Abstract
Xanthophyllomyces dendrorhous is a basidiomycete yeast that produces carotenoids, mainly astaxanthin. Astaxanthin is an organic pigment of commercial interest due to its antioxidant and coloring properties. X. dendrorhous has a functional SREBP pathway, and the Sre1 protein is the SREBP homolog in this yeast. However, how sterol regulatory element (Sre)1 promotes the biosynthesis of sterols and carotenoids in X. dendrorhous is unknown. In this work, comparative RNA-sequencing analysis between modified X. dendrorhous strains that have an active Sre1 protein and the WT was performed to identify Sre1-dependent genes. In addition, Sre1 direct target genes were identified through ChIP combined with lambda exonuclease digestion (ChIP-exo) assays. SRE motifs were detected in the promoter regions of several Sre1 direct target genes and were consistent with the SREs described in other yeast species. Sre1 directly regulates genes related to ergosterol biosynthesis as well as genes related to the mevalonate (MVA) pathway, which synthesizes the building blocks of isoprenoids, including carotenoids. Two carotenogenic genes, crtE and crtR, were also identified as Sre1 direct target genes. Thus, carotenogenesis in X. dendrorhous is regulated by Sre1 through the regulation of the MVA pathway and the regulation of the crtE and crtR genes. As the crtR gene encodes a cytochrome P450 reductase, Sre1 regulates pathways that include cytochrome P450 enzymes, such as the biosynthesis of carotenoids and sterols. These results demonstrate that Sre1 is a sterol master regulator that is conserved in X. dendrorhous.
Keywords: isoprenoids, nuclear receptors/sterol regulatory element-binding protein, transcription, antioxidants, molecular biology, astaxanthin, sterols, mevalonate pathway, gene regulation, cytochrome P450
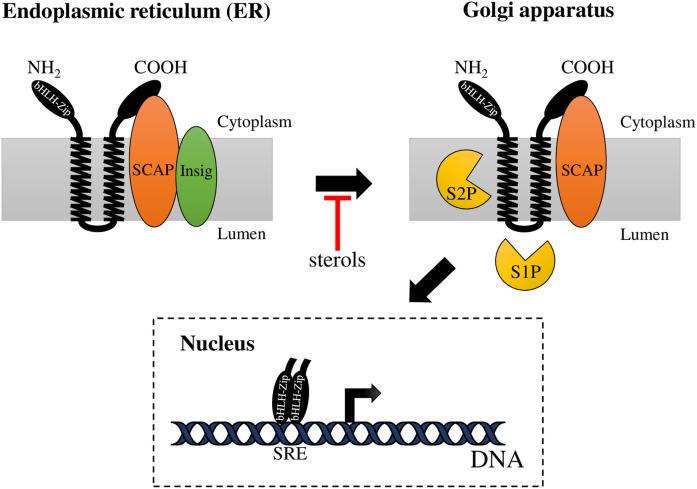
The SREBP pathway regulates sterol metabolism and homeostasis and has been well-studied in mammals (Fig. 1). This pathway includes a family of ER membrane-bound transcription factors (TFs) called SREBPs (1), which are oriented in the ER membrane in a hairpin fashion and contain two transmembrane helices. Both SREBP domains, the N-terminal domain (the TF domain, with a basic-helix-loop-helix leucine zipper, bHLH-LZ) and the C-terminal domain (the regulatory domain), face the cytoplasm (2). In the ER, the regulatory domain of SREBPs binds to the sterol-sensing protein, SREBP cleavage-activating protein (SCAP), which regulates SREBP activation in response to sterol levels. When the intracellular sterol content decreases, SCAP undergoes a conformational change that allows the transport of the SREBP-SCAP complex to the Golgi apparatus. At the Golgi, SREBPs undergo two sequential proteolytic cleavages by site-1 protease (S1P) and site-2 protease (S2P). First, S1P cleaves SREBP within its Golgi luminal loop, and then, S2P cleaves the protein at its first transmembrane segment. Upon cleavage, the N-terminal domain of SREBP is released and translocates to the nucleus where it binds to sterol regulatory elements (SREs) (3) to regulate the transcription of genes that control the uptake and biosynthesis of sterols (4).
Fig. 1.
The mammalian SREBP pathway. When sterol levels are sufficient, the SREBP-SCAP complex is retained at the ER membrane due to SCAP interaction with Insig. When sterol levels are reduced, SCAP transports SREBPs from the ER to the Golgi apparatus, where SREBP undergoes two sequential proteolytic cleavages by S1P and S2P. First, S1P (subtilisin-related serine protease) cleaves SREBP at the hydrophilic loop projected into the lumen of the Golgi apparatus and then, S2P (metallopeptidase) cuts SREBP within the first transmembrane segment. This releases the N-terminal domain of SREBP (bHLH-Zip), which enters the nucleus and binds to a SRE in the enhancer/promoter region of target genes activating their transcription.
The SREBP pathway has also been studied in some fungi, which are organisms that produce ergosterol as the main sterol (5). In Schizosaccharomyces pombe and Cryptococcus neoformans, the SREBP homolog is called Sre1 (6, 7), while in Aspergillus fumigatus, it was named SrbA (8). In these fungal systems, it was shown that, in addition to sterol homeostasis, the SREBP pathway was critical for growth under hypoxic conditions (6–8), and in C. neoformans and A. fumigatus, it was proven that this pathway is also involved in fungal pathogenesis and resistance to antifungal drugs (8, 9). In fungi, the SREBP-like proteins are activated by different mechanisms. In S. pombe and C. neoformans, a SCAP homolog (Scp1) was described (6, 7); however, A. fumigatus lacks a SCAP homolog (10). S. pombe and A. fumigatus lack S1P and S2P protease homologs, and SREBP-like proteins are proteolytically activated by a mechanism involving the Golgi Dsc (defective in Sre1 cleavage) E3 ligase complex (11) and rhomboid protease, Rbd2 (10, 12). Recently, an additional essential protease responsible for SrbA cleavage and activation (signal peptide peptidase, SppA) was described in Aspergillus nidulans (13). In contrast, a S2P protease homolog, called Stp1, was shown to be involved in the SREBP pathway in C. neoformans (7). In summary, fungal SREBP pathways exhibit differences from the mammalian pathways; and more interestingly, there are differences among fungal groups.
The basidiomycetous yeast Xanthophyllomyces dendrorhous [formerly Phaffia rhodozyma (14)] is a natural producer of astaxanthin, which is a carotenoid with antioxidant properties (15, 16). Astaxanthin is also used as a dye in aquaculture species, because the salmonid coloring is perceived as a key quality attribute by consumers (16). Carotenoids are isoprenoid compounds, whose biosynthesis derives from the mevalonate (MVA) pathway, which also supplies the precursor for ergosterol biosynthesis (17) (Fig. 2). In this way, both biosynthetic pathways are related, and several lines of evidence indicate that they share some common regulatory elements. For example, when the ergosterol pathway of X. dendrorhous was interrupted by disrupting the cytochrome P450 (P450) encoding gene, CYP61 (cyp61− mutant), ergosterol production was blocked and carotenoid content increased by approximately 2-fold compared with those of the WT strain (18). Recent studies demonstrated that X. dendrorhous has a functional SREBP pathway in which SREBP and S2P homologs have been described and were named Sre1 (19) and Stp1 (20), respectively. Functional characterization studies of the Sre1 and Stp1 encoding genes showed that Sre1 is involved in the regulation of isoprenoid biosynthesis, and its function depends on Stp1. The sre1− and Δstp1 mutations reduced sterol and carotenoid production in the cyp61− mutant strain to the levels observed in the WT strain, and the expression of only the Sre1 N-terminal domain (Sre1N mutant) increased carotenoid production by more than 2-fold compared with that of the WT strain (19, 20). By ChIP-PCR assays, it was demonstrated that Sre1 binds to the promoter region of the HMG-CoA synthase (HMGS) and HMG-CoA reductase (HMGR) genes, which are related to the MVA pathway (19) and are well-known SREBP gene targets in other organisms (4). In addition, these genes were upregulated in the cyp61− and Sre1N mutants, which could be related to an increase in the precursors used for the biosynthesis of carotenoids and sterols, resulting in the carotenoid-overproducing phenotype in these strains.
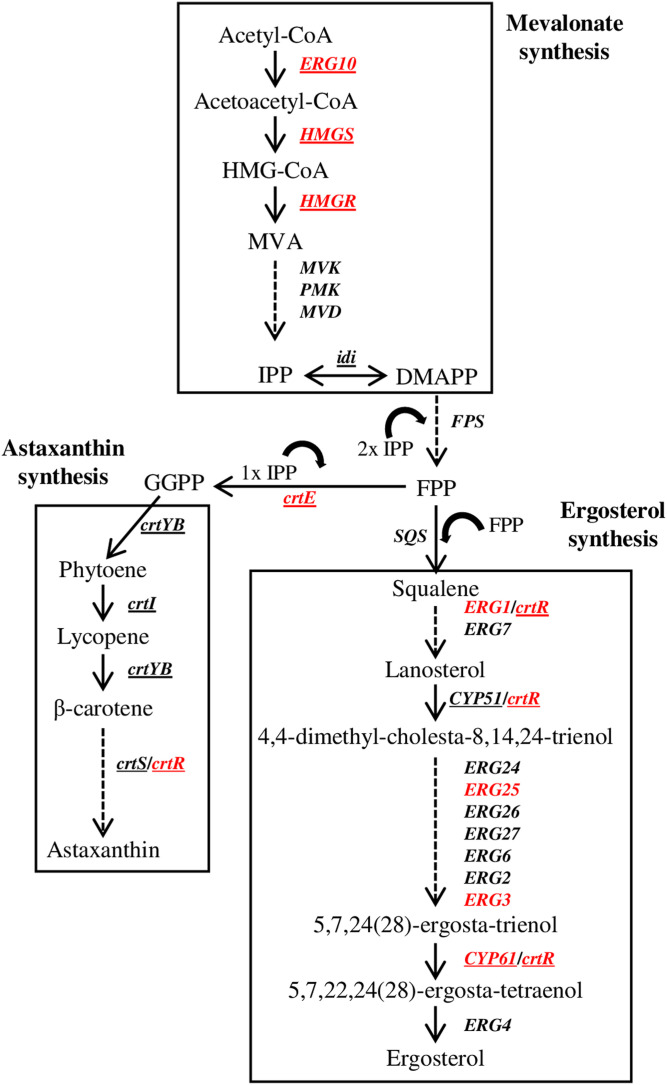
Fig. 2.
Biosynthesis of astaxanthin and ergosterol in X. dendrorhous. The astaxanthin and ergosterol pathways depend on the metabolites produced by the MVA pathway. MVK, mevalonate kinase; PMK, phosphomevalonate kinase; MVD, mevalonate diphosphate decarboxylase; IPP, isopentenyl-pyrophosphate; DMAPP, dimethylallyl-pyrophosphate; FPP, farnesyl-pyrophosphate; GGPP, geranylgeranyl-pyrophosphate. The arrows represent the catalytic step with the corresponding enzyme encoding gene in X. dendrorhous (underlined) or in S. cerevisiae. Genes in red correspond to Sre1 direct targets identified by ChIP-exo in this work (Gene ID in brackets): ERG10 (g1536), acetyl-CoA C-acetyltransferase; HMGS, (g3516); HMGR, (g1377); crtE, GGPP synthase (g5104); crtR (g5928), CPR; ERG1, squalene epoxidase (g3385); ERG25, C-4 methyl sterol oxidase (g602); ERG3, C-5 sterol desaturase (g5794); CYP61, C-22 sterol desaturase (g989). CrtR was included as a redox partner of the P450 monooxygenases Cyp51, Cyp61, and CrtS, and probably of squalene epoxidase (figure adapted from (18).
To gain a more comprehensive understanding of the regulation of gene expression by Sre1 in X. dendrorhous, in this work, comparative RNA-sequencing (RNA-seq) analysis was performed to identify Sre1-dependent genes. Sre1 direct target genes were detected by ChIP combined with lambda exonuclease digestion (ChIP-exo) (21). The results showed that Sre1 mainly regulates genes related to the biological process (BP), “ergosterol biosynthesis process”, including genes related to the MVA pathway. However, two carotenogenic genes, crtE and crtR, were also identified as Sre1 targets. In summary, the results from this work indicate that Sre1 regulates genes related to sterol biosynthesis pathways and the MVA pathway, activating their expression and thereby promoting the biosynthesis of sterols and carotenoids in this yeast.
MATERIALS AND METHODS
Growth conditions
The X. dendrorhous strains (Table 1) were cultured at 22°C with constant agitation in YM medium (0.3% yeast extract, 0.3% malt extract, and 0.5% peptone) supplemented with 1% glucose, until the late exponential phase of growth (36 h of culture) for the extraction of RNA for RNA-seq assays and the performance of ChIP-exo assays. For each assay, three biological replicates of each strain were included.
TABLE 1.
Strains used in this work
| Strains | Description | RNA-seq | ChIP-exo | Reference |
| Untagged strains of X. dendrorhous | ||||
| CBS 6938 | WT strain. | x | ATCC 96594 | |
| CBS.sre1− | Mutant derived from CBS 6938. Gene SRE1 was partially deleted (approximately 90% of the coding region was replaced by the zeocin resistance cassette). | x | x | (19) |
| CBS.cyp61− | Mutant derived from CBS 6938. The single CYP61 locus was interrupted by the hygromycin resistance cassette. | x | (18) | |
| CBS.cyp61−.sre1− | Mutant derived from CBS.cyp61−. The single SRE1 locus was replaced by the zeocin resistance cassette. | x | (19) | |
| FLAG-tagged strains of X. dendrorhous | ||||
| CBS.cyp61−.FLAG.SRE1 | Mutant derived from CBS.cyp61−. The native SRE1 gene was replaced by a gene variant that expresses the Sre1 protein fused to the 3xFLAG epitope at its N-terminus, followed by the hygromycin B resistance cassette. | x | (20) | |
| CBS.FLAG.SRE1N | Mutant derived from CBS 6938. The native SRE1 gene was replaced by a gene version that expresses Sre1N fused to the 3xFLAG epitope at its N-terminal, followed by the zeocin resistance cassette. | x | x | (19) |
An “x” indicates whether the strain was used in RNA-seq and/or ChIP-exo analysis.
X. dendrorhous genome sequencing, assembly, and annotation
DNA from the X. dendrorhous strain CBS 6938 was extracted from spheroplasts according to (22) and shipped in ethanol to the Cold Spring Harbor Laboratory (Cold Spring Harbor, NY) for single-molecule real-time (SMRT) sequencing using the Pacific Biosciences (PacBio) Sequel system (23). The genome was assembled by a hybrid assembly pipeline developed by Cold Spring Harbor Laboratory using the obtained PacBio reads and available reads of the same strain that were obtained by an Illumina HiSeq machine with 100 bp paired-end chemistry (24). PacBio reads were assembled with FALCON (25) and polished by Arrow built in SMRT Link v7.0, with the following configurations: genome_size = 19m; seed_coverage = 30; minimum_read_length = 500; pbalign_option = -minMatch 12 -bestn 10 -minPctSimilarity; min_cov = 5; min_confidence = 40. Then, the PacBio reads-only assembly was used as the primary assembly and the assembly built using Illumina reads (24) was merged to the primary assembly using Metassembler v1.5 (26), with the following configurations: bowtie2 v2.2.3 (27) as Illumina paired-end reads alignment tool; mateAn_A = 100 and mateAn_B = 500 for CE-stat computation parameters; NUCmer in MUMmer version 4.0.0 (28) as whole genome alignment tool with parameters nucmer_l = 50 and nucmer_c = 300. To check the completeness of the assembly, conserved and single copy genes were searched using Benchmarking Universal Single-Copy Orthologs v3.0.2 (BUSCO) (29) with the “snccharomycetales_odb9” database.
For formal genome annotation, the RNA-seq data available from BioProject PRJNA517352 (BioSample SAMN10829289) were mapped to the new genome with HISAT2 (30), and the aligned reads were used for gene prediction with BRAKER1 using default parameters. The coding sequences were compared against proteins of the NCBI Reference Sequence database (RefSeq) using BLASTx software (31) with an E-value threshold of 0.001. Gene Ontology (GO) annotation was performed using the Blast2GO suite (32), and the annotation data were filtered by basidiomycete taxa.
RNA extraction and RNA-seq analysis
Total RNA samples were prepared as follows: cell pellets were suspended in lysis buffer [0.002 M sodium acetate (pH 5.5), 0.5% SDS, 1 mM EDTA, in 0.1% DEPC water)] with 0.5 mm glass beads. The cells were lysed using a Mini-beadbeater-16 for 3 min, followed by the addition of 800 μl of TRI Reagent™ solution (Thermo Fisher Scientific Inc., Waltham, MA) and another 3 min of mechanical rupture. Then, chloroform was added, followed by incubation at room temperature for 10 min and centrifugation at 18,440 g for 10 min at 4°C. The aqueous phase was recovered, and the RNA was precipitated with isopropanol. The RNA quality was evaluated and quantified by spectrophotometry. The RNA samples were shipped to Macrogen (Macrogen Inc., Seoul, South Korea) for library preparation and sequencing. The libraries were prepared using the TruSeq RNA Sample Prep Kit v2, and the sequencing was carried out with the NovaSeq 6000 sequencer. The paired-end reads were obtained from Macrogen, and standard computational analysis of quality and adaptor trimming was performed. The reads were mapped to protein coding genes in the X. dendrorhous strain CBS 6938 genome assembly using the RNA-seq analysis tools from the CLC Genomics Workbench 20 software to determine the count data values. To evaluate the RNA-seq datasets, they were visualized by PCA plots using the PlotPCA function of the DESeq2 package (33). Differentially expressed genes (DEGs) were estimated using the DESeq2 and edgeR (34) packages of Bioconductor using the criteria of an adjusted P-value <0.05, and DEGs common to both analyses (DESeq2 and edgeR) were selected. Additionally, the DEGs were represented as volcano plots using the EnhancedVolcano package (35) to identify the genes whose expression varied the most among the compared strains. For GO enrichment analysis of DEGs, the Log2 fold-change was incorporated. The topGO package (36) was used for the enrichment analysis.
ChIP-exo
Formaldehyde-cross-linked cell pellets were prepared according to a standardized ChIP-PCR protocol described in (19). Then, the samples were shipped on dry ice to Peconic LLC (State College, PA) for the ChIP-exo assay, which is a variation of a ChIP-seq assay and includes lambda exonuclease digestion of sonicated chromatin from the formaldehyde-induced cross-linked protein-DNA complexes (21). The samples were treated by Peconic LLC according to a ChIP-exo protocol adapted for yeasts (37). For immunoprecipitation, the monoclonal antibody, anti-FLAG® M2 (catalog number F3165; Sigma-Aldrich, Saint Louis, MO), was used. Additionally, controls IgG (nonspecific control) and PolII (RNA polymerase II) were included in the service provided by Peconic LLC.
Alignment, peak identification, and motif discovery
The reads obtained from Peconic LLC were mapped to the X. dendrorhous strain CBS 6938 genome assembly using the aligner Bowtie2 v2.3.5.1 (27) with default settings for paired-end reads. The reads were filtered to retain the reads mapped in proper pairs using SAMtools (38). BAM files were obtained for each triplicate: strain CBS.FLAG.SRE1N had 38 million reads mapped on average, and strain CBS.cyp61−.FLAG.SRE1 had nine million mapped reads. To evaluate the ChIP-exo datasets, the reads were visualized by a PCA plot and a heatmap using the plotPCA and the plotCorrelation tools, respectively, from the suit of python tools, deepTools (39). Additionally, coverage tracks were generated with the bamCoverage tool (39) and visualized with the IGV software (40). For the peak calling step, the MACS2 tool was used (41); the size selected was 20 Mb, the peak detection was based on a q-value of 0.05, and the control used for the treatment of BAM files was an IgG BAM file. The intersect function from the BEDtools tool (42) was used to identify overlapping peaks among the MACS2 output files from each triplicate. For comparative purposes, the Epigenetics Analysis tool from the CLC Genomics Workbench 20 software was also used for the peak calling step using a maximum P-value of 0.01. To ensure the generation of a highly reliable dataset, in addition to the standard computational analysis, a manual correction of the associated genes up- and downstream of some peaks was included. Additionally, common peaks between each FLAG-tagged strain and the untagged control strain were discarded.
To map the peaks to the DEGs, the “annotate with nearby gene information” tool from the Epigenetics Analysis package from the CLC Genomics Workbench 20 software was used. For this, the list of peaks that were obtained with each peak caller was used as input, and to track each gene, gene information was taken from the genome annotation. With this tool, peaks were associated to the 5′ or 3′ end of genes, and only DEGs detected by the RNA-seq analysis were retained. Then, only those peaks that were present in the promoter region of the DEGs were kept, considering a promoter region of 5 kb upstream the translation start codon of each gene. To find a sequence motif within the ChIP-exo peaks, 100 bp centered on each of the peaks were selected. The website version of Multiple Em for Motif Elicitation (MEME) version 5.1.1 (43) was run using the classic mode for motif discovery. Additionally, the zoops model was used, and a minimum of 10 and maximum of 11 bp motif widths were selected. The most significative motif was used with the tool GO for Motifs (GOMo) version 5.1.1 (44), and S. pombe was selected as the most suitable microorganism whose promoters were scanned to match the motif provided. Additionally, to retrieve the sequences carrying the selected motif, the Find Individual Motif Occurrences (FIMO) version 5.1.1 tool (45) was used. The topGO package (36) of Bioconductor was used for enrichment analysis for GO of genes identified by ChIP-exo.
RESULTS
To gain further insight into the role of Sre1 in the biosynthesis of isoprenoid compounds in X. dendrorhous, RNA-seq analysis and ChIP-exo assays were performed using different X. dendrorhous strains. Strains CBS.FLAG.SRE1N and CBS.cyp61−/CBS.cyp61−.FLAG.SRE1 (Table 1) were selected, as these strains have an active SREBP pathway (20). Strain CBS.FLAG.SRE1N expresses the portion of the SRE1 gene that encodes Sre1N fused to the FLAG epitope, and this strain exhibits increased production of carotenoids and sterols compared with that of the WT strain (19). Similarly, the production of carotenoid and sterol intermediates is also enhanced in the mutant CBS.cyp61− strain compared with that in the WT strain (18), and by FLAG-tagging Sre1 in the latter strain (resulting in strain CBS.cyp61−.FLAG.SRE1), it was demonstrated by Western blot that, unlike the WT strain, this strain has higher levels of the active form of Sre1 (Sre1N) (20), indicating that strain CBS.cyp61− provides the conditions to activate the SREBP pathway. Therefore, the selected strains were chosen as models to evaluate the effect of the TF Sre1 at the transcriptional level in X. dendrorhous.
Genome sequencing, assembly, and annotation
To perform RNA-seq and ChIP-exo assays, genome sequencing and assembly of the X. dendrorhous WT strain CBS 6938 was performed. To this end, the strain genome was obtained by SMRT sequencing, developed by PacBio (23), and was annotated as described in the Materials and Methods. The genome assembly has a total length of 20,076,274 bases that are organized in 22 contigs, and the unsupervised annotation using BRAKER1 (46) with RNA-seq data identified 6,469 genes and 6,674 transcripts, and 205 alternative transcripts were identified for 187 genes (Table 2).
TABLE 2.
Assembly and annotation quality report
| Statistics Assembly | Values | Statistics Annotation | Values |
| Number of contigs | 22 | Number of genes | 6,469 |
| Min contig length (bp) | 3,969 | Number of transcripts | 6,674 |
| Max contig length (bp) | 2,651,629 | Number of transcripts with a BLAST hit | 5,678 |
| Median contig length (bp) | 1,107,845 | ||
| Mean contig length (bp) | 912,557.909 | ||
| Std contig length (bp) | 982,703.5 | ||
| Total length (bp) | 20,076,274 | ||
| N50 (bp) | 1,858,312 | ||
| L50 | 5 | ||
| Number of Ns per 100 kbp | 0 | ||
| GC content (%) | 47.59 |
The sequencing and assembly of the X. dendrorhous genome was carried out with SMRT sequencing developed by PacBio. The annotation was carried out with BRAKER1. Number of Ns per 100 kbp, average number of uncalled bases (Ns) per 100,000 assembly bases; N50, minimum contig length needed to cover 50% of the assembly size; L50, minimum number of contigs accounting for at least half of the assembly; GC content (%), percentage of G and C nucleotides in the assembly.
Transcriptomic analysis
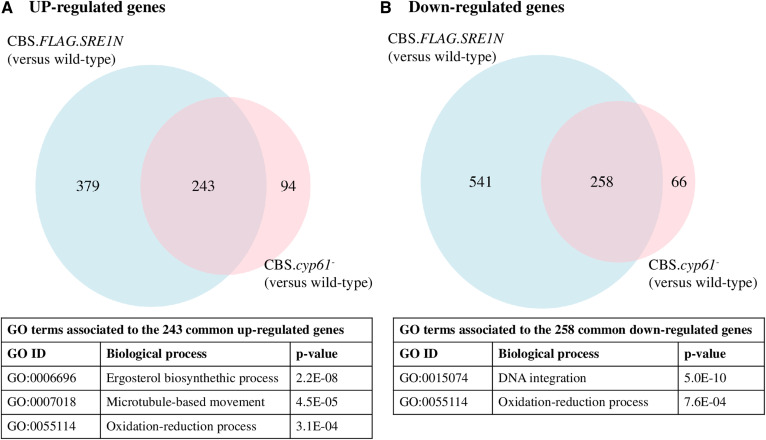
A comparative transcriptomic analysis between each mutant, CBS.FLAG.SRE1N or CBS.cyp61−, and the WT strain CBS 6938 was performed. For this analysis, strains were cultivated in triplicate under the same growth conditions, and RNA was extracted after 36 h of growth (late exponential phase) when high levels of both carotenoids and sterols are observed (47). RNA-seq reads were aligned to the X. dendrorhous genome to perform a differential expression analysis among the strains. The expression profiles of strain CBS.FLAG.SRE1N versus the WT and between strain CBS.cyp61− versus the WT are represented in PCA plots (supplemental Fig. S1A). A total of 1,421 DEGs (799 downregulated and 622 upregulated) were identified between strains CBS.FLAG.SRE1N and the WT strain, while only 661 DEGs (324 downregulated and 337 upregulated) were identified between strain CBS.cyp61− and the WT strain (supplemental Fig. S1B). Because strains CBS.FLAG.SRE1N and CBS.cyp61− share a similar phenotype (overproduction of sterols and carotenoids), a GO term enrichment analysis by BP of the common up- and downregulated genes identified in both strains compared with the WT was performed using topGO (36) (Fig. 3). A total of 243 common upregulated genes were associated with three BPs, with the ergosterol biosynthesis process (GO:0006696) being the most significant BP (Fig. 3A), while a total of 258 common downregulated genes were associated with “DNA integration” (GO:0015074) as the most significant BP (Fig. 3B).
Fig. 3.
Number and associated BPs of up- and downregulated genes in X. dendrorhous strains CBS.FLAG.SRE1N and CBS.cyp61−. Venn diagrams of upregulated (A) and downregulated (B) genes in strains CBS.FLAG.SRE1N and CBS.cyp61− when compared with the WT strain CBS 6938. DEGs were estimated using the DESeq2 and edgeR packages using the |log2 fold change| ≥ 1, P-adjusted < 0.05 (DESeq2) or FDR < 0.05 (edgeR) as criteria. DEGs common to both analyses (DESeq2 and edgeR) were selected and plotted. The GO term enrichment analysis was performed with topGO using the “weight01” algorithm and “fisher” statistic to take the GO hierarchy into account (cut-off P-value < 0.001).
Next, a GO term enrichment analysis was performed independently for both strains considering the genes that were up- and downregulated compared with the WT strain (Table 3). Again, the ergosterol biosynthesis process was the most significant BP among the upregulated genes in both comparisons. This result was expected as there were 243 common genes related to this BP (Fig. 3A) and, at the phenotype level, strains CBS.FLAG.SRE1N and CBS.cyp61− produce more sterols and carotenoids than the WT strain (19). It is important to note that the ergosterol biosynthesis process BP, represented by upregulated genes in CBS.FLAG.SRE1N and CBS.cyp61− (Table 3), includes genes related to the MVA pathway, such as the HMGS and HMGR genes (supplemental Table S1). The MVA pathway supplies common metabolites to both carotenoid and ergosterol biosynthesis in X. dendrorhous (Fig. 2), which could explain the overproducer phenotype of sterols and carotenoids in strains CBS.FLAG.SRE1N and CBS.cyp61−. Additionally, the ergosterol biosynthesis process BP includes the P450 reductase (CPR) encoding gene crtR, which is also involved in both sterol synthesis and carotenogenesis (Fig. 2). Thus, genes of the MVA pathway and crtR should also be considered as carotenogenic genes.
TABLE 3.
GO term enrichment analysis (by BP) of up- and downregulated genes identified by comparative RNA-seq analysis in X. dendrorhous strains
| GO ID | BP | Annotated | Significant | Expected | Rank in Classic Fisher | P (classic) | P (weight01) | |
| RNA-seq analysis of strain CBS.FLAG.SRE1N compared with the WT CBS 6938 (DEGs: 1,421 genes. UP: 622, DW: 799) | ||||||||
| UP | ||||||||
| 1 | GO:0006696 | Ergosterol biosynthetic process | 14 | 9 | 1.43 | 3 | 1.3E-06 | 1.3E-06 |
| DW | ||||||||
| 1 | GO:0055114 | Oxidation-reduction process | 409 | 91 | 44.58 | 1 | 6.8E-13 | 4.9E-13 |
| 2 | GO:0055085 | Transmembrane transport | 349 | 64 | 38.04 | 3 | 7.6E-06 | 8.1E-08 |
| 3 | GO:0015074 | DNA integration | 52 | 18 | 5.67 | 2 | 4.0E-06 | 4.0E-06 |
| RNA-seq analysis of strain CBS.cyp61− compared with the WT CBS 6938 (DEGs: 661 genes. UP: 337, DW: 324) | ||||||||
| UP | ||||||||
| 1 | GO:0006696 | Ergosterol biosynthetic process | 14 | 8 | 0.83 | 4 | 3.0E-07 | 3.0E-07 |
| 2 | GO:0006334 | Nucleosome assembly | 11 | 5 | 0.66 | 33 | 2.5E-04 | 2.5E-04 |
| 3 | GO:0007018 | Microtubule-based movement | 17 | 6 | 1.01 | 34 | 2.9E-04 | 2.9E-04 |
| 4 | GO:0055114 | Oxidation-reduction process | 409 | 41 | 24.36 | 36 | 3.8E-04 | 7.4E-04 |
| DW | ||||||||
| 1 | GO:0015074 | DNA integration | 52 | 15 | 2.08 | 1 | 6.0E-10 | 6.0E-10 |
| 2 | GO:0055114 | Oxidation-reduction process | 409 | 32 | 16.32 | 3 | 9.2E-05 | 2.0E-04 |
| RNA-seq analysis of strain CBS.cyp61−.sre1− compared with strain CBS.cyp61− (DEGs: 469 genes. UP: 230, DW: 239) | ||||||||
| UP | ||||||||
| 1 | GO:0055114 | Oxidation-reduction process | 409 | 31 | 11.21 | 1 | 3.8E-08 | 4.1E-08 |
| 2 | GO:0055085 | Transmembrane transport | 349 | 22 | 9.56 | 2 | 1.2E-04 | 2.2E-06 |
| DW | ||||||||
| 1 | GO:0006696 | Ergosterol biosynthetic process | 14 | 8 | 0.57 | 5 | 1.5E-08 | 1.5E-08 |
| 2 | GO:0007018 | Microtubule-based movement | 17 | 6 | 0.69 | 20 | 3.5E-05 | 3.5E-05 |
| 3 | GO:0055114 | Oxidation-reduction process | 409 | 31 | 16.69 | 26 | 3.4E-04 | 1.8E-04 |
| 4 | GO:0008610 | Lipid biosynthetic process | 92 | 18 | 3.75 | 4 | 1.5E-08 | 2.0E-04 |
| RNA-seq analysis of strain CBS.sre1− compared with the WT CBS 6938 (DEGs: 13 genes. UP: 3, DW: 10) | ||||||||
| DW | ||||||||
| 1 | GO:0006696 | Ergosterol biosynthetic process | 14 | 5 | 0.04 | 2 | 7.0E-11 | 7.0E-11 |
| 2 | GO:0008610 | Lipid biosynthetic process | 92 | 8 | 0.25 | 1 | 2.1E-12 | 1.0E-06 |
The enrichment analysis was performed with the topGO package. The table shows GO terms according to the “classic” and “weight01” algorithms and the “fisher” statistic; a P < 0.001 cut-off was used for the “weight01” algorithm. GO terms in the table were sorted by the P associated with the “weight01” algorithm. No significant results were found for the upregulated genes in strain CBS.sre1− compared with the WT CBS 6938. UP, upregulated genes; DW, downregulated genes.
As a control, the transcriptomic effect of the sre1− mutation was also evaluated by comparing strain CBS.sre1− versus the WT strain and strain CBS.cyp61−.sre1− versus CBS.cyp61−, including a GO term enrichment analysis of up- and downregulated genes (Table 3). The sre1− mutation in strain CBS.cyp61− reduces carotenoid and sterol production to WT levels (19), and these processes (sterol biosynthesis and carotenogenesis) were represented in the enrichment analysis of the downregulated genes in the sre1− mutant strains (Table 3). In an enrichment analysis, a gene can have more than one GO term associated; therefore, a gene can be associated with more than one BP. In our analysis, several genes related to the biosynthesis of carotenoids and to the biosynthesis of sterols were also associated with the “oxidation-reduction process” (GO:0055114) BP. These genes included HMGR, HMGS, crtR, and other sterol enzyme-encoding genes (supplemental Table S1).
The quality of the dataset of strain CBS.sre1− only allowed a significant enrichment analysis of the downregulated genes (Table 3). The sre1− mutation in the WT context affected BPs related to the biosynthesis of sterols, suggesting that there are basal levels of Sre1N in the WT strain that regulate sterol biosynthesis in X. dendrorhous.
ChIP-exo analysis
To evaluate which of the DEGs could be Sre1 direct targets in X. dendrorhous, a ChIP-exo assay was performed. The quality of the immunoprecipitation using the same antibody against the FLAG epitope fused to Sre1 was previously confirmed by ChIP-PCR (19). The ChIP-exo assay was performed using FLAG.Sre1-tagged strains CBS.FLAG.SRE1N and CBS.cyp61−.FLAG.SRE1, and strain CBS.sre1− was included as a control, as it lacks the SRE1 gene and the FLAG encoding sequence (Table 1). These strains were cultivated in triplicate under the same conditions used for RNA extraction in the transcriptomic analysis. The variability and correlation among datasets are represented as a PCA plot and heatmap, respectively (supplemental Fig. S1C, D). The results showed variability among strains and correlation between replicates, with only slight discrepancies in one of the replicates of each of the strains CBS.FLAG.SRE1N and CBS.cyp61−.FLAG.SRE1 (supplemental Fig. S1C, D; replicates “I” and “H,” respectively). The reads were mapped to the X. dendrorhous genome, but only significant peaks present in at least two of the three biological replicates from each strain (CBS.FLAG.SRE1N or CBS.cyp61−.FLAG.SRE1) were selected. The peaks that were also present in the control strain CBS.sre1− were discarded. For comparative purposes, two peak callers were used, namely, MACS2 (41) and the peak caller from the CLC Genomics Workbench 20 software for ChIP-seq analysis. The coinciding peaks that were identified with both peak callers (peaks in which the summit differed in 300 nucleotides at most when comparing both peak callers) or peaks that were identified with only one of the peak callers were associated with the DEGs detected by the RNA-seq comparative analysis of strains CBS.FLAG.SRE1N and CBS.cyp61−. The ChIP-exo results of each of the X. dendrorhous FLAG.Sre1-tagged strains are presented below.
CBS.FLAG.SRE1N.
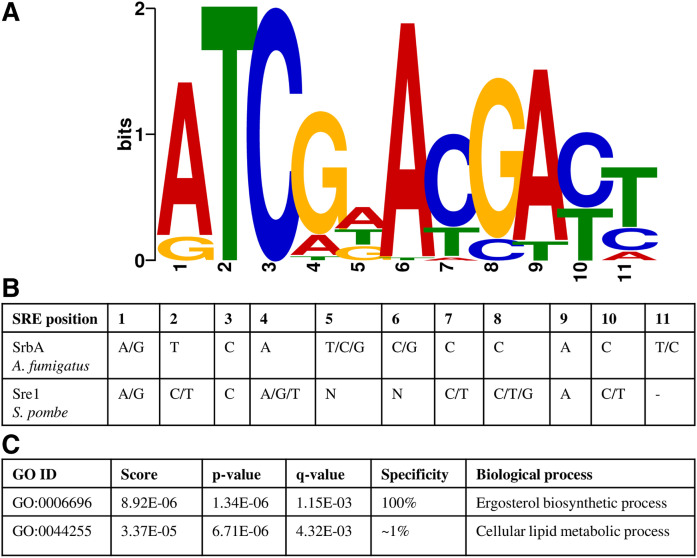
Among replicates of strain CBS.FLAG.SRE1N, approximately 600 peaks were associated with DEGs when using the MACS2 peak caller. To determine the motif associated with the DNA binding site of Sre1, a region of 100 nucleotides spanning each peak summit (±50 nucleotides from summit position) was selected. These DNA sequences were analyzed with the MEME tool, which identifies DNA sequence motifs among a collection of sequences (43). A potential SRE motif was found (supplemental Fig. S2A1), but this motif was not present in all of the DNA sequences associated with peaks close to known Sre1 targets in other organisms, such as the SRE gene itself, whose autoregulation has been reported in mammals and fungi (48, 49). SRE motif searches were then performed using only DNA sequences derived from peaks associated with up- or downregulated genes among the DEGs obtained from the RNA-seq analysis of strain CBS.FLAG.SRE1N compared with the WT. In this way, a characteristic SRE motif was detected in the DNA sequences from the peaks associated with upregulated genes (Fig. 4A), which on average was located at 18 ± 10 nucleotides from the peak summit position. The MEME tool identified 56 sites that contributed to the construction of the SRE motif (an 11 bp binding region) using 335 DNA sequences (associated with 335 peaks) as input. The constructed SRE motif (MEME E-value 2.3E-028) resembled the binding motif described for SrbA of A. fumigatus [5′-(A/G)TCA(T/C/G)(C/G)CCAC(T/C)-3′], with positions 1, 2, and 3 dominated by ATC, a cytosine in position 7, and an adenine in position 9 (49) (Fig. 4B). Additionally, the X. dendrorhous SRE motif retains the conserved cytosine at the third position, which was reported in the SRE motif of A. fumigatus (49), in the predicted SRE motif of S. pombe identified from the promoter regions of Sre1-dependent genes [5′-(A/G)(C/T)C(A/G/T)NN(C/T)(C/T/G)A(C/T)-3′] (50) (Fig. 4B), and in the SRE-1 motif of mammals (5′-ATCACCCCAC-3′) reported in gene promoters of the LDL receptor (51, 52), HMGS (53), and HMGR (54). We searched for associations between the identified SRE motif of X. dendrorhous and GO terms using the GOMo tool, which predicts the role of a TF by detecting associations between a user-specified DNA regulatory motif (expressed as a position weight matrix, PWM) and GO terms (44). Using the S. pombe database as the most suitable among those available to search GO terms associated with a motif, the GOMo tool associated the X. dendrorhous SRE motif with the BPs “ergosterol biosynthetic process” (with 100% specificity) and “cellular lipid metabolic process” (with ∼1% specificity) (Fig. 4C). These results emphasize the role of Sre1 in sterol biosynthesis regulation in X. dendrorhous.
Fig. 4.
Analysis of the X. dendrorhous SRE motif. A: The SRE motif of X. dendrorhous identified with the MEME tool (MEME E-value 2.3E-028). The 335 DNA sequences associated with peaks (determined with MACS2) close to upregulated genes detected by comparative RNA-seq analysis of strain CBS.FLAG.SRE1N versus the WT CBS 6938 were used as input. B: SRE motifs described in fungi. C: Association of the X. dendrorhous SRE motif with GO terms. The X. dendrorhous SRE motif was used as input for the GOMo tool.
On the other hand, a motif that resembles an E-box motif (CANNTG), recognized by bHLH TFs (55), was detected in sequences from peaks associated to downregulated genes (supplemental Fig. S2C1), which on average was located at 16 ± 14 nucleotides from the peak summit position. Although these genes were downregulated in strain CBS.FLAG.SRE1N when compared with the WT, they did not show to be differentially expressed in strains CBS.cyp61−.sre1− (when compared with CBS.cyp61−) or CBS.sre1− (when compared with the WT strain), where they should be overexpressed due to the sre1− mutation to support the potential role of Sre1 as a repressor.
The identified X. dendrorhous SRE motif was detected in DNA sequences from peaks that were close to 42 genes (supplemental Table S2). In approximately 80% of these genes, the peak was located within 1 kb upstream the translation start codon and about 10% of them had the peak located beyond 1 kb, but less than 2 kb upstream of the translation start codon. On the other hand, the peak caller from the CLC Genomics Workbench 20 software detected approximately 570 peaks associated with DEGs among the biological replicates of strain CBS.FLAG.SRE1N, and among these peaks, 274 peaks were associated with upregulated genes. The identified SRE motif was very similar to that identified with the MACS2 peak caller (supplemental Fig. S2B1), and the motif was associated to the DNA sequences from peaks located mostly within 1 kb upstream the translation start codon of the genes (supplemental Table S2). In summary, 27 genes were identified with both peak callers (coinciding peaks associated with the same genes), and 29 genes were associated with a unique peak identified with only one of the peak callers in strain CBS.FLAG.SRE1N.
CBS.cyp61−.FLAG.SRE1.
Previously, it was shown that the sre1− and Δstp1 mutations in a WT genetic context did not strongly affect sterol or carotenoid production. However, these mutations reduced the production of both types of metabolites to WT levels in strain CBS.cyp61−, which is an ergosterol biosynthesis mutant that overproduces sterols and carotenoids (18). Additionally, the active form of Sre1 was detected by western blot in a CBS.cyp61− genetic context (in strain CBS.cyp61−.FLAG.SRE1), demonstrating that the SREBP pathway is active in this mutant (20). Thus, a ChIP-exo assay w′s performed using strain CBS.cyp61−.FLAG.SRE1 cultured under the same conditions as strains CBS.FLAG.SRE1N and CBS.sre1−. In this analysis, in addition to the DEGs detected by RNA-seq of strain CBS.cyp61− versus the WT strain, the DEGs of the strain CBS.FLAG.SRE1N versus the WT strain were also included, as the SRE1 gene fold-change in strain CBS.cyp61− compared with the WT did not satisfy the RNA-seq analysis criteria (|log2 fold change| ≥ 1) despite having a peak associated with an SRE motif. Thus, compared with strain CBS.FLAG.SRE1N, fewer common peaks associated with DEGs were found among replicates of strain CBS.cyp61−.FLAG.SRE1 (supplemental Table S3). This result could be attributed to the “natural” processing of Sre1 as Sre1 is normally activated in strain CBS.cyp61−.FLAG.SRE1, unlike in strain CBS.FLAG.SRE1N where the active Sre1 form (Sre1N) is independent of proteolytic cleavage by Stp1. Despite these differences, the identified SRE motif in the ChIP-exo samples from strain CBS.cyp61−.FLAG.SRE1, which was located at 15 ± 11 nucleotides from the peak summit position, was very similar to that identified in strain CBS.FLAG.SRE1N. Unlike strain CBS.FLAG.SRE1N, a similar SRE motif was identified in strain CBS.cyp61−.FLAG.SRE1 when using DNA sequences associated with peaks close to DEGs as well as when using peaks only associated to upregulated genes (supplemental Fig. S2A2, B2). This difference could be related to the different number of DEGs identified in both strains. Strain CBS.FLAG.SRE1N has more DEGs than strain CBS.cyp61−.FLAG.SRE1, probably because the first expresses a nonregulated form of Sre1 so it represents a less physiological strain than strain CBS.cyp61−.FLAG.SRE1 and thus may yield false positives. Interestingly, like in strain CBS.FLAG.SRE1N, a motif that resembles an E-box motif (located at 19 ± 15 nucleotides from the peak summit position) was also found in DNA sequences associated with peaks close to downregulated genes in strain CBS.cyp61−.FLAG.SRE1 (supplemental Fig. S2C1, C2). However, also the RNA-seq analysis of strains CBS.cyp61−.sre1− (versus CBS.cyp61−) and CBS.sre1− (versus the WT strain) did not provide evidence enough to support the potential role of Sre1 as a repressor. In the identified SRE motif, positions 1, 2, and 3 (5′-ATC-3′), 7 (C), and 9 (A) are preserved, and the small variations that were observed were related to the number of sequences used as input. Additionally, genes with an SRE motif at the promoter region coincide with those identified in strain CBS.FLAG.SRE1N (supplemental Table S3). About 90% of the genes had the peak located within 1 kb upstream the translation start codon. The presence of an SRE in the peak located in the promoter region of the SRE1 gene, which has the same peak distribution as that observed in the SRE1 gene from strain CBS.FLAG.SRE1N, implies that the SREBP pathway is activated in strain CBS.cyp61−.FLAG.SRE1 (Fig. 5). Despite the fact that several affected BPs are observed in mutant strain CBS.cyp61− compared with the WT strain (Table 3), Sre1 is only linked to processes related to the biosynthesis of sterols.
Fig. 5.
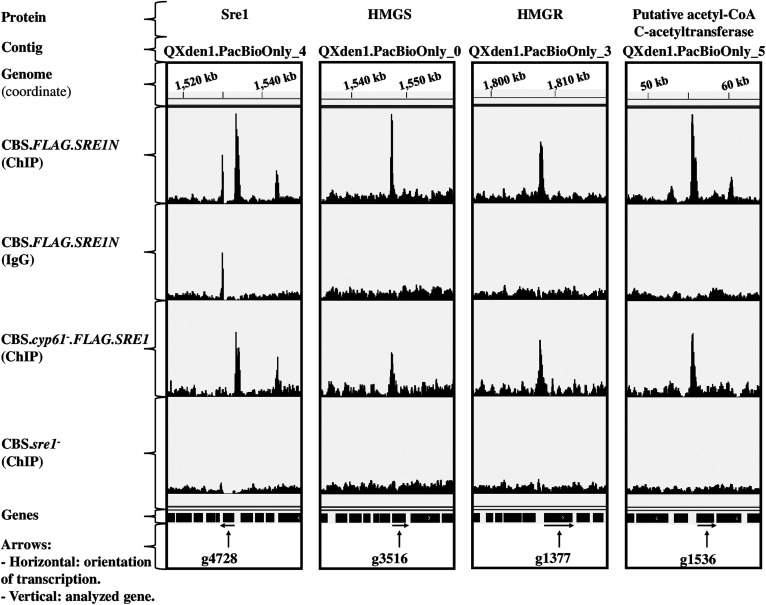
Distribution of peaks associated with Sre1 target genes that encode proteins related to carotenoid and sterol biosynthesis. The figure shows tracks (coverage bigWid files) of the ChIP-exo assay of strains CBS.FLAG.SRE1N, CBS.cyp61−.FLAG.SRE1, and CBS.sre1− using the IGV software. ChIP: Immunoprecipitation assay with anti-FLAG antibody against FLAG-tagged Sre1. IgG: Isotype control assay with IgG. Peaks were located in the promoter regions of the genes that encode (Gene ID in brackets) Sre1 (g4728), HMGS (g3516), HMGR (g1377), and putative acetyl-CoA C-acetyltransferase (g1536).
Sre1 is a conserved sterol master regulator in X. dendrorhous
Taking together the common ChIP-exo results from strains CBS.FLAG.SRE1N and CBS.cyp61−.FLAG.SRE1, a list of Sre1 direct target genes in X. dendrorhous was created (Table 4). All these genes were associated with upregulated genes from the RNA-seq experiment of strains CBS.FLAG.SRE1N and CBS.cyp61−, versus the WT, and with downregulated genes from the RNA-seq analysis of the sre1− strains (Table 4). Interestingly, the GO term enrichment analysis of these genes with TopGO (36) showed that the BPs associated with the sterols’ biosynthesis processes were the most significant (Table 5), among them the ergosterol biosynthetic process BP was also a significant BP in the enrichment analysis of the upregulated genes as in the RNA-seq analysis of strains CBS.FLAG.SRE1N and CBS.cyp61−.FLAG.SRE1, compared with the WT strain. These results imply that in general, the identified genes are associated with the sterol biosynthesis process, and these results emphasize the role of Sre1 in the regulation of sterol biosynthesis in X. dendrorhous, as expected for a homolog of the master regulators of lipid homeostasis, namely, the SREBP TFs (56). As in the RNA-seq enrichment analysis, the GO terms associated with the sterol biosynthesis processes included the HMGR and HMGS genes related to the MVA pathway and the crtR gene, which in X. dendrorhous are also involved in carotenoid biosynthesis (Table 5).
TABLE 4.
ChIP-exo results from upregulated genes detected in both strains, CBS.FLAG.SRE1N and CBS.cyp61−.FLAG.SRE1, using two peak callers
| Contig | Gene ID | Product | RNA-seq (Log2 Fold-Change; Adjusted P < 0.05) | MEME Tool | ||||
| CBS.FLAG.SRE1N versus WT | CBS.cyp61− versus WT | CBS.cyp61−.sre1− versus CBS.cyp61− | CBS.sre1−versus WT | SRE Motif | DNA Strand | |||
| QXden1.PacBioOnly_0 | g3516 | HMGS | 4.6 | 3.2 | −4.5 | −1.5 | ATCGGACGACT | + |
| QXden1.PacBioOnly_0 | g3385 | SE-domain-containing protein (squalene epoxidase) | 2.7 | 1.7 | −2.3 | ND | ATCGTACGATC | + |
| QXden1.PacBioOnly_0 | g3176 | Putative brefeldin A resistance protein | 2.6 | 1.4 | −1.3 | ND | ATCGAATGATT | + |
| QXden1.PacBioOnly_0 | g3515 | WD40 repeat domain 85 | 1.7 | 1.2 | −1.4 | ND | ATCGGACGACT | + |
| QXden1.PacBioOnly_1 | g602 | C4-methyl sterol oxidase | 1.8 | 1.4 | −2.7 | −1.6 | ATCGAACGATT/ATCGTTCGATC | +/− |
| QXden1.PacBioOnly_2 | g5168 | Glyoxalase I | 2.3 | 2.2 | −2.3 | ND | ATCGAACCACA | + |
| QXden1.PacBioOnly_2 | g5104 | Terpenoid synthase (CrtE) | 1.8 | 0.6 | −0.8 | ND | GTCGAACCACC | − |
| QXden1.PacBioOnly_2 | g5169 | Multidrug resistance-associated ABC transporter | 2.7 | 0.9 | −1.8 | ND | ATCGAACCACA | + |
| QXden1.PacBioOnly_3 | g944 | RTA1-domain-containing protein | 4.6 | 3.1 | −2.9 | ND | ATCGAACGTCA | + |
| QXden1.PacBioOnly_3 | g1193 | NA | 4.5 | 3.4 | −4.4 | ND | ATCGTACGACA/GTCGTACGATA | +/− |
| QXden1.PacBioOnly_3 | g943 | NAD-P-binding protein | 2.9 | 2.6 | −1.7 | ND | ATCGAACGTCA | + |
| QXden1.PacBioOnly_3 | g905* | Choline kinase | 1.4 | 0.7 | −0.9 | ND | GTCGAACGACT | + |
| QXden1.PacBioOnly_3 | g1192 | Mandelate racemase/muconate lactonizing enzyme domain-containing protein | 1.3 | 0.4 | −0.8 | ND | ATCGTACGACA/GTCGTACGATA | +/− |
| QXden1.PacBioOnly_3 | g1377 | HMGR (NADPH) | 2.5 | 1.3 | −2.2 | −1.0 | ATCGTCCCAC | − |
| QXden1.PacBioOnly_3 | g989 | C-22 sterol desaturase (Cyp61) | 1.6 | −1.8 | −0.9 | ND | ATCGACCCAC | + |
| QXden1.PacBioOnly_3 | g806 | Hypothetical protein MELLADRAFT_117746 | 1.3 | 1.4 | −1.8 | ND | AACGTATGAC | + |
| QXden1.PacBioOnly_3 | g904 | Delta-sterol C-methyltransferase (C-24 methyl transferase) | 4.8 | 4.2 | −6.0 | −1.9 | GTCGAACGACT | + |
| QXden1.PacBioOnly_4 | g4709 | Aquaporin-like protein | 2.8 | 2.5 | −1.2 | ND | ATCGTACGTTT | + |
| QXden1.PacBioOnly_4 | g4728 | Sterol regulatory element-binding protein 1-like (Sre1) | 1.3 | 0.4 | −1.2 | −2.5 | ATCGTACGTTT | + |
| QXden1.PacBioOnly_4 | g4312 | NA | 1.1 | ND | −0.4 | ND | GTCGGACGACT | − |
| QXden1.PacBioOnly_4 | g4710 | Predicted protein | 1.1 | 0.4 | −0.6 | ND | ATCGTACGTTT | + |
| QXden1.PacBioOnly_5 | g1536 | Putative acetyl-CoA C-acetyltransferase (Erg10p) | 2.6 | 2.2 | −1.8 | ND | ATCGTACGATT/ATCGTACGATC | +/− |
| QXden1.PacBioOnly_6 | g5758 | Multidrug resistance-associated ABC transporter | 2.6 | 1.6 | −1.6 | ND | ATCGTATGACC/GTCATACGATT | +/− |
| QXden1.PacBioOnly_6 | g5928 | Cytochrome P450 oxidoreductase (CrtR) | 2.2 | 1.7 | −2.4 | ND | ATCGAACGACT | + |
| QXden1.PacBioOnly_6 | g5794* | Fatty acid hydroxylase (C-5 desaturase) | 2.0 | ND | −1.9 | −1.2 | ATCGAACCACT | − |
| QXden1.PacBioOnly_6 | g5613 | Conserved hypothetical protein | 3.3 | 2.4 | −3.0 | ND | ATCGAAAGACT | − |
| QXden1.PacBioOnly_7 | g2548* | MFS general substrate transporter | 3.8 | 2.6 | −2.7 | ND | ATCGTACGACA/GTCGTACGATT | −/+ |
| QXden1.PacBioOnly_9 | g4112 | NA | 2.3 | 1.6 | −2.0 | ND | ATCGAACGACC | + |
| QXden1.PacBioOnly_9 | g4000 | Alternative cyclin Pcl12 | 1.5 | 0.5 | −0.8 | ND | ATCGATCGATC/ATCGATCGATC | +/− |
| QXden1.PacBioOnly_9 | g4113 | Phospholipid-translocating ATPase | 1.5 | 0.5 | −0.9 | ND | ATCGAACGACC | + |
The table shows genes associated with peaks identified with the MACS2 peak caller and/or the peak caller from the CLC Genomics Workbench 20 software. The table includes genes that satisfy the following criteria: peak located in the promoter region within 2 kb of the 5′ end of the translation start codon, and Log2 Fold-Change with an adjusted P < 0.05 in the RNA-seq experiments. The table shows the SRE motif (Fig. 4A) associated to the peaks identified with the MACS2 peak caller, except for genes g1377, g989, and g806, where the SRE motif (supplemental Fig. S2B1) was associated to peaks identified with the peak caller from the CLC Genomics Workbench 20 software. ND, not detected; NA, no annotation associated. An “*” in the Gene ID designates peak located close to the translation start codon.
TABLE 5.
GO term enrichment analysis (by BP) applied to genes detected in both strains, CBS.FLAG.SRE1N and CBS.cyp61−.FLAG.SRE1, associated with ChIP-exo peaks
| GO ID | BP and Associated Genes | Annotated | Significant | Expected | Rank in Classic Fisher | P (classic) | P (weight01) |
| GO:0006696 | Ergosterol biosynthetic process | 14 | 6 | 0.08 | 1 | 2.7E-11 | 2.7E-11 |
| g1377: HMGR | |||||||
| g3385: SE-domain-containing protein (Squalene epoxidase) | |||||||
| g3516: HMGS | |||||||
| g5928: Cytochrome P450 oxidoreductase (CrtR) | |||||||
| g904: Delta-sterol C-methyltransferase | |||||||
| g989: C-22 sterol desaturase (Cyp61) | |||||||
| GO:0008299 | Isoprenoid biosynthetic process | 13 | 3 | 0.07 | 21 | 3.6E-05 | 6.7E-04 |
| g1377: HMGR | |||||||
| g3516: HMGS | |||||||
| g5104: Terpenoid synthase (CrtE) | |||||||
| GO:0008610 | Lipid biosynthetic process | 92 | 9 | 0.49 | 13 | 2.3E-10 | 2.3E-03 |
| g1377: HMGR | |||||||
| g3385: SE-domain-containing protein (Squalene epoxidase) | |||||||
| g3516: HMGS | |||||||
| g5104: Terpenoid synthase (CrtE) | |||||||
| g5794: Fatty acid hydroxylase | |||||||
| g5928: Cytochrome P450 oxidoreductase (CrtR) | |||||||
| g602: C4-methyl sterol oxidase | |||||||
| g904: Delta-sterol C-methyltransferase | |||||||
| g989: C-22 sterol desaturase (Cyp61) | |||||||
| GO:0055114 | Oxidation-reduction process | 409 | 7 | 2.19 | 27 | 3.6E-03 | 3.6E-03 |
| g1377: HMGR | |||||||
| g3385: SE-domain-containing protein (Squalene epoxidase) | |||||||
| g5794: Fatty acid hydroxylase | |||||||
| g5928: Cytochrome P450 oxidoreductase (CrtR) | |||||||
| g602: C4-methyl sterol oxidase | |||||||
| g943: NAD-P-binding protein | |||||||
| g989: C-22 sterol desaturase (Cyp61) | |||||||
| GO:0010142 | Farnesyl diphosphate biosynthetic process | 1 | 1 | 0.01 | 28 | 5.4E-03 | 5.4E-03 |
| g3516: HMGS |
The enrichment analysis was performed with the topGO package, using as input the list of Sre1 direct target genes included in Table 4. Table shows GO terms according to the “classic” and “weight01” algorithms and the “fisher” statistic. Significant GO terms were selected and sorted by the P associated with the algorithm “weight01”.
Genes with SREs in their promoter regions (Table 4, supplemental Tables S2, S3) include the Sre1 encoding gene (g4728) itself, which coincides with mammals as both mammalian SREBP encoding genes are among the over 30 genes regulated by SREBP in mammals (57). In X. dendrorhous, two P450 monooxygenases involved in ergosterol biosynthesis have been characterized, Cyp51 (58) and Cyp61 (18). The peak callers detected peaks associated to SRE motif in the promoter region of the Cyp51 and Cyp61 encoding genes (supplemental Fig. S3), but it is important to highlight that the Cyp51 encoding gene was not included in the list of Sre1 direct target genes (Table 4), because the peak callers did not detect the peak shown by the IGV software in the promoter region of this gene in the strain CBS.cyp61−.FLAG.SRE1 (supplemental Fig. S3). Other Sre1 direct target genes correspond to the acetyl-CoA C-acetyltransferase, HMGS, and HMGR encoding genes (Table 4, g1536, g3516, g1377; Fig. 5) of the MVA pathway. In addition, it was demonstrated by ChIP-PCR that, in X. dendrorhous, Sre1 binds to the promoter region of the HMGS and HMGR encoding genes (19). An interesting Sre1 direct target gene corresponds to the CPR encoding gene (Table 4, g5928; Fig. 6), which is involved in ergosterol biosynthesis as well as in carotenoid biosynthesis in X. dendrorhous (47, 59). Among specific genes of the ergosterol biosynthesis, genes ERG25 and ERG3 (Table 4, g602, g5794) were detected as Sre1 direct target. Gene ERG3 was recently characterized in X. dendrorhous and its mutation blocked ergosterol biosynthesis (60). Another gene that probably is involved in the sterol biosynthesis pathway includes the aquaporin-like protein encoding gene (Table 4, g4709), as it was reported that aquaporin-8 participates in the SREBP pathway favoring the cholesterol synthesis in liver cells (61). Interestingly, results also suggest that Sre1 is involved in the synthesis of phospholipids through the regulation of the choline kinase encoding gene (Table 4, g905), which is involved in the synthesis of phosphatidylcholine, an abundant glycerophospholipid present in eukaryotic cells’ membranes (62). Also, through the regulation of the phospholipid-translocating ATPase encoding gene (Table 4, g4113), Sre1 would be involved in phospholipid localization in membranes.
Fig. 6.
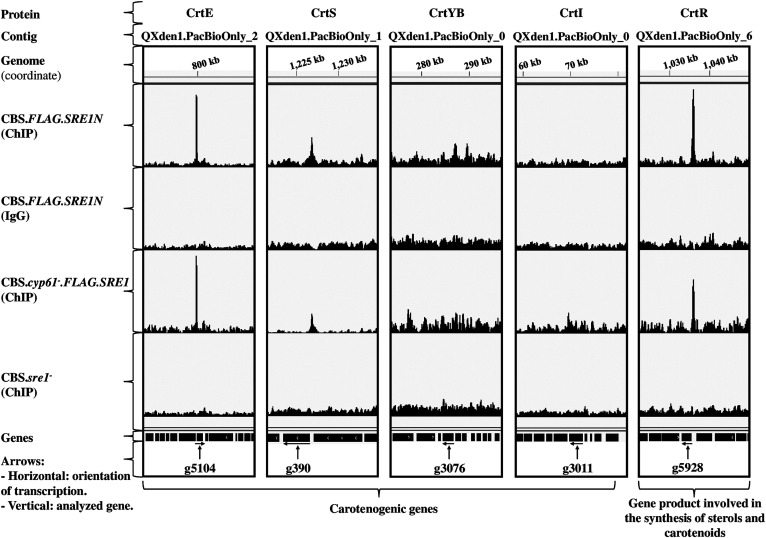
Distribution of peaks associated with Sre1 target genes that encode proteins involved in carotenoid biosynthesis. The figure shows tracks (coverage bigWid files) of ChIP-exo assays of strains CBS.FLAG.SRE1N, CBS.cyp61−.FLAG.SRE1, and CBS.sre1− using the IGV software. ChIP: Immunoprecipitation assay with anti-FLAG antibody against FLAG-tagged Sre1. IgG: Isotype control assay with IgG. Peaks were located in the promoter regions of the genes that encode (Gene ID in brackets) CrtE (g5104), CrtS (g390), CrtYB (g3076), CrtI (g3011), and CrtR (g5928). The peak shown by the IGV software in the promoter region of gene g390 (CrtS) was not detected by the peak callers in strain CBS.cyp61−.FLAG.SRE1.
Sre1 and its role in the carotenogenesis of X. dendrorhous
The obtained results indicate that Sre1 regulates carotenogenesis by regulating genes related to the MVA pathway and the genes crtE (Table 4, g5104; Fig. 6) and crtR (Table 4, g5928; Fig. 6). The latter two genes had a peak with the SRE motif, showed upregulation in the RNA-seq comparative analysis of strains CBS.FLAG.SRE1N or CBS.cyp61−.FLAG.SRE1 compared with the WT strain, and downregulation in strain CBS.cyp61−.sre1− compared with strain CBS.cyp61−. In X. dendrorhous, one of the redox partners of the crtR gene product corresponds to astaxanthin synthase (CrtS) (63), which converts β-carotene to astaxanthin. Small peaks in the promoter region of the crtS gene were observed when using the IGV software (Fig. 6); however, the peak callers only detected peaks in the promoter region of this gene in strain CBS.FLAG.SRE1N. For this reason, the promoter region of the crtS gene was manually analyzed, but still a potential SRE motif based on our ChIP-exo results was not found. Nevertheless, the transcriptional profile of crtS obtained in the RNA-seq analysis was similar to that of the crtE and crtR genes. No ChIP-exo peaks were detected close to the carotenogenic genes crtYB and crtI (Fig. 6).
Other possible functions of Sre1 in X. dendrorhous
Most likely, Sre1 is involved in other BPs in addition to the biosynthesis of sterols in X. dendrorhous. For example, a gene potentially encoding a mandelate racemase homolog showed to be a Sre1 direct target gene in X. dendrorhous (Table 4, g1192). In a recent work, using ascomycete yeasts, it was proposed that the mandelate pathway would be related to the synthesis of benzenoid compounds (64). Also, the major facilitator superfamily (MFS) general substrate transporter encoding gene (Table 4, g2548) was identified as a Sre1 direct target gene; the MFS transports a wide spectrum of substrates across membranes and plays a pivotal role in multiple physiological processes (65). The alternative cyclin Pcl12 encoding gene (Table 4, g4000), which is a cyclin that interacts with Cdk5 (66), also showed to be a Sre1 direct target. Sre1 could also be involved in the regulation of the response to toxic compounds, as the glyoxalase I and a RTA1-domain-containing protein encoding genes (Table 4, g5168, g944) are Sre1 direct targets. Glyoxalase I is involved in the glyoxalase system, which maintains tolerable levels of methylglyoxal (67) and in Saccharomyces cerevisiae, Rta1p confers resistance to 7-aminocholesterol, a toxic ergosterol biosynthesis inhibitor (68). Also, a putative brefeldin A resistance protein encoding gene was detected as a Sre1 direct target. This protein could be involved in the resistance to drugs like brefeldin A, which is a fungal antibacterial reagent reported to inhibit the transport of proteins out of the ER (69). Finally, other Sre1 direct target genes that are involved in undetermined functions were identified. These correspond to genes encoding hypothetical proteins, genes encoding proteins without annotation associated, and genes encoding proteins associated to domains like the W40 repeat domain and NADP-binding domain (Table 4, g1193, g806, g4710, g4312, g4112, g5613, g3515, g943).
DISCUSSION
In this study, direct target genes of the TF Sre1 in the SREBP pathway in the carotenogenic yeast X. dendrorhous were identified. Sre1 is a master TF involved in sterol homeostasis, demonstrating the conservation of this regulator in X. dendrorhous. To identify Sre1-dependent genes, a comparative RNA-seq analysis between modified strains having an active Sre1 protein and the WT was performed; then, Sre1 direct target genes were identified through ChIP-exo assays. For this, a new genome assembly of the X. dendrorhous WT strain CBS 6938 was performed. The first report of the genome sequencing and assembly of this strain was carried out by the Illumina sequencing technology (24), and, particularly, the genome used in our current work was assembled through a hybrid assembly pipeline using the available Illumina reads (24) and reads obtained by SMRT sequencing (PacBio). Thus, the number of contigs was reduced from 775 to 22 contigs resulting in a genome assembly of about 20 Mb where 50% of the genome is represented in only five contigs (sum of contigs: 11.5 Mb). Also, about 300 more coding sequences (CDS) than previously reported were identified in the annotation of this new genome from X. dendrorhous strain CBS 6938. Thus, alignments of RNA-seq and ChIP-exo assays reads from this work were performed using the genome assembly currently reported.
SRE motifs were detected in the promoter regions of several Sre1 direct target genes and were consistent with SREs described in other organisms. The differences observed when searching for an SRE motif using the input DNA sequences associated with peaks close to DEGs, or when using only peaks associated with up- or downregulated genes detected by the RNA-seq analysis of strains CBS.FLAG.SRE1N and CBS.cyp61− versus the WT strain, could be due to slight differences between the SRE motif used by Sre1 to activate genes exclusively involved in sterol biosynthesis and the SRE motif used by this factor to regulate other target genes as observed in mammals (70). In the RNA-seq analysis of these strains, genes involved in sterol biosynthesis were upregulated (Table 3), and this result could explain why the SRE motif that was found when using only the DNA sequences from the peaks associated with upregulated genes is more similar to the SRE motifs in other organisms and associated with the ergosterol biosynthetic process BP. In DNA sequences associated with peaks detected in the promoters of downregulated genes in both strains, no SRE motifs were identified. In contrast, E-box like sequences that resemble the E-box motif 5′-CANNTG-3′ that are recognized by bHLH proteins (55) were found in these genes. However, the DNA sequences that were used as input when using the MEME tool, were less enriched in E-box like sequences than the DNA sequences from peaks associated with upregulated genes with the SRE motif. Probably, Sre1 binds to these E-box like sequences because Sre1 is a bHLH protein, but the tyrosine that replaces the arginine in the bHLH domain of Sre1, a characteristic feature of SREBPs (71), allows Sre1 to also recognize SRE sequences. In addition, it was demonstrated that SREBP binds E-box sequences in vitro (72), but sterol regulation is mediated through SREBP binding to SRE elements and not to E-boxes (73). Importantly, our results demonstrate that several of the identified Sre1 direct target genes in X. dendrorhous, belong to the sterol biosynthesis pathway and had an SRE motif in their promoter region.
The SRE motif was found predominantly in the promoter region of genes, which is a finding that coincides with other studies; for example, the mammal SREBP has a strong binding preference proximal to promoter regions, and through ChIP-seq assays, it was detected that most SREs are within 2 kb of the 5′ end of the target genes (74). Similarly, a ChIP-seq assay in A. fumigatus showed that SREs are preferentially located within 2 kb of the 5′ start codon of a regulated gene (49).
Some of the genes regulated by Sre1 also influence carotenogenesis. In X. dendrorhous, the MVA pathway generates the universal isoprenoid building blocks, isopentenyl-pyrophosphate (IPP) and its isomer, dimethylallyl-pyrophosphate (DMAPP) (17), which are required in X. dendrorhous for both carotenoid and sterol biosynthesis (Fig. 2). In this work, we detected three direct gene targets of Sre1 related to the MVA pathway of X. dendrorhous: ERG10 (encoding acetyl-CoA acetyltransferase; g1536), HMGS (encoding HMG-CoA synthase; g3516), and HMGR (encoding HMG-CoA reductase; g1377); in mammals, these genes are targets of the isoform SREBP-2 (4). In other works, it was observed that overexpression of these three genes in X. dendrorhous increased astaxanthin production (75), indicating that modifications at the MVA pathway level directly affect carotenogenesis. These genes are not exclusively carotenogenic; thus, the SREBP pathway additionally regulates carotenogenesis in X. dendrorhous by regulating genes of the MVA pathway.
Sre1 in X. dendrorhous regulates genes encoding enzymes of the sterol biosynthesis pathway, including C-4 methyl sterol oxidase (Erg25; g602) and the P450 enzymes lanosterol 14-α demethylase (Cyp51; g190) and C22-sterol desaturase (Cyp61; g989), which are also targets of the TF SrbA of A. fumigatus (49, 76). These genes are also regulated by Sre1 in the yeasts S. pombe (6) and C. neoformans (7). Cyp51 is conserved in mammals and fungi (77), and the Cyp51encoding gene is regulated by the isoform SREBP-1a, through an SRE in the promoter region, in mammals (77). Considering this background, it is very possible that the Cyp51 encoding gene is regulated by Sre1 in X. dendrorhous, despite the fact that the peak callers only detected peaks in the promoter region of this gene in the ChIP-exo analysis of strain CBS.FLAG.SRE1N. P450 enzymes require an electron donor, which in class II eukaryotic P450s is usually a CPR that mediates electron transfer from NADPH (78). In mammals, the mutation of the CPR encoding gene activates the SREBP pathway, as indicated by increased expression of SREBP gene targets (79). CPR also supplies electrons to squalene epoxidase, which is the second rate-limiting enzyme in cholesterol biosynthesis (80). Evidence of the presence of SREs and the binding of SREBP-2 to the promoter of the squalene epoxidase encoding gene has been reported (81). The X. dendrorhous squalene epoxidase encoding gene (Table 4, g3385) was also shown to be an Sre1 gene target; in fact, this gene was one of the most highly upregulated genes detected by RNA-seq analysis in strain CBS.FLAG.SRE1N compared with that in the WT strain. The crtR gene in X. dendrorhous encodes the single yeast CPR, and the results showed that crtR is a Sre1 direct target gene. It is likely that CrtR also transfers electrons to squalene epoxidase in X. dendrorhous (Fig. 2, ERG1), because in fungi, CPR partners are not limited to P450 monooxygenases and include squalene epoxidase (82).
It was observed that mutation of the STP1 gene, which encodes a protease responsible for the proteolytical activation of Sre1, reduced the crtR transcript levels in X. dendrorhous (20), emphasizing that crtR expression is regulated by Sre1. In the opposite case, the expression of only the active form of Sre1 (Sre1N) in strain CBS.FLAG.SRE1N upregulated crtR. Similar results were observed in C. neoformans, in which the expression of the CPR encoding gene also depends on Sre1 and Stp1 (7). X. dendrorhous CrtR is involved in the biosynthesis of both carotenoids and sterols. CrtR is essential for the production of astaxanthin from β-carotene (59), but crtR− mutants still produce ergosterol, although in a lower proportion (47). These findings indicate that the P450s from ergosterol biosynthesis may use alternative electron donors, while CrtS exclusively uses CrtR as an electron donor. Therefore, by regulating the expression of the crtR gene, Sre1 directly regulates carotenogenesis in X. dendrorhous at the steps involved in the production of astaxanthin from β-carotene. In X. dendrorhous, a total of 13 P450 encoding genes were identified, but to date, only three have been functionally described. In several of these genes, SRE sequences were predicted at their promoter regions (83). Even though these genes were not identified as DEGs by the RNA-seq analysis, it is important to note that some P450 genes are induced under specific conditions (for example, in the presence of the enzyme substrate) (84). Thus, it is possible that under the conditions used in this study, it was not possible to evaluate the Sre1 contribution to the regulation of these P450 genes in such a way as to elucidate the role of Sre1 in the regulation of other pathways involving P450s.
Previously, it was demonstrated that Sre1 is activated and induced in X. dendrorhous strain CBS.cyp61− (20), and Sre1 activation depends on alterations in steros composition rather than on the absence of ergosterol (60). However, the mechanism that senses sterol levels and/or sterol composition changes is still unknown in X. dendrorhous, as this yeast does not have a SCAP homolog. Despite this, results from this current work support that Sre1 is activated in strain CBS.cyp61−, because the ChIP-exo assays showed that the SRE1 gene itself is an Sre1 target (Fig. 5), which was upregulated in strain CBS.FLAG.SRE1N. These results indicate that Sre1 in X. dendrorhous activates its own gene transcription in an autoregulatory loop, as observed in mammals (48) and fungi (49).
Results from this work also suggest that Sre1 regulates the biosynthesis of phospholipid in X. dendrorhous through the regulation of the choline kinase encoding gene, which participates in the phosphatidylcholine synthesis pathway. This pathway has been described in yeasts (85), and in mammals, the choline kinase α isoform encoding gene is regulated under hypoxia conditions by the hypoxia-inducible factor 1α (HIF-1α) (86). In addition, Sre1 probably regulates the alternative cyclin Pcl12 encoding gene, which is a cyclin that interacts with Cdk5 and has a regulatory role in the morphogenesis of the basidiomycete Ustilago maydis (66). This is an interesting finding, as the A. fumigatus SREBP homolog, named as SrbA, was proven to be involved in the maintenance of cell polarity and hyphal morphogenesis (8). Moreover, noncanonical functions of Cdk/cyclins involved in the SREBP pathway have been reported; for example, Cdk8/cyclin C induces phosphorylation of SREBP-1c, which enhanced SREBP-1c ubiquitination and protein degradation (87). Another Sre1 direct target in X. dendrorhous is the glyoxalase I encoding gene. Glyoxalase I is involved in the maintenance of methylglyoxal levels that derive from glycolysis (67) and glyoxalase I cooperates with fatty acid synthase to protect against sugar toxicity (88). Interestingly, transcriptomic analysis from kidneys of methylglyoxal-treated rats compared with controls, showed downregulation of the SREBP1 encoding gene (89). This suggests that regulation of the glyoxalase I encoding gene in X. dendrorhous by Sre1 could be an important regulatory loop because, through glyoxalase I, Sre1 would regulate methylglyoxal levels, which is a compound that negatively affects Sre1 gene transcription. However, further studies are required, as the functionality of the glyoxalase I encoding gene has not yet been proven in X. dendrorhous. Another Sre1 direct target gene that could be involved in the response to toxic compounds is the RTA1-domain-containing protein encoding gene. In S. cerevisiae, Rta1p is involved in 7-aminocholesterol resistance (68), which is a cholesterol derivative with strong antifungal and antibacterial activity that inhibits the Δ8→Δ7-sterol isomerase (Erg2p) and C-14 sterol reductase (Erg24p) (90). Another interesting finding is that a potential mandelate racemase encoding gene was shown to be a Sre1 direct target in X. dendrorhous. Recently, it was proposed that some yeasts may use the mandelate pathway to synthetize benzenoid compounds as an alternative to the phenylalanine ammonia lyase pathway (64). Some basidiomycetes use the phenylalanine ammonia lyase pathway to synthesize benzenoids (64, 91); however, there is no evidence to exclude the involvement of the mandelate pathway in the synthesis of benzenoid-derived metabolites in X. dendrorhous, but further studies are required in this topic.
In summary, the results presented in this work demonstrate that Sre1 is mainly involved in the regulation of genes of the ergosterol biosynthesis pathway in X. dendrorhous. In addition, Sre1 also regulates carotenogenesis in X. dendrorhous, mainly by regulating genes related to the MVA pathway that produce the carotenoid precursors and by regulating the CPR gene (crtR) and crtE gene expression.
Data availability
This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession number JACGZH000000000. The version described in this work is version JACGZH010000000. The annotation associated to the genome assembly was included in the supplemental information as supplemental Table S4. The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (92) and are accessible through GEO Series accession number GSE152739.
Supplementary Material
Footnotes
Author contributions—M.G. formal analysis; M.G. validation; M.G., S.C., M.S.G., D.S., S.B., M.B., and V.C. investigation; M.G., S.C., D.S., and V.C. methodology; M.G. and J.A. writing-original draft; J.A. conceptualization; J.A. resources; J.A. supervision; J.A. funding acquisition; J.A. project administration.
Funding and additional information—This research was supported by FONDECYT Grant 1160202 and by the graduate scholarships ANID-PFCHA/Doctorado Nacional/2017-21170613 to M.G., ANID-PFCHA/Doctorado Nacional/2014-21130807 to M.S.G., and PEEI/Facultad de Ciencias, Universidad de Chile to S.C.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
Abbreviations—
- BP
- biological process
- ChIP-exo
- ChIP combined with lambda exonuclease digestion
- CPR
- cytochrome P450 reductase
- CrtS
- astaxanthin synthase
- DEG
- differentially expressed gene
- GO
- Gene Ontology
- GOMo
- Gene Ontology for Motifs
- HMGR
- HMG-CoA reductase
- HMGS
- HMG-CoA synthase
- MEME
- Multiple Em for Motif Elicitation
- MFS
- major facilitator superfamily
- MVA
- mevalonate
- P450
- cytochrome P450
- PacBio
- Pacific Biosciences
- RNA-seq
- RNA-sequencing
- SCAP
- SREBP cleavage-activating protein
- S1P
- site-1 protease
- S2P
- site-2 protease
- SMRT
- single-molecule real-time
- SRE
- sterol regulatory element
- Sre1N
- N-terminal domain of Sre1
- TF
- transcription factor
Manuscript received June 12, 2020, and in revised form September 2, 2020. Published, JLR Papers in Press, September 15, 2020, DOI 10.1194/jlr.RA120000975.
REFERENCES
- 1.Brown M. S., and Goldstein J. L.. 1997. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 89: 331–340. [DOI] [PubMed] [Google Scholar]
- 2.Goldstein J. L., DeBose-Boyd R. A., and Brown M. S.. 2006. Protein sensors for membrane sterols. Cell. 124: 35–46. [DOI] [PubMed] [Google Scholar]
- 3.Yokoyama C., Wang X., Briggs M. R., Admon A., Wu J., Hua X., Goldstein J. L., and Brown M. S.. 1993. SREBP-1, a basic-helix-loop-helix-leucine zipper protein that controls transcription of the low density lipoprotein receptor gene. Cell. 75: 187–197. [PubMed] [Google Scholar]
- 4.Horton J. D., Goldstein J. L., and Brown M. S.. 2002. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 109: 1125–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parks L. W., and Casey W. M.. 1995. Physiological implications of sterol biosynthesis in yeast. Annu. Rev. Microbiol. 49: 95–116. [DOI] [PubMed] [Google Scholar]
- 6.Hughes A. L., Todd B. L., and Espenshade P. J.. 2005. SREBP pathway responds to sterols and functions as an oxygen sensor in fission yeast. Cell. 120: 831–842. [DOI] [PubMed] [Google Scholar]
- 7.Chang Y. C., Bien C. M., Lee H., Espenshade P. J., and Kwon-Chung K. J.. 2007. Sre1p, a regulator of oxygen sensing and sterol homeostasis, is required for virulence in Cryptococcus neoformans. Mol. Microbiol. 64: 614–629. [DOI] [PubMed] [Google Scholar]
- 8.Willger S. D., Puttikamonkul S., Kim K. H., Burritt J. B., Grahl N., Metzler L. J., Barbuch R., Bard M., Lawrence C. B., and Cramer R. A. J.. 2008. A sterol-regulatory element binding protein is required for cell polarity, hypoxia adaptation, azole drug resistance, and virulence in Aspergillus fumigatus. PLoS Pathog. 4: e1000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chun C. D., Liu O. W., and Madhani H. D.. 2007. A link between virulence and homeostatic responses to hypoxia during infection by the human fungal pathogen Cryptococcus neoformans. PLoS Pathog. 3: e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dhingra S., and Cramer R. A.. 2017. Regulation of sterol biosynthesis in the human fungal pathogen Aspergillus fumigatus: opportunities for therapeutic development. Front. Microbiol. 8: 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheung R., and Espenshade P. J.. 2013. Structural requirements for sterol regulatory element-binding protein (SREBP) cleavage in fission yeast. J. Biol. Chem. 288: 20351–20360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hwang J., Ribbens D., Raychaudhuri S., Cairns L., Gu H., Frost A., Urban S., and Espenshade P. J.. 2016. A Golgi rhomboid protease Rbd2 recruits Cdc48 to cleave yeast SREBP. EMBO J. 35: 2332–2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bat-Ochir C., Kwak J. Y., Koh S. K., Jeon M. H., Chung D., Lee Y. W., and Chae S. K.. 2016. The signal peptide peptidase SppA is involved in sterol regulatory element-binding protein cleavage and hypoxia adaptation in Aspergillus nidulans. Mol. Microbiol. 100: 635–655. [DOI] [PubMed] [Google Scholar]
- 14.Golubev W. I. 1995. Perfect state of Rhodomyces dendrorhous (Phaffia rhodozyma). Yeast. 11: 101–110. [DOI] [PubMed] [Google Scholar]
- 15.Mortensen A., Skibsted L. H., and Truscott T. G.. 2001. The interaction of dietary carotenoids with radical species. Arch. Biochem. Biophys. 385: 13–19. [DOI] [PubMed] [Google Scholar]
- 16.Higuera-Ciapara I., Félix-Valenzuela L., and Goycoolea F. M.. 2006. Astaxanthin: a review of its chemistry and applications. Crit. Rev. Food Sci. Nutr. 46: 185–196. [DOI] [PubMed] [Google Scholar]
- 17.Schmidt I., Schewe H., Gassel S., Jin C., Buckingham J., Hümbelin M., Sandmann G., and Schrader J.. 2011. Biotechnological production of astaxanthin with Phaffia rhodozyma/Xanthophyllomyces dendrorhous. Appl. Microbiol. Biotechnol. 89: 555–571. [DOI] [PubMed] [Google Scholar]
- 18.Loto I., Gutiérrez M. S., Barahona S., Sepúlveda D., Martínez-Moya P., Baeza M., Cifuentes V., and Alcaíno J.. 2012. Enhancement of carotenoid production by disrupting the C22-sterol desaturase gene (CYP61) in Xanthophyllomyces dendrorhous. BMC Microbiol. 12: 235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gutiérrez M. S., Campusano S., González A. M., Gómez M., Barahona S., Sepúlveda D., Espenshade P. J., Fernández-Lobato M., Baeza M., Cifuentes V., et al. 2019. Sterol regulatory element-binding protein (Sre1) promotes the synthesis of carotenoids and sterols in Xanthophyllomyces dendrorhous. Front. Microbiol. 10: 586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gómez M., Gutiérrez M. S., González A. M., Gárate-Castro C., Sepúlveda D., Barahona S., Baeza M., Cifuentes V., and Alcaíno J.. 2020. Metallopeptidase Stp1 activates the transcription factor Sre1 in the carotenogenic yeast Xanthophyllomyces dendrorhous. J. Lipid Res. 61: 229–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rhee H. S., and Pugh B. F.. 2011. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell. 147: 1408–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cifuentes V., Hermosilla G., Martínez C., León R., Pincheira G., and Jiménez A.. 1997. Genetics and electrophoretic karyotyping of wild-type and astaxanthin mutant strains of Phaffia rhodozyma. Antonie van Leeuwenhoek. 72: 111–117. [DOI] [PubMed] [Google Scholar]
- 23.Eid J., Fehr A., Gray J., Luong K., Lyle J., Otto G., Peluso P., Rank D., Baybayan P., and Bettman B.. 2009. Real-time DNA sequencing from single polymerase molecules. Science. 323: 133–138. [DOI] [PubMed] [Google Scholar]
- 24.Sharma R., Gassel S., Steiger S., Xia X., Bauer R., Sandmann G., and Thines M.. 2015. The genome of the basal agaricomycete Xanthophyllomyces dendrorhous provides insights into the organization of its acetyl-CoA derived pathways and the evolution of Agaricomycotina. BMC Genomics. 16: 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chin C-S., Peluso P., Sedlazeck F. J., Nattestad M., Concepcion G. T., Clum A., Dunn C., O’Malley R., Figueroa-Balderas R., Morales-Cruz A., et al. 2016. Phased diploid genome assembly with single-molecule real-time sequencing. Nat. Methods. 13: 1050–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wences A. H., and Schatz M. C.. 2015. Metassembler: merging and optimizing de novo genome assemblies. Genome Biol. 16: 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Langmead B., and Salzberg S. L.. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marçais G., Delcher A. L., Phillippy A. M., Coston R., Salzberg S. L., and Zimin A.. 2018. MUMmer4: A fast and versatile genome alignment system. PLOS Comput. Biol. 14: e1005944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waterhouse R. M., Seppey M., Simão F. A., Manni M., Ioannidis P., Klioutchnikov G., Kriventseva E. V., and Zdobnov E. M.. 2018. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol. Biol. Evol. 35: 543–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim D., Paggi J. M., Park C., Bennett C., and Salzberg S. L.. 2019. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37: 907–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., and Madden T. L.. 2009. BLAST+: architecture and applications. BMC Bioinformatics. 10: 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Götz S., García-Gómez J. M., Terol J., Williams T. D., Nagaraj S. H., Nueda M. J., Robles M., Talon M., Dopazo J., and Conesa A.. 2008. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 36: 3420–3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Love M. I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robinson M. D., McCarthy D. J., and Smyth G. K.. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blighe K., Rana S., and Lewis M.. 2019. EnhancedVolcano: Publication-ready volcano plots with enhanced colouring and labeling. R package version 1.4.0. [Google Scholar]
- 36.Alexa A., Rahnenführer J., and Lengauer T.. 2006. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics. 22: 1600–1607. [DOI] [PubMed] [Google Scholar]
- 37.Rossi M. J., Lai W. K. M., and Pugh B. F.. 2018. Simplified ChIP-exo assays. Nat. Commun. 9: 2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., and 1000 Genome Project Data Processing Subgroup . 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramírez F., Ryan D. P., Grüning B., Bhardwaj V., Kilpert F., Richter A. S., Heyne S., Dündar F., and Manke T.. 2016. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44: W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robinson J. T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E. S., Getz G., and Mesirov J. P.. 2011. Integrative genomics viewer. Nat. Biotechnol. 29: 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., Bernstein B. E., Nusbaum C., Myers R. M., Brown M., Li W., et al. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quinlan A. R., and Hall I. M.. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bailey T. L., and Elkan C.. 1994. Fitting a mixture model by expectation maximization to discover motifs in bipolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2: 28–36. [PubMed] [Google Scholar]
- 44.Buske F. A., Bodén M., Bauer D. C., and Bailey T. L.. 2010. Assigning roles to DNA regulatory motifs using comparative genomics. Bioinformatics. 26: 860–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grant C. E., Bailey T. L., and Noble W. S.. 2011. FIMO: scanning for occurrences of a given motif. Bioinformatics. 27: 1017–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hoff K. J., Lange S., Lomsadze A., Borodovsky M., and Stanke M.. 2016. BRAKER1: Unsupervised RNA-Seq-based genome annotation with GeneMark-ET and AUGUSTUS. Bioinformatics. 32: 767–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gutiérrez M. S., Rojas M. C., Sepúlveda D., Baeza M., Cifuentes V., and Alcaíno J.. 2015. Molecular characterization and functional analysis of cytochrome b5 reductase (CBR) encoding genes from the carotenogenic yeast Xanthophyllomyces dendrorhous. PLoS One. 10: e0140424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reed B. D., Charos A. E., Szekely A. M., Weissman S. M., and Snyder M.. 2008. Genome-wide occupancy of SREBP1 and its partners NFY and SP1 reveals novel functional roles and combinatorial regulation of distinct classes of genes. PLoS Genet. 4: e1000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chung D., Barker B. M., Carey C. C., Merriman B., Werner E. R., Lechner B. E., Dhingra S., Cheng C., Xu W., Blosser S. J., et al. 2014. ChIP-seq and in vivo transcriptome analyses of the Aspergillus fumigatus SREBP SrbA reveals a new regulator of the fungal hypoxia response and virulence. PLoS Pathog. 10: e1004487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Todd B. L., Stewart E. V., Burg J. S., Hughes A. L., and Espenshade P. J.. 2006. Sterol regulatory element binding protein is a principal regulator of anaerobic gene expression in fission yeast. Mol. Cell. Biol. 26: 2817–2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dawson P. A., Hofmann S. L., Van der Westhuyzen D. R., Südhof T. C., Brown M. S., and Goldstein J. L.. 1988. Sterol-dependent repression of low density lipoprotein receptor promoter mediated by 16-base pair sequence adjacent to binding site for transcription factor Sp1. J. Biol. Chem. 263: 3372–3379. [PubMed] [Google Scholar]
- 52.Smith J. R., Osborne T. F., Goldstein J. L., and Brown M. S.. 1990. Identification of nucleotides responsible for enhancer activity of sterol regulatory element in low density lipoprotein receptor gene. J. Biol. Chem. 265: 2306–2310. [PubMed] [Google Scholar]
- 53.Smith J. R., Osborne T. F., Brown M. S., Goldstein J. L., and Gil G.. 1988. Multiple sterol regulatory elements in promoter for hamster 3-hydroxy-3-methylglutaryl-coenzyme A synthase. J. Biol. Chem. 263: 18480–18487. [PubMed] [Google Scholar]
- 54.Osborne T. F., Gil G., Goldstein J. L., and Brown M. S.. 1988. Operator constitutive mutation of 3-hydroxy-3-methylglutaryl coenzyme A reductase promoter abolishes protein binding to sterol regulatory element. J. Biol. Chem. 263: 3380–3387. [PubMed] [Google Scholar]
- 55.Atchley W. R., and Fitch W. M.. 1997. A natural classification of the basic helix–loop–helix class of transcription factors. Proc. Natl. Acad. Sci. USA. 94: 5172–5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eberlé D., Hegarty B., Bossard P., Ferré P., and Foufelle F.. 2004. SREBP transcription factors: master regulators of lipid homeostasis. Biochimie. 86: 839–848. [DOI] [PubMed] [Google Scholar]
- 57.McPherson R., and Gauthier A.. 2004. Molecular regulation of SREBP function: the Insig-SCAP connection and isoform-specific modulation of lipid synthesis. Biochem. Cell Biol. 82: 201–211. [DOI] [PubMed] [Google Scholar]
- 58.Leiva K., Werner N., Sepúlveda D., Barahona S., Baeza M., Cifuentes V., and Alcaíno J.. 2015. Identification and functional characterization of the CYP51 gene from the yeast Xanthophyllomyces dendrorhous that is involved in ergosterol biosynthesis. BMC Microbiol. 15: 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alcaíno J., Barahona S., Carmona M., Lozano C., Marcoleta A., Niklitschek M., Sepúlveda D., Baeza M., and Cifuentes V.. 2008. Cloning of the cytochrome P450 reductase (crtR) gene and its involvement in the astaxanthin biosynthesis of Xanthophyllomyces dendrorhous. BMC Microbiol. 8: 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Venegas M., Barahona S., González A. M., Sepúlveda D., Zúñiga G. E., Baeza M., Cifuentes V., and Alcaíno J.. 2020. Phenotypic analysis of mutants of ergosterol biosynthesis genes (ERG3 and ERG4) in the red yeast Xanthophyllomyces dendrorhous. Front. Microbiol. 11: 1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Danielli M., Marrone J., Capiglioni A. M., and Marinelli R. A.. 2019. Mitochondrial aquaporin-8 is involved in SREBP-controlled hepatocyte cholesterol biosynthesis. Free Radic. Biol. Med. 131: 370–375. [DOI] [PubMed] [Google Scholar]
- 62.Boumann H. A., Gubbens J., Koorengevel M. C., Oh C-S., Martin C. E., Heck A. J. R., Patton-Vogt J., Henry S. A., De Kruijff B., and de Kroon A. I. P. M.. 2006. Depletion of phosphatidylcholine in yeast induces shortening and increased saturation of the lipid acyl chains: evidence for regulation of intrinsic membrane curvature in a eukaryote. Mol. Biol. Cell. 17: 1006–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ojima K., Breitenbach J., Visser H., Setoguchi Y., Tabata K., Hoshino T., van den Berg J., and Sandmann G.. 2006. Cloning of the astaxanthin synthase gene from Xanthophyllomyces dendrorhous (Phaffia rhodozyma) and its assignment as a β-carotene 3-hydroxylase/4-ketolase. Mol. Genet. Genomics. 275: 148–158. [DOI] [PubMed] [Google Scholar]
- 64.Valera M. J., Boido E., Ramos J. C., Manta E., Radi R., Dellacassa E., and Carrau F.. 2020. The mandelate pathway, an alternative to the phenylalanine ammonia lyase pathway for the synthesis of benzenoids in ascomycete yeast. Appl. Environ. Microbiol. 86: e00701-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yan N. 2013. Structural advances for the major facilitator superfamily (MFS) transporters. Trends Biochem. Sci. 38: 151–159. [DOI] [PubMed] [Google Scholar]
- 66.Flor-Parra I., Castillo-Lluva S., and Pérez-Martín J.. 2007. Polar growth in the infectious hyphae of the phytopathogen Ustilago maydis depends on a virulence-specific cyclin. Plant Cell. 19: 3280–3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thornalley P. J. 1993. The glyoxalase system in health and disease. Mol. Aspects Med. 14: 287–371. [DOI] [PubMed] [Google Scholar]
- 68.Soustre I., Letourneux Y., and Karst F.. 1996. Characterization of the Saccharomyces cerevisiae RTA1 gene involved in 7-aminocholesterol resistance. Curr. Genet. 30: 121–125. [DOI] [PubMed] [Google Scholar]
- 69.Lippincott-Schwartz J., Yuan L. C., Bonifacino J. S., and Klausner R. D.. 1989. Rapid redistribution of Golgi proteins into the ER in cells treated with brefeldin A: evidence for membrane cycling from Golgi to ER. Cell. 56: 801–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sharpe L. J., and Brown A. J.. 2013. Controlling cholesterol synthesis beyond 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR). J. Biol. Chem. 288: 18707–18715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Párraga A., Bellsolell L., Ferre-D’Amare A. R., and Burley S. K.. 1998. Co-crystal structure of sterol regulatory element binding protein 1a at 2.3 Å resolution. Structure. 6: 661–672. [DOI] [PubMed] [Google Scholar]
- 72.Kim J. B., Spotts G. D., Halvorsen Y-D., Shih H-M., Ellenberger T., Towle H. C., and Spiegelman B. M.. 1995. Dual DNA binding specificity of ADD1/SREBP1 controlled by a single amino acid in the basic helix-loop-helix domain. Mol. Cell. Biol. 15: 2582–2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Athanikar J. N., and Osborne T. F.. 1998. Specificity in cholesterol regulation of gene expression by coevolution of sterol regulatory DNA element and its binding protein. Proc. Natl. Acad. Sci. USA. 95: 4935–4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Seo Y. K., Jeon T. I., Chong H. K., Biesinger J., Xie X., and Osborne T. F.. 2011. Genome-wide localization of SREBP-2 in hepatic chromatin predicts a role in autophagy. Cell Metab. 13: 367–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hara K. Y., Morita T., Mochizuki M., Yamamoto K., Ogino C., Araki M., and Kondo A.. 2014. Development of a multi-gene expression system in Xanthophyllomyces dendrorhous. Microb. Cell Fact. 13: 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Blatzer M., Barker B. M., Willger S. D., Beckmann N., Blosser S. J., Cornish E. J., Mazurie A., Grahl N., Haas H., and Cramer R. A.. 2011. SREBP coordinates iron and ergosterol homeostasis to mediate triazole drug and hypoxia responses in the human fungal pathogen Aspergillus fumigatus. PLoS Genet. 7: e1002374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Aoyama Y., Funae Y., Noshiro M., Horiuchi T., and Yoshida Y.. 1994. Occurrence of a P450 showing high homology to yeast lanosterol 14-demethylase (P45014DM) in the rat liver. Biochem. Biophys. Res. Commun. 201: 1320–1326. [DOI] [PubMed] [Google Scholar]
- 78.Iyanagi T., Xia C., and Kim J-J. P.. 2012. NADPH–cytochrome P450 oxidoreductase: prototypic member of the diflavin reductase family. Arch. Biochem. Biophys. 528: 72–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Weng Y., DiRusso C. C., Reilly A. A., Black P. N., and Ding X.. 2005. Hepatic gene expression changes in mouse models with liver-specific deletion or global suppression of the NADPH-cytochrome P450 reductase gene. Mechanistic implications for the regulation of microsomal cytochrome P450 and the fatty liver phenotype. J. Biol. Chem. 280: 31686–31698. [DOI] [PubMed] [Google Scholar]
- 80.Padyana A. K., Gross S., Jin L., Cianchetta G., Narayanaswamy R., Wang F., Wang R., Fang C., Lv X., Biller S. A., et al. 2019. Structure and inhibition mechanism of the catalytic domain of human squalene epoxidase. Nat. Commun. 10: 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Howe V., Sharpe L. J., Prabhu A. V., and Brown A. J.. 2017. New insights into cellular cholesterol acquisition: promoter analysis of human HMGCR and SQLE, two key control enzymes in cholesterol synthesis. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 1862: 647–657. [DOI] [PubMed] [Google Scholar]
- 82.Yoshida Y. 1988. Cytochrome P450 of fungi: primary target for azole antifungal agents. In Current Topics in Medical Mycology. M. R. McGinnis, editor. Springer, New York. 388–418. [DOI] [PubMed] [Google Scholar]
- 83.Córdova P., González A-M., Nelson D. R., Gutiérrez M-S., Baeza M., Cifuentes V., and Alcaíno J.. 2017. Characterization of the cytochrome P450 monooxygenase genes (P450ome) from the carotenogenic yeast Xanthophyllomyces dendrorhous. BMC Genomics. 18: 540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zordoky B. N. M., and El-Kadi A. O. S.. 2008. Induction of several cytochrome P450 genes by doxorubicin in H9c2 cells. Vascul. Pharmacol. 49: 166–172. [DOI] [PubMed] [Google Scholar]
- 85.Gibellini F., and Smith T. K.. 2010. The Kennedy pathway–de novo synthesis of phosphatidylethanolamine and phosphatidylcholine. IUBMB Life. 862: 414–428. [DOI] [PubMed] [Google Scholar]
- 86.Bansal A., Harris R. A., and DeGrado T. R.. 2012. Choline phosphorylation and regulation of transcription of choline kinase α in hypoxia. J. Lipid Res. 53: 149–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhao X., Feng D., Wang Q., Abdulla A., Xie X. J., Zhou J., Sun Y., Yang E. S., Liu L. P., Vaitheesvaran B., et al. 2012. Regulation of lipogenesis by cyclin-dependent kinase 8-mediated control of SREBP-1. J. Clin. Invest. 122: 2417–2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Garrido D., Rubin T., Poidevin M., Maroni B., Le Rouzic A., Parvy J. P., and Montagne J.. 2015. Fatty acid synthase cooperates with glyoxalase 1 to protect against sugar toxicity. PLoS Genet. 11: e1004995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Markova I., Hüttl M., Oliyarnyk O., Kacerova T., Haluzik M., Kacer P., Seda O., and Malinska H.. 2019. The effect of dicarbonyl stress on the development of kidney dysfunction in metabolic syndrome - a transcriptomic and proteomic approach. Nutr. Metab. (Lond.). 16: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Elkihel L., Soustre I., Karst F., and Letourneux Y.. 1994. Amino-and aminomethylcholesterol derivatives with fungicidal acitivity. FEMS Microbiol. Lett. 120: 163–167. [DOI] [PubMed] [Google Scholar]
- 91.Hyun M. W., Yun Y. H., Kim J. Y., and Kim S. H.. 2011. Fungal and plant phenylalanine ammonia-lyase. Mycobiology. 39: 257–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Edgar R., Domrachev M., and Lash A. E.. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30: 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession number JACGZH000000000. The version described in this work is version JACGZH010000000. The annotation associated to the genome assembly was included in the supplemental information as supplemental Table S4. The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (92) and are accessible through GEO Series accession number GSE152739.