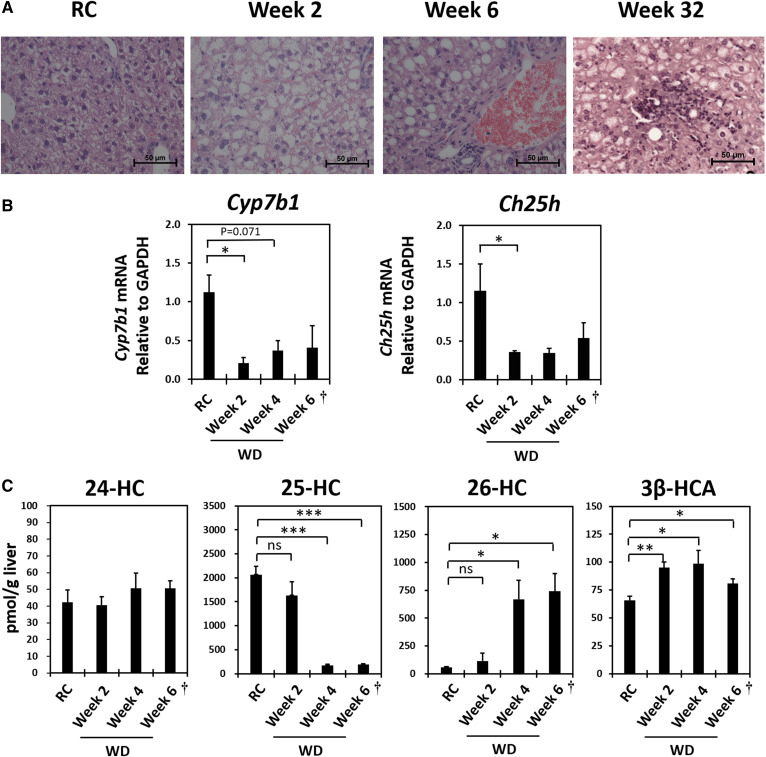
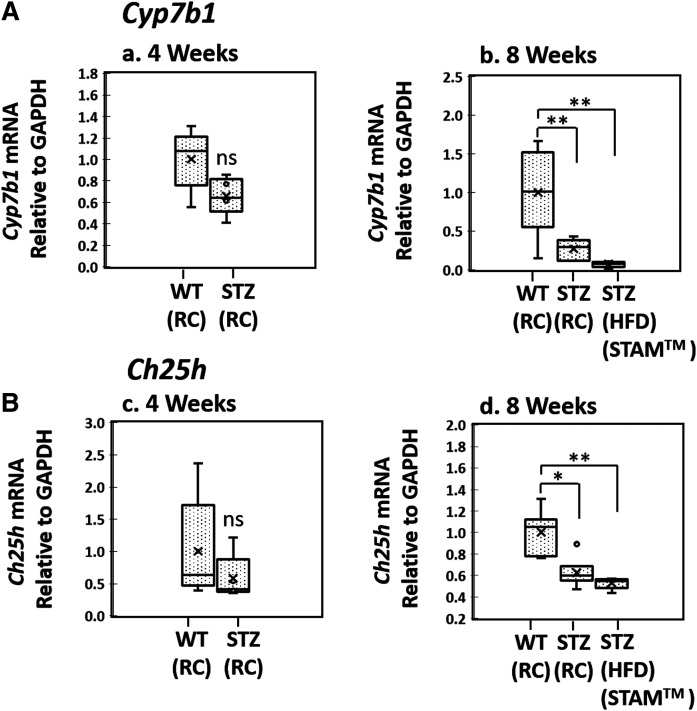
Effect of WD feeding (2–6 weeks) on key hepatic oxysterols, Cyp7b1 and Ch25h mRNA expression, and histological inflammation in B6/129 mice. A: Representative liver histology (H&E staining, ×40) of RC-fed control and timed WD-fed B6/129 mice. B: Relative hepatic Cyp7b1 and Ch25h mRNA expression levels of B6/129 mice fed WD. C: Hepatic key oxysterol and 3β-HCA levels of WD-fed B6/129 mice. All mice were 12 weeks old when euthanized except for the 6 week WD-fed group (marked as “†”), which was 15 weeks old when euthanized. The 12-week-old B6/129 mice fed with RC were used as controls. RC, irradiated Teklad LM-485; WD, Harlan-Teklad TD.88137. RC control, n = 5; 2 Week WD, n = 5; 4 Week WD, n = 4; 6 Week WD, n = 3. Welch’s t-tests were performed between RC-fed animals and each group of WD-fed animals. Significance is indicated by *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001.
The hepatic Cyp7b1 mRNA level of expression in 2 week WD-fed B6/129 male mice was only 20% of control RC-fed B6/129 mice (Fig. 4B). This suppression in Cyp7b1 persisted with continued WD feeding. As previously demonstrated by our laboratory (29), changes in Cyp7b1 protein level correlated to changes in the mRNA level. Cyp7b1 protein level of 2 week WD-fed B6/129 mice was ∼45% of that of RC-fed controls (supplemental Fig. S2). Liver oxysterol and cholestenoic acid analyses revealed that 26-HC and 3β-HCA levels became elevated in a time-dependent manner over the 6 week WD feeding (Fig. 4C). The accumulation of these two metabolites correlated with Cyp7b1 mRNA repression.
Table 2 presents liver bile acid compositions of the timed WD-fed B6/129 mice. As in the StarD1 overexpression model, the hepatic total bile acid concentration was unchanged with WD feeding at all measured time points. In addition, as compared with mice fed RC, the elevated composition of βMCA in WD-fed mice was more hydrophilic. A similar change in liver bile acid composition without altering total concentration was recently reported in C57Bl/6J mice fed a colin-deficient HFD (30). CA derivatives were lowered in 2 week WD-fed B6/129 mice (Table 2) with corresponding lowered Cyp8b1 mRNA expression (∼25% of control RC-fed mice, P = 0.015, n = 5) (data not shown). Therefore, bile acid toxicity was an unlikely cause of hepatic inflammation in these mice. Of note, accumulation of 26-HC and 3β-HCA, which occurred in the 2 week WD-fed mice, presented with a >2-fold elevation in ALT and AST (Table 2) at a time when no histologic inflammatory infiltrate was visualized. In summary, the data strongly indicate that excess accumulation of 26-HC and 3β-HCA associated with chronic suppression of Cyp7b1 contribute to hepatic inflammation (Fig. 4A).
TABLE 2.
Liver bile acid compositions of the timed WD-fed B6/129 mice
| RC (Control) (n = 5) | WD | |||
| Week 2 (n = 5) | Week 4 (n = 4) | Week 6 (n = 3) | ||
| Total bile acid | 210 ± 67.0 | 195 ± 36.4 | 278 ± 191 | 131 ± 12.5 |
| TCA | 92.1 ± 35.2 | 55.1 ± 14.3 | 112 ± 79.4 | 49.7 ± 6.26 |
| CA | 2.38 ± 0.64 | 1.67 ± 0.26 | 1.88 ± 0.99 | 1.66 ± 0.417 |
| TCA-7S | 2.02 ± 0.401 | 2.53 ± 0.181 | 3.08 ± 0.911 | 2.07 ± 0.209 |
| TCDCA | 8.12 ± 2.92 | 7.29 ± 1.29 | 8.62 ± 4.35 | 6.70 ± 0.683 |
| CDCA | 1.45 ± 0.29 | 0.67 ± 0.04 | 0.58 ± 0.171 | 0.48 ± 0.162 |
| TUDCA | 1.93 ± 0.59 | 2.08 ± 0.44 | 3.41 ± 2.14 | 1.55 ± 0.176 |
| UDCA | 0.60 ± 0.15 | 1.48 ± 0.14a | 1.19 ± 0.61 | 0.79 ± 0.233 |
| TDCA | 3.90 ± 1.09 | 1.39 ± 0.23 | 2.48 ± 1.13 | 3.60 ± 0.324 |
| THDCA | 1.59 ± 0.43 | nd | nd | nd |
| T-αMCA | 4.92 ± 1.70 | 4.14 ± 1.40 | 5.11 ± 2.93 | 4.07 ± 1.21 |
| αMCA | 3.32 ± 0.85 | 5.13 ± 0.75 | 3.10 ± 1.34 | 2.24 ± 0.190 |
| T-βMCA | 54.5 ± 18.1 | 43.6 ± 10.8 | 83.9 ± 66.7 | 22.4 ± 5.85 |
| βMCA | 15.9 ± 2.78 | 62.4 ± 8.04a | 46.5 ± 28.0 | 28.7 ± 0.687b |
| T-ωMCA | 11.4 ± 3.28 | 2.20 ± 0.661b | 2.27 ± 1.50b | 1.97 ± 0.531 |
| ωMCA | 6.19 ± 0.83 | 3.88 ± 0.62 | 2.87 ± 1.34 | 3.71 ± 0.939 |
| T-Δ22-βMCA | 1.16 ± 0.37 | 1.53 ± 0.34 | 1.95 ± 0.92 | 1.49 ± 0.661 |
| MDCA | nd | nd | 0.46 ± 0.13 | nd |
| TCA-6α-ol | 0.62 ± 0.16 | nd | nd | nd |
| T-isoLCA-Δ5-3S | nd | 0.38 ± 0.08 | nd | nd |
Values are expressed as mean ± SE in nanomoles per gram of tissue. Total bile acid represents sum of 124 bile acids (supplemental Table S1) measured. Only detected bile acids are presented above. nd, not detected; THDCA, taurohyodeoxycholic acid; T-Δ22-βMCA, tauro 3α,6β,7β-trihydroxychol-5,22-diene-24-oic acid; MDCA, murideoxycholic acid; TCA-6α-ol, tauro 6α-hydroxycholic acid; T-isoLCA-Δ5-3S, tauro 3β-sulfoxy-5-cholen-24-oic acid.
P < 0.01 versus control group (RC), by Welch’s t-test.
P < 0.05 versus control group (RC), by Welch’s t-test.
To establish whether the suppression of Cyp7b1 was specific for WD feeding or the B6/129 strain, WT (C57Bl/6J) mice were fed a WD and/or a high-fructose corn syrup diet, a diet also known to induce fatty liver (supplemental Fig. S3). As C57Bl/6J mice have been shown to develop fatty liver at a slower rate than B6/129 mice, mice were fed for 4 weeks. Either together or alone, WD and high-fructose corn syrup diets led to equal, but lower than RC-fed, levels of Cyp7b1 mRNA expression.
To bring human relevance to mouse findings, a small cohort of human samples determined as normal liver or live with steatosis without inflammation was obtained for analysis from NIH-sponsored LTCDS. CYP7B1 mRNA levels were lower in livers with steatosis. The levels of CYP7B1 mRNA expression correlated with tissue level of 26-HC (supplemental Fig. S4).
WD feeding in B6/129 male mice lead to lower hepatic Ch25h mRNA level of expression and lower intracellular level of 25-HC
Unlike the StarD1 overexpression mouse model, WD feeding did not lead to an elevation in hepatic 24-HC levels in B6/129 mice. And, in contrast to children with CYP7B1 deficiency (15) and in StarD1-overexpressed mouse liver (14), liver 25-HC level was lower with WD feeding in B6/129 mice (Fig. 4C). The decrease of 25-HC appeared due to impaired synthesis of 25-HC with WD feeding, following the observed lower mRNA expression level of cholesterol 25-hydroxylase (Ch25h) (Fig. 4B). As compared with mitochondrial Cyp27a1, Ch25h in the ER (31) normally plays a larger role in cholesterol metabolism to 25-HC in both humans and rodents. Therefore, it was not unexpected to see a decrease in 25-HC levels with a lower level of Ch25h mRNA expression.
Developing insulin resistance correlates with lower levels of Cyp7b1 and Ch25h mRNA expression in male B6/129 mice
With WD feeding, serum glucose and insulin levels became elevated in B6/129 mice (Table 1). A homeostatic model assessment of insulin resistance (HOMA-IR) score showed that these mice had developed insulin resistance with just 2 weeks of WD feeding. There are reports showing that the insulin signaling pathway is strongly linked to Cyp7b1 and Ch25h expression. For example, a significantly lower Cyp7b1 mRNA expression was reported in selective liver-specific insulin receptor knockout mice (18). Reduced Cyp7b1 (32, 33) and Ch25h (34) expression was also observed in insulin-resistant mice. Therefore, the reduced hepatic insulin sensitivity due to WD feeding appeared to impair downstream Cyp7b1 and Ch25h mRNA expression in B6/129 mice.
In summary, insulin resistance induced with WD feeding in B6/129 mice led to the chronic suppression of Cyp7b1 mRNA and accumulation of a toxic concentration of 26-HC and 3β-HCA in liver, initiating inflammation. 25-HC, which can be rapidly metabolized to 25-hydroxycholesterol-3β-sulfate (25HC3S) (35), has previously been shown to have both pro- and anti-inflammatory effects on immune response and broadly suppresses viral infection in a cell-intrinsic manner (36, 37). Suppression of Ch25h with insulin resistance led to lower levels of 25-HC.
WD feeding of B6/129 female mice led to similar changes as those observed in WD-fed B6/129 male mice
As shown in supplemental Fig. S5, female B6/129 mice expressed markedly lower basal levels of Cyp7b1 mRNA than their male age-matched counterparts. And, like their male counterparts, the female mice showed a lower Cyp7b1 mRNA level of expression with WD feeding. Supplemental Table S3 shows that all other metabolic parameter changes with WD feeding in female B6/129 mice were similar to their male counterparts.
Expression levels of Cyp7b1 and Ch25h mRNA in STZ-induced diabetic mice
To further investigate the linking of insulin signaling with Cyp7b1-mediated hepatocyte inflammation, we obtained liver tissues from STAM™ mice (20). The STAM™ model represents a previously well-described NASH mouse model where STZ-induced insulin resistance is used to accelerate the transition from fatty liver to NASH with HFD feeding. The 2-day-old pups of C57Bl/6J mice are given STZ, which reliably destroys 60% of β cells, as described in the Methods (supplemental Fig. S1). When stressed with HFD feeding starting at age 4 weeks, the mice develop insulin resistance with NASH by 8 weeks (20). Attempting to minimize the effects of HFD feeding yet explore STZ pancreatic β cell reduction, we studied STZ male mice fed RC first. Cyp7b1 mRNA levels in 4- and 8-week-old STZ mice were 70% and 30% of age-matched littermates who did not receive STZ, respectively (Fig. 5A). Ch25h mRNA expression in 4-week-old WT and STZ mice were not statistically different, but Ch25h expression levels of 8-week-old STZ mice were 60% of control mice. Table 3 presents the plasma biochemical markers of the 8-week-old STZ-injected mice fed a RC diet. The STZ-injected mice had markedly elevated plasma glucose and insulin levels as compared with those of age-matched uninjected littermates. The significantly elevated HOMA-IR score demonstrated that the mice had developed insulin resistance by 8 weeks of age without the need for HFD feeding. These findings suggested the development of early insulin resistance with a compensatory mechanism of insulin overproduction in the remaining β cells. Similar findings with increased serum insulin levels were previously reported in STZ-treated RC-fed rats (38). Interestingly, TG was also elevated 1.5-fold compared with controls. As in HFD-fed B6/129 mice, the ALT was increased 2.5-fold as compared with control mice, demonstrating early biochemical inflammation. The results suggest that insufficient intracellular insulin signaling with insulin resistance as the likely cause of the lower levels of Cyp7b1 and Ch25h mRNA expression. In the traditional STAM™ model (STZ-injected mice fed HFD), Cyp7b1 and Ch25h mRNA expression levels in 8-week-old mice were 10% and 55%, respectively, of age-matched uninjected controls fed a RC diet (Fig. 5). Thus, like the B6/129 WD feeding model, the STZ-injected mice with/without HFD feeding models also provide in vivo findings correlating insulin resistance to lower levels of Cyp7b1 and Ch25h mRNA expression and hepatocyte inflammatory responses.
Fig. 5.
Hepatic Cyp7b1 and Ch25h mRNA expression in the insulin-resistant mouse models: The 2-day-old male pups of C57Bl/6J mice were given low dose (200 μg/mouse) STZ to partially destroy β cells as described in the Methods. The mice were fed a low-fat RC diet (CE-2, CLEA Japan Inc.) until 4 weeks of age (STZ-RC), and then fed an ad libitum HFD (HFD32, CLEA Japan Inc.) until age 8 weeks (STZ-HFD, STAM™). One group of mice was continued on their RC diet for an additional 4 weeks (8-week-old STZ-RC). Controls (WT-RC) were 4- or 8-week-old untreated C57Bl/6J mice fed RC. STZ-RC (4 weeks old, n = 5; 8 weeks old, n = 6), STZ-HFD (8 weeks old, STAM™, n = 5), WT-RC (4 weeks old, n = 5; 8 weeks old, n = 6). Welch’s t-tests were performed between WT and STZ-treated animals of the same age. Significance is indicated by *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001.
TABLE 3.
Plasma parameters of 8-week-old STZ-injected mice fed a RC diet
| Control | STZ-RC | |
| TG (mg/dl) | 109 ± 12.1 | 167 ± 13.4a |
| ALT (U/ml) | 19.5 ± 2.37 | 53 ± 8.04b |
| AST (U/ml) | 53.1 ± 5.26 | 83.1 ± 8.91a |
| Glucose (mg/dl) | 212 ± 16.8 | 654 ± 15.2c |
| Insulin (ng/ml) | 0.527 ± 0.0823 | 1.16 ± 0.0774c |
| Glucagon (pg/ml) | 36.1 ± 4.32 | 129 ± 20.8b |
| HOMA-IR | 0.279 ± 0.0478 | 1.88 ± 0.146c |
Values are expressed as mean ± SE. All mice were 8 weeks old when euthanized (n = 6 for each group). Controls was age-matched C57Bl/6J mice fed a RC diet.
P < 0.05 versus control group (RC), by Welch’s t-test.
P < 0.01 versus control group (RC), by Welch’s t-test.
P < 0.001 versus control group (RC), by Welch’s t-test.
Insulin coordinately regulates hepatic 26-HC and 25-HC levels
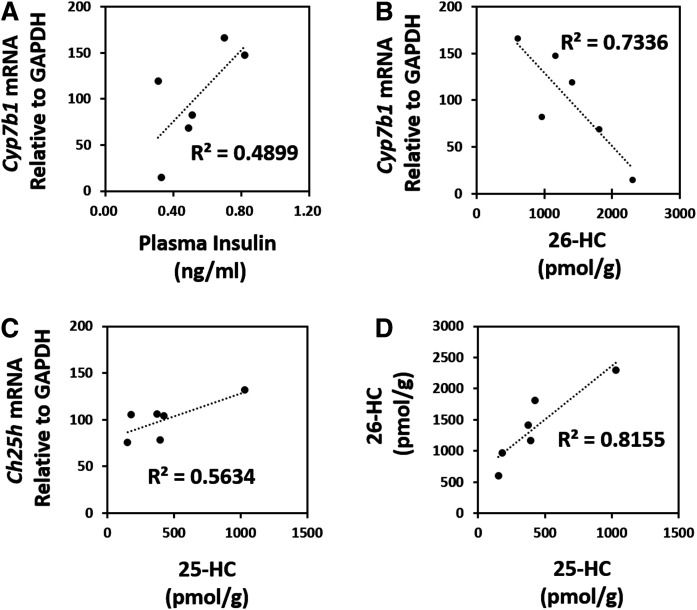
Hypothesizing insulin as a prime regulator of Cyp7b1 and Ch25h levels of expression, one could speculate a coordinated Cyp7b1-Ch25h response to be associated with fluctuations in serum/plasma insulin levels under physiologic conditions. The data in Fig. 6 demonstrate the correlation among plasma insulin, hepatic Cyp7b1 and Ch25h mRNA expression, and tissue 26-HC and 25-HC levels in individual WT (C57Bl/6J) mice fed a RC diet. With mice having free access to food until being euthanized, there was a large variation in the plasma insulin levels of individual mice (0.38–0.82 ng/ml). As hypothesized, the plasma insulin level of each individual mouse was relatively correlated to its hepatic Cyp7b1 mRNA level of expression (R2 = 0.490), and the Cyp7b1 mRNA expression level was well-correlated to the hepatic 26-HC (R2 = 0.734). Also, the hepatic Ch25h mRNA expression level of individual mice was well-correlated to the hepatic 25-HC level (R2 = 0.563). Thus, hepatic 26-HC levels were strongly correlated to their 25-HC levels in WT mouse liver (R2 = 0.816).
Fig. 6.
Coordinated regulation of hepatic 25-HC and 26-HC levels in WT mice. Eight-week-old C57Bl/6J mice were harvested, and their hepatic mRNA, oxysterols, and serum insulin were determined. The mice had free access to RC diet (CE-2, CLEA Japan Inc) until being euthanized. Each dot in the graphs represents one mouse (n = 6). A: Correlation of plasma insulin level to hepatic Cyp7b1 mRNA expression level of freely fed C57Bl/6J mice. B: Correlation of hepatic 26-HC to Cyp7b1 mRNA expression level. C: Correlation hepatic 25-HC level to Ch25h mRNA expression level. D: Correlation of hepatic 26-HC and 25-HC levels.
Supplemental Fig. S6 compares how levels of insulin in a fasted and fed state correlate with Cyp7b1 mRNA level of expression. Unlike the data presented in Fig. 6 where physiologic dietary conditions were employed, B6/129 mice were fed a WD for 2 weeks prior to being euthanized in either the fed or the 4 h-presacrifice fasted state. As previously shown in Fig. 4B, the basal Cyp7b1 mRNA level of nonfasted WD-fed male mice was 50% that of RC-fed mice in the presence of early insulin resistance (Fig. 4, Table 1). Despite the presence of WD feeding leading to lower Cyp7b1 mRNA expression in mice exhibiting evidence of early insulin resistance, a lower serum insulin level with fasting led to even lower expression of Cyp7b1 mRNA. Once again, these in vivo findings provide evidence for insulin signaling as an important regulator of Cyp7b1 level of expression.
Levels of Cyp7b1 and Ch25h mRNA expression in B6/129 and C57Bl/6J mice under physiologic conditions
As shown in supplemental Fig. S7, RC-fed B6/129 mice have a markedly higher level of Ch25h mRNA expression compared with that of RC-fed C57Bl/6J mice; changes which corresponded with trends in 25-HC levels. No significant difference was seen in levels of Cyp7b1 mRNA expression between the two strains. Refer to Figs. 4 and 6 for the relative liver oxysterol levels of these mice.
Glucagon acts as a complimentary regulator of Cyp7b1
Based upon the studies of Hylemon et al. (39) demonstrating the ability of glucagon and cAMP to downregulate the expression of cholesterol 7α-hydroxylase (Cyp7a1) at both the mRNA and activity levels, we had previously explored the regulation of Cyp7b1 in primary rat hepatocytes (29). Based upon other regulatory similarities between Cyp7a1 and Cyp7b1, we observed glucagon to downregulate Cyp7b1 activity in vitro, but to a lesser degree (40% as opposed to the >80% found in Cyp7a1). Supplemental Fig. S8 demonstrates the in vivo existence of an inverse level of regulation between Cyp7b1 mRNA and plasma glucagon in STZ-treated diabetic mice fed a RC diet. This correlation is supportive of our previous in vitro primary hepatocyte findings (29). It is well-documented that with T2DM, a paucity of insulin signaling within the cell with advancing insulin resistance leads to a paradoxical increase in levels of cell and serum glucagon. Whether glucagon is able to further suppress Cyp7b1 in an insulin resistant state is yet unknown. However, GLP-1 agonists have demonstrated beneficial effects in NAFLD treatment (40).
DISCUSSION
With the transition from NAFLD to NASH, there exists an early coexistence of pro-inflammatory and fibrotic processes that generally occur in a complicated metabolic setting of obesity, hyperlipidemia, and/or insulin resistance. As a result, an understanding of what drives the transition from benign NAFLD to NASH in humans remains poorly defined. It is therefore not surprising that various treatment protocols for NASH have met with limited success. The findings of the current study provide strong evidence that insulin resistance-mediated dysregulation of CYP7B1 establishes a cellular mechanism for the accumulation of “toxic” intracellular cholesterol metabolites that may initiate/perpetuate fatty liver to NASH progression. The lack of hepatic inflammatory cell infiltrate found in the presence of early markers of hepatocellular inflammation is supportive of toxicity originating within the hepatocyte.
Hepatocyte ER stress as promoting NASH has become a pervasive theory on the transition from fatty liver to NASH. In times of cell stress, the ER is capable of activating an evolutionarily conserved survival pathway termed the unfolded protein response in order to gain control of protein and lipid homeostasis (41). However, metabolically induced ER stress due to chronic excess of cholesterol in the ER leads to hepatocyte lipid accumulation in the presence of existing surplus, which may contribute to an inflammatory response as in NASH. Similarly, alone or in combination with excess ER cholesterol, activation of the unfolded protein response by excess FFA has long been proposed as contributing to the NASH inflammatory response (42).
The WD-fed B6/129 NASH model appears to differ from ER stress models in several important ways. In this regard, as outlined in the introduction and previous publications (11, 14), the alternative pathway of bile acid synthesis is designed for maintaining hepatocellular cholesterol and lipid homeostasis. Our data show that insulin resistance alters this pathway by leading to chronic lower expression of Cyp7b1. As a result, the chronic accumulation of oxysterols appears not only to directly provide substrate for toxic metabolites but also to alter the regulation of pathways such as mitochondrial cholesterol uptake and FFA synthesis (11). Additionally, the increase in oxysterols appears to continue to drive lipoprotein cholesterol out of the liver, as demonstrated in the described StarD1 overexpression model (Fig. 1F). In contrast, the ER stress response differs in that cholesterol synthesis continues upregulated while lipid export is dramatically repressed, i.e., contributing to the further increase in hepatocellular lipid accumulation and impaired lipid metabolism (43). Additionally, levels of ER stress markers, BiP and CHOP, were unchanged in B6/129 mice that showed histologic and biochemical evidence of early NASH following 2 weeks of WD feeding (supplemental Fig. S2).
It is clear that early WD feeding with associated Cyp7b1 dysregulation secondary to insulin resistance, as described in this paper, and chemically induced ER stress are both able to induce steatohepatitis (44). Despite their differences, both processes may either independently or collaboratively perpetuate liver disease. However, in the WD feeding model, Cyp7b1 dysregulation is a very early process leading to the onset of NASH. This model can also help explain why type 1 diabetes mellitus (10), lean individuals (8, 9), and the genetic absence of CYP7B1 in children (16) all develop NASH.
The increase in hepatic B cells seen in the StarD1 overexpression model (Fig. 3) may reflect a role for these cells in liver inflammation or an early pathway to a fibrotic phenotype. It has been previously shown that while T cell-deficient animals can still get liver fibrosis, B cell-deficient animals are protected (45). High eosinophil levels in the liver are associated with increased type 2 liver inflammation that can lead to eventual fibrosis (46, 47). We also found increased Ccl24 gene expression, which is a pro-inflammatory mediator seen to be increased in NAFLD and NASH models (48). Increased type 2 inflammation is also associated with increased Chi3l3. We observed a decrease in the Arg1 mRNA level. Arg1 is associated with tissue repair responses (49) and can be a suppressive mediator in liver inflammation by reducing Nos2 responses (50). Nos2 is additionally associated with increased liver inflammation and fibrosis (51). An increase in the mRNA levels of the transcription factor, Gata3, was observed. Interestingly, Gata3 can block against a T regulatory cell conversion, further exacerbating an inflammatory phenotype (52). These results all suggest that the B6/129 mice overexpressing StarD1 with early signs of inflammation in the liver were consistent with oxysterol-induced toxicity, which may eventually lead to a fibrotic phenotype.
It should be noted that 7α-hydroxylation of cholesterol or oxysterols not only lessens any further metabolite’s ability to serve as a regulatory molecule but also decreases its hydrophobicity and potential as a toxic metabolite. This becomes particularly relevant with CYP7B1. CYP7B1 is not only capable of 7α-hydroxylation of 24-, 25-, and 26-HC but is also able to 7α-hydroxylate toxic 3β-HCA (53, 54). Therefore, in the presence of low levels of CYP7B1 expression, toxic unsaturated cholestenoic acids have the propensity to accumulate and induce tissue injury, as observed with neonatal liver disease where there is an absence of CYP7B1 (15, 16). Of note, mutation in CYP7B1 develops spastic paraplegia type 5 (SPG5) in later life, an autosomal recessive inherited spastic paraplegia. These patients do not develop liver failure in their childhood, but prolonged neonatal cholestasis has been reported (55, 56). It should also be noted that significant variation in oxysterol levels exists in described SPG5 patients, and serum levels of 26-HC and 3β-HCA in patients with SPG5 (57, 58) are elevated compared with healthy individuals, but not as high as those of the neonates with significant liver failure (15). It remains unclear whether one or the combination of several accumulating metabolites contribute to the early toxicity. The differences in the level, type, and rate of oxysterols and their metabolites’ accumulations are likely reasons as to why this gene mutation can develop different phenotypes. In this context, there is an evidence that the liver failure due to the Cyp7b1 mutation was successfully treated with supplemental bile acid therapy (16). This patient had lower serum oxysterols (or 3β-HCA) than those who progressed to the end stage liver disease (15). Moreover, an early trial has provided early evidence for the possible use of bile acid therapy for the treatment of SPG5 patients (57).
Interestingly, Raselli et al. (59) recently determined hepatic oxysterols in NASH patients, and found only 24- and 7-hydroxylated sterols to be elevated. Furthermore, with 20 weeks of HFD feeding of Cyp27a1−/− and Cyp7a1−/− mice, they were able to induce hepatic steatosis, inflammation, and a similar oxysterol profile as seen in their NASH patients. This differs from our in vivo findings in hepatic tissues of mice and of humans where 26-HC was the predominate elevated oxysterol. The reason for this discrepancy is currently unclear. However, Na et al. (60) found a significant reduction of hepatic 3β,7α-dihydroxy-5-cholestenoic acid (3β,7α-diHCA) with decreased Cyp7b1 gene expression in C57Bl/6J mice fed a HFD for 11 weeks; observations supportive of our findings.
Cyp7b1−/− mice accumulate increased levels of 24-, 25-, and 26-HC (14, 61) similar to humans with the absence of CYP7B1. However, Cyp7b1−/− mice seem resistant to the “fatty liver → steatohepatitis → fibrosis” phenotype observed in humans or mouse models such as those outlined in this report. Although previously well-characterized, until recently the model has not been challenged with aging or crossed with an existing phenotypic genetic mouse model to amplify a phenotypic presentation. In cleverly designed studies, Umetani et al. (62) utilized Cyp7b1−/− mice crossed with ApoE−/− mice and allowed the mice to age a full 12 months, which enabled them to demonstrate that elevated tissue 26-HC was associated with inflammation and atherogenesis. The required aging and utilization of the ApoE−/− mice suggests that, as with other mouse models of cholesterol and lipid metabolism lacking a distinct human phenotype (i.e., Cyp27a1−/− and cerebrotendinous xanthomatosis), compensatory survival mechanisms can ensue. However, with more focused metabolite study, the Cyp7b1−/− mouse may yet be used to uncover a clearer understanding of the responsible metabolite(s) and the compensatory pathways (61) that may be protective. As a function of their chemical structure and membrane disruptive properties, the in vitro addition of oxysterols or their metabolites is fraught with difficulty in interpreting their ability to disrupt cell function as occurs in a NAFLD setting. The toxicity found with their endogenous in vivo production in the StarD1 overexpression model (Fig. 1) coupled to the findings of patients with CYP7B1 deficiency (15, 16) provides for a correlative “cause and effect” model, which is currently undergoing further exploration in our laboratory and others.
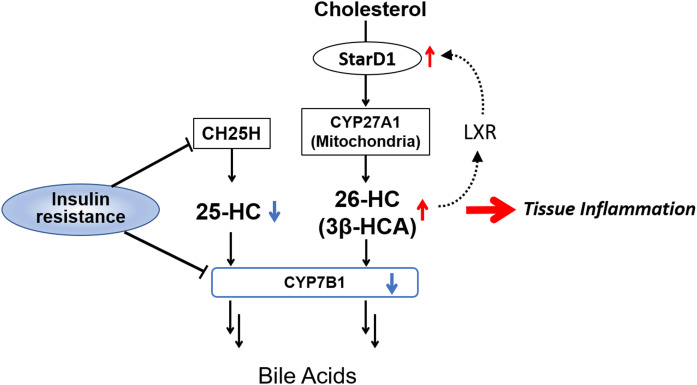
Figure 7 outlines the role of Cyp7b1 in a mitochondrial feed-forward response to excess cellular cholesterol. Like LXR, StarD1, a mitochondrial cholesterol delivery protein, has been shown to be upregulated in a feed-forward manner by oxysterols (14, 63). Under normal physiologic conditions, this works to transiently increase oxysterol synthesis, which regulate HMGR-controlled cholesterol synthesis and LXR-driven FFA synthesis to control cholesterol and lipid homeostasis. However, with chronically elevated exogenously delivered cell cholesterol and persistently elevated oxysterols, this can be envisioned to become a “feed-forward” pathway of excess cholesterol delivery into the mitochondria. Therefore, an inability to adequately prevent accumulation of surplus mitochondrial cholesterol, cholesterol which is also capable of being oxidized within the mitochondria, could become toxic. The recent findings of Torres et al. (64) suggest StarD1 to be upregulated in ER stress, findings that appear to couple ER stress-increased mitochondrial cholesterol delivery to toxicity. The toxicity associated with increased oxysterol levels driving StarD1 expression, as shown in this work, is further evidence. Furthermore, the findings of this work and of supportive human data, support our hypothesis that toxic cholesterol metabolites are able to initiate and promote NASH.
Fig. 7.
Proposed mechanism promoting fatty liver to inflammation: Chronic persistent increases in 26-HC and 3β-HCA as shown in this work with StarD1 overexpression, WD feeding, and T2DM models (STZ-treated mice) leads to accumulation of oxysterols and their lipotoxic metabolites, as occurs in humans in the absence of CYP7B1. In early fatty liver, lowered hepatocellular insulin levels due to the insulin-resistant condition suppressed Cyp7b1 gene expression. Interestingly, Ch25h was also repressed in the insulin-resistant condition, leading to lower hepatocellular 25-HC.
Previously, we showed that the in vitro addition of glucagon (cAMP) to primary rat hepatocyte culture led to downregulation of Cyp7b1 mRNA (29). With the early development of insulin resistance in our WD-fed B6/129 mouse model (Table 1), no significant increase in serum glucagon levels was found, as is usually found with more advanced T2DM. These limited findings clearly do not rule out glucagon as a downregulator of Cyp7b1, but rather suggest the importance of insulin signaling for Cyp7b1 expression (Fig. 7).
Our findings also provide evidence that insulin acts to coordinately regulate Cyp7b1 and Ch25h mRNA expression in the liver. This represents a novel observation with a yet unclear understanding that was uncovered as a function of the observed disparity in 25-HC and 26-HC in B6/129 mice fed a WD. In the present study, the hepatic Ch25h mRNA level was found to be lower in WD-fed B6/129 and STAM™ mice, both of which demonstrate a level of insulin resistance. Noebauer et al. (34) also previously reported similar findings in obese insulin-resistant C57Bl/6J mice where significantly lower hepatic Ch25h mRNA and protein levels were found as compared with WT mice. More specifically, they demonstrated that adenovirus-mediated Ch25h overexpression improved glucose tolerance and insulin sensitivity, lowering the HOMA-IR in these insulin-resistant mice. As observed in the studies presented in this report, their observations suggest Ch25h’s regulation, like Cyp7b1, to be tied to insulin and play a role in homeostasis of glucose.
Under physiologic conditions, the majority of 25-HC is produced by Ch25h within the ER of cells (31). Traditionally, 25-HC has been viewed as a cholesterol metabolite with cholesterol and lipid regulatory properties, but 25-HC also possesses a range of inflammatory and anti-inflammatory effects (36, 37, 65–67). With LPS treatment, Ch25h−/− mice secrete higher levels of the cytokines, interleukin (IL)-1α, IL-1β, and IL-18 (66), with increased sensitivity to septic shock and a greater ability to repress bacterial growth (66). More specifically, 25-HC is known to repress IL-1β expression via antagonizing SREBPs, which creates a negative feedback pathway for interferon-induced inflammation (66). 25-HC also amplifies secretion of inflammatory cytokines such as macrophage colony-stimulating factor, IL-6, and IL-8 (65, 68). Thus, suppression of Ch25h with insulin resistance could lead to lower levels of potentially protective 25-HC, potentiating the toxic effects of 26-HC and 3β-HCA. Conversely, our laboratories have provided evidence suggesting that 25-HC induces a proinflammatory response, but for the subsequent sulfated metabolite (25HC3S), by translocating to the nucleus to initiate PPAR-γ anti-inflammatory pathways. This finding suggests a more intricate regulation is involved in 25-HC mediated inflammatory pathways (69). Moreover, 25-HC has recently been shown to be a high-affinity activator of DNA methyltransferase 1, and an important epigenetic regulator of numerous genes (70). However, how this unique coordinate response of hepatic Ch25h and Cyp7b1 to insulin resistance contributes to liver inflammation remains unclear.
In summary, the inability to upregulate Cyp7b1 in the setting of insufficient insulin signaling appears to establish hepatocyte accumulation of toxic intracellular cholesterol metabolites; metabolites that then initiate/perpetuate hepatocyte injury. More specifically, findings by Tang, Pettersson, and Norlin (17) suggest that the suppression of Cyp7b1 appears to be explained by a paucity of “cellular” insulin, which under most circumstances presents as insulin resistance; a state where “serum” insulin and glucagon levels are elevated. This hypothesis is further supported by the fact that focused treatment of diabetes with insulin sensitization and glucagon relief has been the most successful medicinal treatment to date. As this cellular regulatory pathway is found in most cells in the body, it may be relevant for our understanding of insulin resistance-related inflammatory pathways outside the liver that promote diseases such as atherosclerosis.
Data availability
Essential data are contained in the article. All other data are available in the supplemental figures and tables.
Supplementary Material
Acknowledgments
The authors acknowledge excellent technical help from Ms. Emily Gurley, Ms. Xuan Wang, and Ms. Patsy Cooper.
Footnotes
This article contains supplemental data.
Author contributions—G.K. study design; G.K. and W.M.P supervision; G.K. data analysis; G.K. and W.M.P writing-original draft. D.M., D.R-A., and S.A.L. in vivo experiments on B6/129; D.M., D.R-A., and S.A.L. Western blot and qPCR analyses of the STZ treated mice livers. R.M. flow cytometry and qPCR data on the nonparenchymal cells from B6/129 mice liver. T.H. in vivo studies on the STZ treated mice. X.L. and R.G. data acquistion on the WD/HFCS fed C57/B6J mice. H.T. and H.N. bile acids and oxysterols measurements. T.M. authentic standards of bile acids and oxysterols for LC-MS analyses. S.E. provided breeding pairs of B6/129 mice used in this study. G.G., M.F., M.S., P.B.H. and writing-review and editing. W.M.P study concept.
Funding and additional information—This work was supported by Gilead Sciences Liver Research Award 2016 to G.K. and Veterans Administration Veterans Affairs Merit Award I01 BX000197-07 to W.M.P. NIH-sponsored Liver Tissue Distribution Center at the University of Minnesota supplied presented human liver tissues used for analysis.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
Abbreviations—
- ALT
- alanine aminotransferase
- Arg1
- arginase 1
- AST
- aspartate aminotransferase
- CA
- cholic acid
- Ch25h
- cholesterol 25-hydroxylase
- CYP7A1
- cholesterol 7α-hydroxylase (Cyp7a1 denotes murine)
- CYP7B1
- oxysterol 7α-hydroxylase (Cyp7b1 denotes murine)
- CYP27A1
- sterol 27-hydroxylase (Cyp27a1 denotes murine)
- FC
- free cholesterol
- β-Gal
- β-galactosidase
- Gata3
- GATA binding protein 3
- 24-HC
- 24(S)-hydroxycholesterol
- 25-HC
- 25-hydroxycholesterol
- 26-HC
- (25R)-26-hydroxycholestetol
- 3β-HCA
- 3β-hydroxy-5-cholestenoic acid
- HOMA-IR
- homeostatic model assessment of insulin resistance
- IL
- interleukin
- IS
- internal standard
- MCA
- muricholic acid
- NAFL
- nonalcoholic fatty liver
- NIH
- National Institutes of Health
- Nos2
- nitric oxide synthase 2
- RC
- regular chow
- SPG5
- spastic paraplegia type 5
- StarD1
- steroidogenic acute regulatory protein
- STZ
- streptozotocin
- TC
- total cholesterol
- T2DM
- type 2 diabetes mellitus
Manuscript received May 22, 2020, and in revised form September 14, 2020. Published, JLR Papers in Press, October 2, 2020, DOI 10.1194/jlr.RA120000924.
REFERENCES
- 1.Younossi Z. M., Marchesini G., Pinto-Cortez H., and Petta S.. 2019. Epidemiology of nonalcoholic fatty liver disease and nonalcoholic steatohepatitis: implications for liver transplantation. Transplantation. 103: 22–27. [DOI] [PubMed] [Google Scholar]
- 2.Younossi Z., Tacke F., Arrese M., Chander Sharma B., Mostafa I., Bugianesi E., Wai-Sun Wong V., Yilmaz Y., George J., Fan J., et al. . 2019. Global perspectives on nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Hepatology. 69: 2672–2682. [DOI] [PubMed] [Google Scholar]
- 3.Reddy Y. K., Marella H. K., Jiang Y., Ganguli S., Snell P., Podila P. S. B., Maliakkal B., and Satapathy S. K.. 2020. Natural history of non-alcoholic fatty liver disease: a study with paired liver biopsies. J. Clin. Exp. Hepatol. 10: 245–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De A., and Duseja A.. 2020. Natural history of simple steatosis or nonalcoholic fatty liver. J. Clin. Exp. Hepatol. 10: 255–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eslam M., Newsome P. N., Sarin S. K., Anstee Q. M., Targher G., Romero-Gomez M., Zelber-Sagi S., Wai-Sun Wong V., Dufour J. F., Schattenberg J. M., et al. . 2020. A new definition for metabolic dysfunction-associated fatty liver disease: an international expert consensus statement. J. Hepatol. 73: 202–209. [DOI] [PubMed] [Google Scholar]
- 6.Fon Tacer K., and Rozman D.. 2011. Nonalcoholic fatty liver disease: focus on lipoprotein and lipid deregulation. J. Lipids. 2011: 783976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Browning J. D., and Horton J. D.. 2004. Molecular mediators of hepatic steatosis and liver injury. J. Clin. Invest. 114: 147–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Satapathy S. K., and Sanyal A. J.. 2015. Epidemiology and natural history of nonalcoholic fatty liver disease. Semin. Liver Dis. 35: 221–235. [DOI] [PubMed] [Google Scholar]
- 9.Younes R., and Bugianesi E.. 2019. NASH in lean individuals. Semin. Liver Dis. 39: 86–95. [DOI] [PubMed] [Google Scholar]
- 10.Bhatt H. B., and Smith R. J.. 2015. Fatty liver disease in diabetes mellitus. Hepatobiliary Surg. Nutr. 4: 101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pandak W. M., and Kakiyama G.. 2019. The acidic pathway of bile acid synthesis: not just an alternative pathway. Liver Res. 3: 88–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Javitt N. B. 2002. 25R,26-Hydroxycholesterol revisited: synthesis, metabolism, and biologic roles. J. Lipid Res. 43: 665–670. [PubMed] [Google Scholar]
- 13.Arnon R., Yoshimura T., Reiss A., Budai K., Lefkowitch J. H., and Javitt N. B.. 1998. Cholesterol 7-hydroxylase knockout mouse: a model for monohydroxy bile acid-related neonatal cholestasis. Gastroenterology. 115: 1223–1228. [DOI] [PubMed] [Google Scholar]
- 14.Kakiyama G., Marques D., Takei H., Nittono H., Erickson S., Fuchs M., Rodriguez-Agudo D., Gil G., Hylemon P. B., Zhou H., et al. . 2019. Mitochondrial oxysterol biosynthetic pathway gives evidence for CYP7B1 as controller of regulatory oxysterols. J. Steroid Biochem. Mol. Biol. 189: 36–47. [DOI] [PubMed] [Google Scholar]
- 15.Setchell K. D., Schwarz M., O’Connell N. C., Lund E. G., Davis D. L., Lathe R., Thompson H. R., Weslie Tyson R., Sokol R. J., and Russell D. W.. 1998. Identification of a new inborn error in bile acid synthesis: mutation of the oxysterol 7alpha-hydroxylase gene causes severe neonatal liver disease. J. Clin. Invest. 102: 1690–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dai D., Mills P. B., Footitt E., Gissen P., McClean P., Stahlschmidt J., Coupry I., Lavie J., Mochel F., Goizet C., et al. . 2014. Liver disease in infancy caused by oxysterol 7 alpha-hydroxylase deficiency: successful treatment with chenodeoxycholic acid. J. Inherit. Metab. Dis. 37: 851–861. [DOI] [PubMed] [Google Scholar]
- 17.Tang W., Pettersson H., and Norlin M.. 2008. Involvement of the PI3K/Akt pathway in estrogen-mediated regulation of human CYP7B1: identification of CYP7B1 as a novel target for PI3K/Akt and MAPK signalling. J. Steroid Biochem. Mol. Biol. 112: 63–73. [DOI] [PubMed] [Google Scholar]
- 18.Biddinger S. B., Haas J. T., Yu B. B., Bezy O., Jing E., Zhang W., Unterman T. G., Carey M. C., and Kahn C. R.. 2008. Hepatic insulin resistance directly promotes formation of cholesterol gallstones. Nat. Med. 14: 778–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pandak W. M., Schwarz C., Hylemon P. B., Mallonee D., Valerie K., Heuman D. M., Fisher R. A., Redford K., and Vlahcevic Z. R.. 2001. Effects of CYP7A1 overexpression on cholesterol and bile acid homeostasis. Am. J. Physiol. Gastrointest. Liver Physiol. 281: G878–G889. [DOI] [PubMed] [Google Scholar]
- 20.Fujii M., Shibazaki Y., Wakamatsu K., Honda Y., Kawauchi Y., Suzuki K., Arumugam S., Watanabe K., Ichida T., Asakura H., et al. . 2013. A murine model for non-alcoholic steatohepatitis showing evidence of association between diabetes and hepatocellular carcinoma. Med. Mol. Morphol. 46: 141–152. [DOI] [PubMed] [Google Scholar]
- 21.Jiao S., Cole T. G., Kitchens R. T., Pfleger B., and Schonfeld G.. 1990. Genetic heterogeneity of lipoproteins in inbred strains of mice: analysis by gel-permeation chromatography. Metabolism. 39: 155–160. [DOI] [PubMed] [Google Scholar]
- 22.Pandak W. M., Bohdan P., Franklund C., Mallonee D. H., Eggertsen G., Bjorkhem I., Gil G., Vlahcevic Z. R., and Hylemon P. B.. 2001. Expression of sterol 12alpha-hydroxylase alters bile acid pool composition in primary rat hepatocytes and in vivo. Gastroenterology. 120: 1801–1809. [DOI] [PubMed] [Google Scholar]
- 23.Lownik J. C., Conrad D. H., and Martin R. K.. 2019. A disintegrin and metalloproteinase 17 is required for ILC2 responses to IL-33. Biochem. Biophys. Res. Commun. 512: 723–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Livak K. J., and Schmittgen T. D.. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 25.Sidhu R., Jiang H., Farhat N. Y., Carrillo-Carrasco N., Woolery M., Ottinger E., Porter F. D., Schaffer J. E., Ory D. S., and Jiang X.. 2015. A validated LC-MS/MS assay for quantification of 24(S)-hydroxycholesterol in plasma and cerebrospinal fluid. J. Lipid Res. 56: 1222–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kakiyama G., Muto A., Takei H., Nittono H., Murai T., Kurosawa T., Hofmann A. F., Pandak W. M., and Bajaj J. S.. 2014. A simple and accurate HPLC method for fecal bile acid profile in healthy and cirrhotic subjects: validation by GC-MS and LC-MS. J. Lipid Res. 55: 978–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ning Y., Bai Q., Lu H., Li X., Pandak W. M., Zhao F., Chen S., Ren S., and Yin L.. 2009. Overexpression of mitochondrial cholesterol delivery protein, StAR, decreases intracellular lipids and inflammatory factors secretion in macrophages. Atherosclerosis. 204: 114–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ren S., Hylemon P. B., Marques D., Gurley E., Bodhan P., Hall E., Redford K., Gil G., and Pandak W. M.. 2004. Overexpression of cholesterol transporter StAR increases in vivo rates of bile acid synthesis in the rat and mouse. Hepatology. 40: 910–917. [DOI] [PubMed] [Google Scholar]
- 29.Pandak W. M., Hylemon P. B., Ren S., Marques D., Gil G., Redford K., Mallonee D., and Vlahcevic Z. R.. 2002. Regulation of oxysterol 7alpha-hydroxylase (CYP7B1) in primary cultures of rat hepatocytes. Hepatology. 35: 1400–1408. [DOI] [PubMed] [Google Scholar]
- 30.Suga T., Yamaguchi H., Ogura J., Shoji S., Maekawa M., and Mano N.. 2019. Altered bile acid composition and disposition in a mouse model of non-alcoholic steatohepatitis. Toxicol. Appl. Pharmacol. 379: 114664. [DOI] [PubMed] [Google Scholar]
- 31.Diczfalusy U. 2013. On the formation and possible biological role of 25-hydroxycholesterol. Biochimie. 95: 455–460. [DOI] [PubMed] [Google Scholar]
- 32.Chen C., Hu B., Wu T., Zhang Y., Xu Y., Feng Y., and Jiang H.. 2016. Bile acid profiles in diabetic (db/db) mice and their wild type littermates. J. Pharm. Biomed. Anal. 131: 473–481. [DOI] [PubMed] [Google Scholar]
- 33.Nojima K., Sugimoto K., Ueda H., Babaya N., Ikegami H., and Rakugi H.. 2013. Analysis of hepatic gene expression profile in a spontaneous mouse model of type 2 diabetes under a high sucrose diet. Endocr. J. 60: 261–274. [DOI] [PubMed] [Google Scholar]
- 34.Noebauer B., Jais A., Todoric J., Gossens K., Sutterluty-Fall H., and Einwallner E.. 2017. Hepatic cholesterol-25-hydroxylase overexpression improves systemic insulin sensitivity in mice. J. Diabetes Res. 2017: 4108768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li X., Pandak W. M., Erickson S. K., Ma Y., Yin L., Hylemon P., and Ren S.. 2007. Biosynthesis of the regulatory oxysterol, 5-cholesten-3beta,25-diol 3-sulfate, in hepatocytes. J. Lipid Res. 48: 2587–2596. [DOI] [PubMed] [Google Scholar]
- 36.Blanc M., Hsieh W. Y., Robertson K. A., Kropp K. A., Forster T., Shui G., Lacaze P., Watterson S., Griffiths S. J., Spann N. J., et al. . 2013. The transcription factor STAT-1 couples macrophage synthesis of 25-hydroxycholesterol to the interferon antiviral response. Immunity. 38: 106–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu S. Y., Aliyari R., Chikere K., Li G., Marsden M. D., Smith J. K., Pernet O., Guo H., Nusbaum R., Zack J. A., et al. . 2013. Interferon-inducible cholesterol-25-hydroxylase broadly inhibits viral entry by production of 25-hydroxycholesterol. Immunity. 38: 92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Irako T., Akamizu T., Hosoda H., Iwakura H., Ariyasu H., Tojo K., Tajima N., and Kangawa K.. 2006. Ghrelin prevents development of diabetes at adult age in streptozotocin-treated newborn rats. Diabetologia. 49: 1264–1273. [DOI] [PubMed] [Google Scholar]
- 39.Hylemon P. B., Gurley E. C., Stravitz R. T., Litz J. S., Pandak W. M., Chiang J. Y., and Vlahcevic Z. R.. 1992. Hormonal regulation of cholesterol 7 alpha-hydroxylase mRNA levels and transcriptional activity in primary rat hepatocyte cultures. J. Biol. Chem. 267: 16866–16871. [PubMed] [Google Scholar]
- 40.Khan R. S., Bril F., Cusi K., and Newsome P. N.. 2019. Modulation of insulin resistance in nonalcoholic fatty liver disease. Hepatology. 70: 711–724. [DOI] [PubMed] [Google Scholar]
- 41.Lebeaupin C., Vallee D., Hazari Y., Hetz C., Chevet E., and Bailly-Maitre B.. 2018. Endoplasmic reticulum stress signalling and the pathogenesis of non-alcoholic fatty liver disease. J. Hepatol. 69: 927–947. [DOI] [PubMed] [Google Scholar]
- 42.Schaffer J. E. 2020. Death by lipids: the role of small nucleolar RNAs in metabolic stress. J. Biol. Chem. 295: 8628–8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rutkowski D. T., Wu J., Back S. H., Callaghan M. U., Ferris S. P., Iqbal J., Clark R., Miao H., Hassler J. R., Fornek J., et al. . 2008. UPR pathways combine to prevent hepatic steatosis caused by ER stress-mediated suppression of transcriptional master regulators. Dev. Cell. 15: 829–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamamoto K., Takahara K., Oyadomari S., Okada T., Sato T., Harada A., and Mori K.. 2010. Induction of liver steatosis and lipid droplet formation in ATF6alpha-knockout mice burdened with pharmacological endoplasmic reticulum stress. Mol. Biol. Cell. 21: 2975–2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bhogal R. K., and Bona C. A.. 2005. B cells: no longer bystanders in liver fibrosis. J. Clin. Invest. 115: 2962–2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Starling S. 2017. Type 2 immunity: Hero turns villain in fatty liver. Nat. Rev. Immunol. 17: 466–467. [DOI] [PubMed] [Google Scholar]
- 47.Hart K. M., Fabre T., Sciurba J. C., Gieseck R. L. 3rd, Borthwick L. A., Vannella K. M., Acciani T. H., de Queiroz Prado R., Thompson R. W., White S., et al. . 2017. Type 2 immunity is protective in metabolic disease but exacerbates NAFLD collaboratively with TGF-beta. Sci. Transl. Med. 9: eaal3694. [DOI] [PubMed] [Google Scholar]
- 48.Maor Y., Haberman D., Segal M., Katav A., Hashmueli S., George J., and Mor A.. 2018. Expression of the chemokine CCL24 and its receptor in the sera and livers of patients with non-alcoholic fatty liver disease. J. Hepatol. 68: S341. [Google Scholar]
- 49.Thomas A. C., and Mattila J. T.. 2014. “Of mice and men”: arginine metabolism in macrophages. Front. Immunol. 5: 479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pesce J. T., Ramalingam T. R., Mentink-Kane M. M., Wilson M. S., El Kasmi K. C., Smith A. M., Thompson R. W., Cheever A. W., Murray P. J., and Wynn T. A.. 2009. Arginase-1-expressing macrophages suppress Th2 cytokine-driven inflammation and fibrosis. PLoS Pathog. 5: e1000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Iwakiri Y. 2015. Nitric oxide in liver fibrosis: the role of inducible nitric oxide synthase. Clin. Mol. Hepatol. 21: 319–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boettler T., and Thimme R.. 2018. Lost in inflammation: the functional conversion of regulatory T cells in acute hepatitis A virus infection. Gastroenterology. 154: 798–800. [DOI] [PubMed] [Google Scholar]
- 53.Li-Hawkins J., Lund E. G., Turley S. D., and Russell D. W.. 2000. Disruption of the oxysterol 7alpha-hydroxylase gene in mice. J. Biol. Chem. 275: 16536–16542. [DOI] [PubMed] [Google Scholar]
- 54.Wu Z., Martin K. O., Javitt N. B., and Chiang J. Y.. 1999. Structure and functions of human oxysterol 7alpha-hydroxylase cDNAs and gene CYP7B1. J. Lipid Res. 40: 2195–2203. [PubMed] [Google Scholar]
- 55.Mignarri A., Malandrini A., Del Puppo M., Magni A., Monti L., Ginanneschi F., Tessa A., Santorelli F. M., Federico A., and Dotti M. T.. 2014. Hereditary spastic paraplegia type 5: a potentially treatable disorder of cholesterol metabolism. J. Neurol. 261: 617–619. [DOI] [PubMed] [Google Scholar]
- 56.Mignarri A., Carecchio M., Del Puppo M., Magistrelli L., Di Bella D., Monti L., and Dotti M. T.. 2017. SPG5 siblings with different phenotypes showing reduction of 27-hydroxycholesterol after simvastatin-ezetimibe treatment. J. Neurol. Sci. 383: 39–41. [DOI] [PubMed] [Google Scholar]
- 57.Marelli C., Lamari F., Rainteau D., Lafourcade A., Banneau G., Humbert L., Monin M. L., Petit E., Debs R., Castelnovo G., et al. . 2018. Plasma oxysterols: biomarkers for diagnosis and treatment in spastic paraplegia type 5. Brain. 141: 72–84. [DOI] [PubMed] [Google Scholar]
- 58.Schüle R., Siddique T., Deng H. X., Yang Y., Donkervoort S., Hansson M., Madrid R. E., Siddique N., Schols L., and Bjorkhem I.. 2010. Marked accumulation of 27-hydroxycholesterol in SPG5 patients with hereditary spastic paresis. J. Lipid Res. 51: 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Raselli T., Hearn T., Wyss A., Atrott K., Peter A., Frey-Wagner I., Spalinger M. R., Maggio E. M., Sailer A. W., Schmitt J., et al. . 2019. Elevated oxysterol levels in human and mouse livers reflect nonalcoholic steatohepatitis. J. Lipid Res. 60: 1270–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Na J., Choi S. A., Khan A., Huh J. Y., Piao L., Hwang I., Ha H., and Park Y. H.. 2019. Integrative omics reveals metabolic and transcriptomic alteration of nonalcoholic fatty liver disease in catalase knockout mice. Biomol. Ther. (Seoul). 27: 134–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meljon A., Crick P. J., Yutuc E., Yau J. L., Seckl J. R., Theofilopoulos S., Arenas E., Wang Y., and Griffiths W. J.. 2019. Mining for oxysterols in Cyp7b1−/− mouse brain and plasma: relevance to spastic paraplegia type 5. Biomolecules. 9: 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Umetani M., Ghosh P., Ishikawa T., Umetani J., Ahmed M., Mineo C., and Shaul P. W.. 2014. The cholesterol metabolite 27-hydroxycholesterol promotes atherosclerosis via proinflammatory processes mediated by estrogen receptor alpha. Cell Metab. 20: 172–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Christenson L. K., McAllister J. M., Martin K. O., Javitt N. B., Osborne T. F., and Strauss J. F. 3rd.. 1998. Oxysterol regulation of steroidogenic acute regulatory protein gene expression. Structural specificity and transcriptional and posttranscriptional actions. J. Biol. Chem. 273: 30729–30735. [DOI] [PubMed] [Google Scholar]
- 64.Torres S., Baulies A., Insausti-Urkia N., Alarcon-Vila C., Fucho R., Solsona-Vilarrasa E., Nunez S., Robles D., Ribas V., Wakefield L., et al. . 2019. Endoplasmic reticulum stress-induced upregulation of STARD1 promotes acetaminophen-induced acute liver failure. Gastroenterology. 157: 552–568. [DOI] [PubMed] [Google Scholar]
- 65.Gold E. S., Diercks A. H., Podolsky I., Podyminogin R. L., Askovich P. S., Treuting P. M., and Aderem A.. 2014. 25-Hydroxycholesterol acts as an amplifier of inflammatory signaling. Proc. Natl. Acad. Sci. USA. 111: 10666–10671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reboldi A., Dang E. V., McDonald J. G., Liang G., Russell D. W., and Cyster J. G.. 2014. Inflammation. 25-Hydroxycholesterol suppresses interleukin-1-driven inflammation downstream of type I interferon. Science. 345: 679–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yi T., Wang X., Kelly L. M., An J., Xu Y., Sailer A. W., Gustafsson J. A., Russell D. W., and Cyster J. G.. 2012. Oxysterol gradient generation by lymphoid stromal cells guides activated B cell movement during humoral responses. Immunity. 37: 535–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Koarai A., Yanagisawa S., Sugiura H., Ichikawa T., Kikuchi T., Furukawa K., Akamatsu K., Hirano T., Nakanishi M., Matsunaga K., et al. . 2012. 25-Hydroxycholesterol enhances cytokine release and Toll-like receptor 3 response in airway epithelial cells. Respir. Res. 13: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu L., Shen S., Ma Y., Kim J. K., Rodriguez-Agudo D., Heuman D. M., Hylemon P. B., Pandak W. M., and Ren S.. 2012. 25-Hydroxycholesterol-3-sulfate attenuates inflammatory response via PPARgamma signaling in human THP-1 macrophages. Am. J. Physiol. Endocrinol. Metab. 302: E788–E799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang Y., Chen L., Pandak W. M., Heuman D., Hylemon P. B., and Ren S.. 2020. High glucose induces lipid accumulation via 25-hydroxycholesterol DNA-CpG methylation. iScience. 23: 101102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Essential data are contained in the article. All other data are available in the supplemental figures and tables.