Abstract
Of the known regulators of atherosclerosis, miRNAs have been demonstrated to play critical roles in lipoprotein homeostasis and plaque formation. Here, we generated a novel animal model of atherosclerosis by knocking in LDLRW483X in C57BL/6 mice, as the W483X mutation in LDLR is considered the most common newly identified pathogenic mutation in Chinese familial hypercholesterolemia (FH) individuals. Using the new in vivo mouse model combined with a well-established atherosclerotic in vitro human cell model, we identified a novel atherosclerosis-related miRNA, miR-23a-3p, by microarray analysis of mouse aortic tissue specimens and human aortic endothelial cells (HAECs). miR-23a-3p was consistently downregulated in both models, which was confirmed by qPCR. Bioinformatics analysis and further validation experiments revealed that the TNFα-induced protein 3 (TNFAIP3) gene was the key target of miR-23a-3p. The miR-23a-3p-related functional pathways were then analyzed in HAECs. Collectively, the present results suggest that miR-23a-3p regulates inflammatory and apoptotic pathways in atherogenesis by targeting TNFAIP3 through the NF-κB and p38/MAPK signaling pathways.
Keywords: micro-ribonucleic acid, inflammation, apoptosis, familial hypercholesterolemia, low density lipoprotein receptor, animal model, endothelial cells, tumor necrosis factor α-induced protein 3
Atherosclerosis is a chronic inflammatory disease of the arteries characterized by plaques built up in the vessels (1–3). Endothelial dysfunction in the vessels represents one of the atherogenic processes in the early stage that leads to the formation of atherosclerotic lesions; at a later stage, plaques are built up inside the arteries leading to a narrowed and restricted blood flow, which increases the influx of LDL into the arterial wall (4), induces the migration of monocytes by activating endothelial cells, and results in lipid accumulation and foam cell formation (5, 6). Among atherosclerosis regulators, miRNAs have been shown to play critical roles in processes such as lipoprotein homeostasis (e.g., miR-122, miR-223, and miR-148a) (7–10), endothelial cell inflammation and plaque progression (e.g., miR-17-3p, miR-31, miR-92a, and miR-146a) (11–15), cytokine response (e.g., miR-181b) (16), and leukocyte recruitment and activation (e.g., miR-26,miR-144, and miR-148a) (10, 13, 17). The latest miRNA database (miRBase 22) released 2,654 human mature miRNAs (18). However, it remains unknown whether novel atherosclerosis-regulating miRNAs could be identified in this updated database.
To investigate the roles of miRNAs in atherosclerosis, both in vitro and in vivo models have commonly been used (14, 19–21). For instance, an in vitro model was established by treating human aortic endothelial cells (HAECs) with oxidized LDL (oxLDL), a well-known atherogenic factor (6, 22). oxLDL is known to initiate atherogenesis by stimulating endothelial cells to overexpress cell surface adhesion molecules [e.g., intercellular adhesion molecule (ICAM)-1] (22–24) and inflammatory cytokines, such as E-selectin (25, 26), and to induce endothelial cell apoptosis by activating caspase-3 and caspase-9 (27). Lectin-like oxLDLR-1 (LOX-1) overexpression in the APOE−/− mouse model has been shown to promote atherogenesis (28). LOX-1 upregulation has also been observed in endothelial cells in human atherosclerotic lesions, especially in the early stage of atherogenesis (29).
The most frequently used in vivo mouse models are APOE−/− and LDLR−/− mice (14, 19, 24, 30). These models differ in how plasma lipoprotein clearance is dysregulated, although both show enhanced atherosclerosis. For example, APOE KO in mice results in a significant decrease of HDL and leads to reduced cholesterol efflux capacity (31). Comparatively, LDLR KO mice have a high percentage of their cholesterol carried in IDL/LDL particles, which closely resembles the condition of dyslipidemic humans (32). However, more than 85% of hypercholesterolemia’s genetic variants have been identified in the LDLR gene. Of these, approximately 46% of the variants are single missense mutations of LDLR (33), suggesting that a LDLR-null or missense mutation model may be more applicable, mimicking the majority of human hypercholesterolemia cases.
In this study, we developed a novel mouse model of atherosclerosis that resembled human familial hypercholesterolemia (FH) by knocking in a single missense mutation of W483STOP in the LDLR gene using the CRISPR/Cas9 technology, as the W483X mutation in LDLR is considered the most prevalent missense mutation in Chinese FH individuals (34). The microarray technology was then used to profile whole-genomic miRNAs and mRNAs; subsequent bioinformatics analysis was undertaken to screen candidates of atherosclerosis-related miRNA/mRNA pairs in the new atherosclerosis mouse model as well as an in vitro human cell model. In both models, a novel miRNA, miR-23a-3p, was identified that acted as a key regulator of atherosclerosis. By targeting TNFα-induced protein 3 (TNFAIP3), miR-23a-3p promoted inflammation and endothelial apoptosis through the NF-κB and p38/MAPK pathways. Taken together, these results indicated that the newly identified atherosclerosis-related miRNA plays an important role in inflammatory pathways.
MATERIALS AND METHODS
Animal use and care
All experimental procedures performed in both Biomodel Organism (Shanghai, China) and Wuxi Apptech, Inc. (Shanghai, China) were in accordance with standard Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health, and abided by the institution-approved animal protocol and the AAALAC accreditation.
Knock-in mouse construction
C57BL/6-LDLRW483STOP knock-in (KI) mice were generated at the Shanghai Biomodel Organism. The single mutation W483STOP in the human LDLR gene was generated through the CRISPR/Cas9 technology. Oligo donor DNA was synthesized by Sango Biotech (Shanghai, China). The Cas9 mRNA and guide RNA (gRNA) were transcribed in vitro using MEGAscript® T7 transcription kit (Thermo Fisher, AM1334) and purified with MEGAclear™ transcription clean-up kit (Thermo Fisher, AM1908). The Cas9 mRNA and gRNA were injected into C57BL/6 mouse zygotes using a microinjection system under standard conditions. The injected zygotes were then cultured with embryos and transferred into the oviducts of recipient mice. After birth, genomic DNA was extracted from tail biopsies of all mice by the standard ethanol precipitation method (35). Genotyping was conducted by PCR (primer forward: 5′-ctctacctgccctgcttcccatcc-3′; reverse: 5′-accagccccctctttctttcttgt-3′) and sequencing. Homozygous (HO) mutant mice were obtained by interbreeding heterozygous (HE) mice; the genotype was confirmed by sequencing and Western blot.
Atherosclerosis mouse model generation
The atherosclerosis mouse model was established at Wuxi Apptech, Inc. (Shanghai, China). C57BL/6-LDLRW483STOP KI mice were fed a HFD (0.2% cholesterol and 21% saturated fat; Harlan Teklad, TD88137) for 12 weeks. Two independent groups of WT mice were included in the study and fed normal diet (ND) and HFD, respectively. APOE KO mice were purchased from the Jackson Laboratory (stock number 002052) and used as controls for atherosclerosis. Mouse body weights were recorded weekly for 12 weeks.
Blood biochemistry analysis
For all biochemical analyses, mouse blood was collected by submandibular bleeding or cardiac puncture at the endpoint. Serum was separated by centrifugation at 1,300 g for 10 min at 4°C and stored at −80°C. Serum total cholesterol (TC), TG, LDL-C, and HDL-C amounts were determined every 2–3 weeks on a Hitachi 7180 biochemistry automatic analyzer (Tokyo, Japan).
Oil red O staining and immunohistochemical analysis
The mouse aorta was removed from the heart, opened longitudinally from the intercostal ostia to the iliac bifurcation, and pinned open. Each aorta was fixed with 4% paraformaldehyde (Affymetrix, 19943) and rinsed with 60% isopropanol (Sigma, I9030). Aorta specimens were then stained with 60% Oil red O (Sigma, O0625) solution in isopropanol and washed with 60% isopropanol. The stained aortic tissues underwent a final wash with deionized water prior to imaging.
For immunohistochemical analysis, mouse aorta samples were first immersed in 4% paraformaldehyde overnight. Then, the tissues were dehydrated through a graded ethanol series and embedded in paraffin. Next, 4 μm paraffin sections were obtained for subsequent staining with hematoxylin (Sigma, GHS132) and eosin (Sigma, HT110132). Stained slides of aortic root sections were evaluated by light microscopy on an Aperio ImageScope v12.3.3.7014 (Leica Biosystems, Nussloch, Germany) to assess plaque formation. In each animal, plaque, open lumen, and total vessel areas were averaged from three separate slides.
Cell culture, treatment, and transfection
HAECs were purchased from the ATCC (PCS-100-011) and cultured in vascular cell basal medium (ATCC, PCS-100-030) supplemented with the endothelial cell growth kit (ATCC, PCS-100-041) and 0.1% penicillin/streptomycin (ATCC, PCS-999-002) at 37°C in an incubator with 5% CO2. HAECs were transiently transfected with miR-23a-3p mimic (Thermo Fisher, 4464067, #MC10644), negative control miRNA (miRNA NC) (5′-UUCUCCGAACGUGUCACGU-3′, Sangon Biotech), TNFAIP3 siRNA (5′-GGCCAAUCAUUGUCAUUUCTT-3′, Thermo Fisher, AM16708, #2510), or miR-23a-3p mimic and its inhibitor (Thermo Fisher, 4464084, #MH10644). All transfections utilized the Lipofectamine 3000 reagent (Invitrogen, L3000015) according to the manufacturer’s instructions. After 24 h of cell transfection, the culture medium was replaced with fresh medium with or without oxLDL (Beijing Xiesheng Bio-Technology); inclusion or exclusion of oxLDL was determined based on the design.
Microarray assay and bioinformatics analysis
Total RNA was separately extracted from aortic endothelial cells and mouse aortic tissue specimens and purified with a mirVana™ miRNA isolation kit without phenol (Ambion, AM1561). Total RNA was amplified and labeled with a Low Input Quick Amp Labeling Kit, one-color (Agilent Technologies, 5190-2305). Individual miRNAs obtained from the extracted total RNA were labeled with Cy3 using a miRNA Complete Labeling and Hyb kit (Agilent Technologies, 5190-0456). Labeled miRNAs were then purified with an RNeasy mini kit (Qiagen, 74106). Each slide was hybridized with the Cy3-labeled miRNAs in a hybridization oven (Agilent Technologies, G2545A) at 55°C and 20 rpm for 20 h. After hybridization, slides were washed in staining dishes with Gene Expression Wash Buffer kit (Agilent Technologies, 5188-5327), scanned on an Agilent microarray scanner (Agilent Technologies), and analyzed with the Feature Extraction software v10.7 (Agilent Technologies).
Gene and miRNA differential expression analysis.
For HAEC samples, mRNAs and miRNAs were profiled using Agilent SurePrint G3 Human Gene Expression 8×60K microarray and Agilent SurePrint Human miRNA microarray (V21.0), respectively. In this analysis, 26,083 genes were included in the gene expression microarray; meanwhile, 2,549 miRNAs were included in the miRNA microarray (supplemental Table S1). Each probe was labeled as P (present, expressed) or A (absent, unexpressed) based on its fluorescence intensity in sample testing. Only the probes labeled as P in all samples from at least one group (control vs. experimental groups in the cell model or WT vs. HO groups in the mouse model) were retained for further analysis (supplemental Table S3). Gene and miRNA expression profiles in all samples were quantile-normalized after a log2-based normalization. The log2-transformed fold-change ratio for each gene or miRNA was then calculated between the treatment and control groups. Genes with absolute log2-transformed fold change ≥1 and P < 0.05 were considered to be differentially expressed mRNAs (DE mRNAs); miRNAs with P < 0.05 were considered to be differentially expressed miRNAs (DE miRNAs).
In tissue samples from mice, mRNAs and miRNAs were profiled with Agilent SurePrint G3 mouse GE V2 8×60K microarray and Agilent Mouse miRNA microarray 8×60K (V21.0), respectively. Totally, 27,122 genes were included in the gene expression microarray; meanwhile, 1,881 miRNAs were included in the miRNA microarray (supplemental Table S1). Subsequent analysis was performed as described above for cell microarray data.
Identification of novel atherosclerosis-related miRNA.
Overlapping DE miRNAs between in vitro and in vivo models were retained for target gene prediction using TargetScan (Release 7.2) (36). These miRNAs were further filtered based on the reverse regulation paradigm of miRNA-mRNA pairs on gene expression. The miRNAs in these pairs were defined as candidate atherosclerosis-related miRNAs. Candidate miRNAs targeting previously reported atherosclerosis-related genes were prioritized for selection. Any miRNAs already reported to be related to atherosclerosis were excluded. The remaining candidate miRNAs were further validated by quantitative PCR.
Quantitative PCR
Total RNA was separately extracted from collected cells and/or tissues samples with RNeasy Plus mini kit (Qiagen, 74134) according to the manufacturer’s instructions. In brief, cDNA was obtained from total RNA using FirstStrand synthesis system (Thermo Fisher, 18091050). Then, qPCR analysis was performed in triplicate using SYBR Green PCR Master Mix (Thermo Fisher, 4367659) with the primers shown in supplemental Table S2. Relative mRNA levels were normalized to the housekeeping gene GAPDH. miRNA expression was determined using TaqMan miRNA reverse transcription kit (Thermo Fisher, 4366597) and TaqMan miRNA assays (Thermo Fisher, 4440886). U6 snRNA was used for normalization.
Luciferase reporter assay
The 3′ untranslated regions (UTRs) of WT and mutant TNFAIP3 were amplified by PCR and cloned into PGL3-CMV-Luc vector using XhoI and MluI restriction sites followed by DNA sequencing verification. Then, 293T cells were cotransfected with luciferase reporter plasmids containing WT TNFAIP3, mutant TNFAIP3, or empty vectors, and miR-23a-3p mimic or miRNA NC, respectively, using Lipofectamine 3000. Luciferase activities were evaluated using a dual-luciferase reporter assay system (Promega, E2940) after 48 h.
Apoptosis assay and flow cytometry
The apoptosis assay was performed with the Annexin V-FITC Apoptosis kit (Sigma, APOAF-50TST). Briefly, after appropriate treatment, cells were collected and washed with Dulbecco’s PBS. Then, cells were resuspended at a density of 1 × 106 cells per milliliter and double stained with FITC-Annexin V and propidium iodide. Cells were finally analyzed on an LSRFortessa X-20 (BD) with the FlowJo v10.1 software.
Immunoassays
For cytokine detection, HAECs at a density of 3 × 105 cells per well were treated with miRNA negative control, miR-23a-3p mimic, or miR-23a-3p inhibitor in 6-well plates. Extracellular levels of ICAM and E-selectin were measured with a customized Luminex human magnetic assay kit (R&D, LXSAHM-04) and ELISA kit (Abcam, ab174445), respectively. For TNFAIP3 protein level detection, cells at a density of 8,000 cells per well in 96-well plates were transfected with miRNAs followed by oxLDL treatment. TNFAIP3 expression was measured with an ELISA kit (Aviva Systems Biology, OKCD08256) following the manufacturer’s instructions.
Cell adhesion assay
Cell adhesion was measured with a leukocyte adhesion assay kit (Cell Biolabs, CBA-210). Briefly, HAECs at a density of 5 × 104 in 96-well plates were transfected with miRNA mimic or its inhibitor for 48 h, followed by oxLDL treatment for 24 h. Then, THP-1 cells at 3 × 105 cells per well with LeukoTracker were added to the endothelial monolayer for an additional 90 min incubation before reading, according to manufacturer’s instructions.
Western blot analysis
Cells were lysed with RIPA (Thermo Fisher, 89901) containing 1% protease inhibitors (Sigma, 78444). After centrifugation, the supernatants were collected and total protein amounts were determined with a BCA kit (Thermo Fisher, 23227). Proteins were then separated by SDS-PAGE and transferred onto PVDF membranes. Membranes were blocked and incubated with primary antibodies against TNFAIP3 (CST, 5630), p38 MAPK (CST, 8690), p-p38 MAPK (CST, 4511), IκBα (CST, 9242), p-IκBα (CST, 2859) and GAPDH (R&D Systems, MAB5718) at 4°C overnight. After incubation with appropriate secondary antibodies, the membranes were scanned on a Gel Documentation System.
Statistical analysis
Statistical analysis was performed with GraphPad Prism 7. Multiple comparisons were performed by ANOVA followed by Tukey’s test (Figs. 1C–F, 4B–E, 5D and E, 6B–D) or Dunnett’s test (Figs. 1H–J, 2B, 5C). Comparison between two groups was carried out by Student’s t-test (Figs. 2C and D, 3D and E, 5B). Differences were considered statistically significant at P < 0.05. All data are mean ± standard deviation.
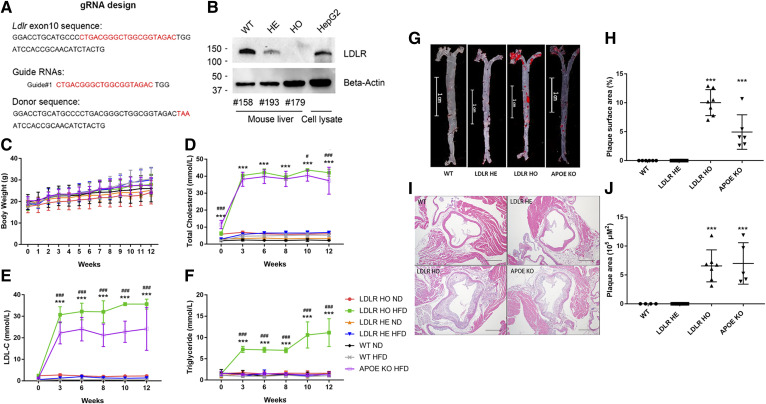
Fig. 1.
HFD induces atherosclerosis in LDLRW483STOP KI mice. A: The gRNA and oligo donor DNA were designed based on LDLR’s exon 10 sequence. B: Western blot analysis of endogenous LDLR from the WT, LDLRW483STOP HO, and LDLRW483STOP HE mice, as well as HepG2 cells. Body weights (C). serum TC (D), LDL-C (E), and TG (F) from WT, LDLR HO, LDLR HE, and APOE KO mice fed ND or HFD for 12 weeks. G: Oil Red O-stained aortic sections from WT, LDLR HE, LDLR HO, and APOE KO mice. Scale bar = 1 cm. H: Percentages of plaque area of the whole aortic surface in WT and other mouse models. I: Hematoxylin and eosin-stained aortic root sections from WT and other mouse models. Scale bar = 500 μm. J: Plaque area quantitation in the aortic roots of WT and other mouse models. Error bars represent standard deviations. There were eight (four male and four female), ten (seven male and three female), ten (five male and five female), and five (three male and two female) mice in the WT, LDLR HO, LDLR HE, and APOE KO groups, respectively. In C, *P < 0.05, ***P < 0.001, multiple comparisons to LDLR HO fed ND. In D–F, ***P < 0.001, multiple comparisons to WT HFD; #P < 0.05, ###P < 0.001, multiple comparisons to APOE KO HFD. Two-way ANOVA followed by post hoc Tukey’s test. In H and J, ***P < 0.001, multiple comparisons to WT, one-way ANOVA followed by post hoc Dunnett’s test.
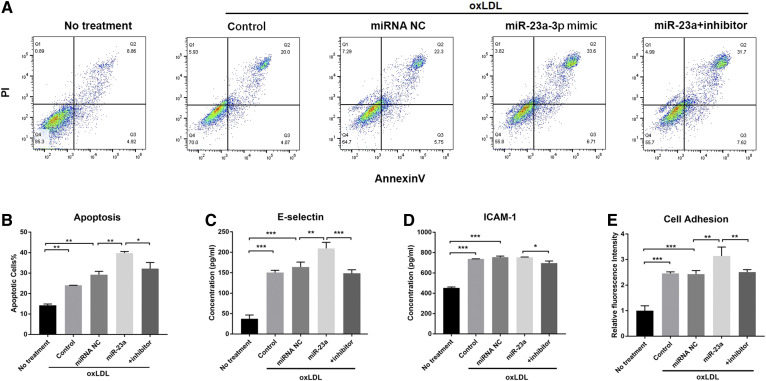
Fig. 4.
miR-23a-3p induces endothelial inflammation and apoptosis in HAECs. A: Flow cytometry analysis of apoptotic HAECs treated with miR-23a-3p and its inhibitor under oxLDL induction. B: Quantification of apoptotic cells in flow cytograms. C: E-selectin expression levels in HAECs treated with miR-23a-3p and its inhibitor under oxLDL induction. D: ICAM-1 expression levels in HAECs treated with miR-23a-3p and its inhibitor under oxLDL induction. E: Cell adhesion measurement by co-incubating THP-1 cells with HAECs pretreated with miR-23a-3p and its inhibitor after oxLDL induction. Cells treated with oxLDL alone served as controls. Error bars represent standard deviations. *P < 0.05, **P < 0.01, ***P < 0.001, multiple comparisons by one-way ANOVA followed by post hoc Tukey’s test.
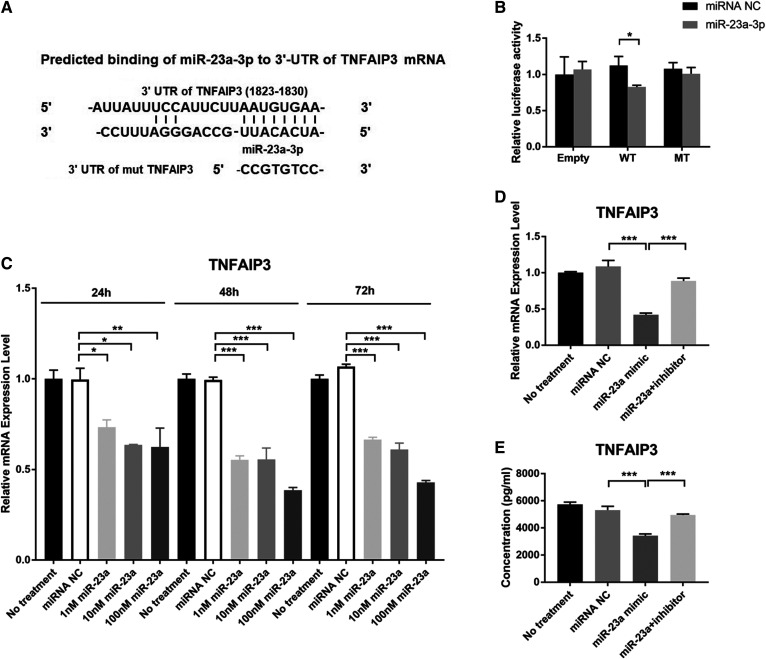
Fig. 5.
miR-23a-3p regulates TNFAIP3 expression in HAECs. A: Prediction of the binding sequence of the human TNFAIP3 gene with miR-23a-3p and design of the mutant TNFAIP3 binding site. B: Luciferase reporter assay of 293T cells cotreated with miR-23a-3p and empty vector or vectors containing WT TNFAIP3 3′UTR or TNFAIP3 3′UTR mutant. C: Effect of miR-23a-3p on the transcription level of TNFAIP3 in HAECs. D: Gene expression of TNFAIP3. E: Protein expression of TNFAIP3. Error bars represent standard deviations. *P < 0.05, **P < 0.01, ***P < 0.001; multiple comparisons were performed by one-way ANOVA followed by post hoc Dunnett’s test (C) and Tukey’s test (D, E). Group pairs were compared by the Student’s t-test (B).
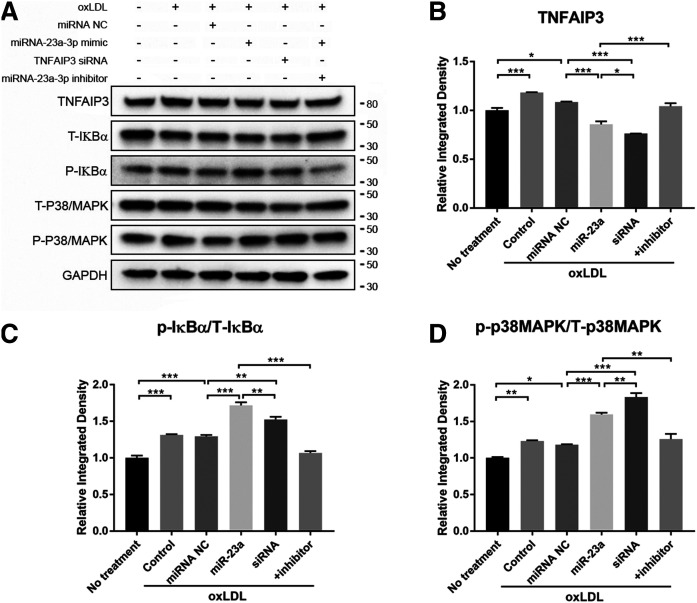
Fig. 6.
miR-23a-3p regulation occurs through the NF-κB and p38/MAPK pathways. A: Western blot analysis of p38/MAPK and IκBα phosphorylation after treatment with miR-23a-3p, miR-23a-3p inhibitor, and TNFAIP3 siRNA under oxLDL induction. Cells were transfected with miRNAs for 24 h followed by oxLDL treatment for 48 h. The + inhibitor indicates co-application of miR-23a-3p and its inhibitor. B: Quantification of TNFAIP3 expression levels. C: Quantification of IκBα phosphorylation. D: Quantification of p38/MAPK phosphorylation. All quantifications were evaluated with ImageJ. p/T, phosphorylation to total protein ratio Error bars represent standard deviations. *P < 0.05, **P < 0.01. Multiple comparisons were performed by one-way ANOVA followed by post hoc Tukey’s test.
Fig. 2.
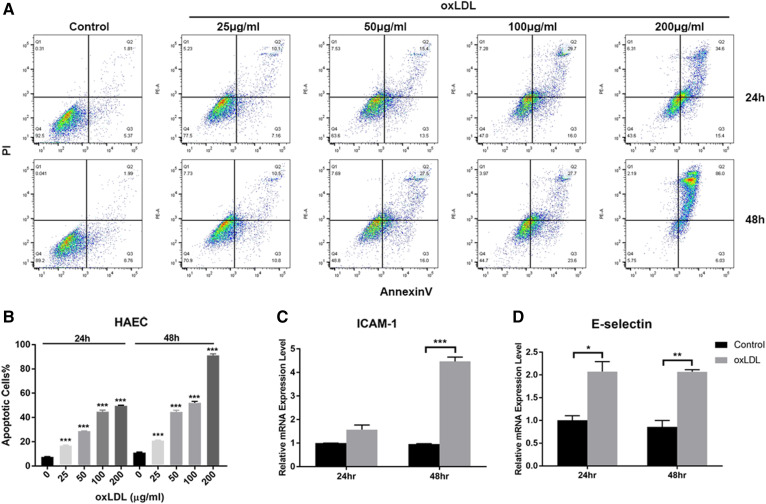
oxLDL induces inflammation and apoptosis in HAECs. A: Flow cytometry analysis of apoptotic HAECs without or with oxLDL treatment (25, 50, 100, and 200 μg/ml, respectively) after 24 and 48 h of treatment. B: Quantification of apoptotic cells after treatment with different oxLDL concentrations. C: Gene expression levels of ICAM-1 in HAECs with or without oxLDL treatment at 50 μg/ml oxLDL for 24 and 48 h. D: Gene expression levels of E-selectin in HAECs with or without oxLDL treatment at 50 μg/ml oxLDL for 24 and 48 h. Controls were untreated cells. Error bars represent standard deviations. *P < 0.05, **P < 0.01, ***P < 0.001, compared with untreated cells, one-way ANOVA followed by post hoc Dunnett’s test (B). Group pairs were compared by the Student’s t-test (C, D). Q8
Fig. 3.
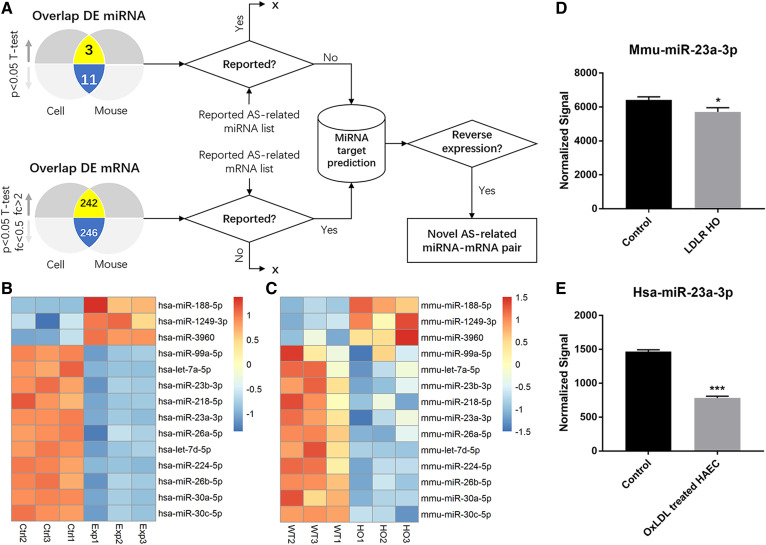
Atherosclerosis-related miRNA screening and profiling. A: Screening flow of novel atherosclerosis (AS)-related miRNAs. B: Heat map of up- and downregulated miRNAs in the in vitro model. C: Heat map of up- and downregulated miRNAs in the in vivo model. D: Microarray analysis of miR-23a-3p in the in vivo model. E: Microarray analysis of miR-23a-3p in the in vitro model. Error bars represent standard deviations. *P < 0.05, ***P < 0.001, compared with control or WT by the Student’s t-test.
RESULTS
Atherosclerosis is induced by HFD in LDLRW483STOP KI mice
Recently, a very common but unique single missense mutation of the LDLR gene (p. W483STOP) has been identified as a pathogenic mutation in Chinese FH individuals (34). The W483 of LDLR is highly conserved across human, mouse, and rat based on the multiple sequence alignment analysis in the LDLR sequences (37). To develop a human mouse model of atherosclerosis that could mimic the human condition mentioned above, we generated a LDLR mutation KI mouse model by the CRISPR-Cas9 technology with gRNA and oligo donor DNA, as shown in Fig. 1A. The chronological steps involved in the generation of C57BL/6-LDLRW483STOP KI mice are highlighted in supplemental Fig. S1. Sequencing results showed that the gRNA was effective (supplemental Fig. S1). Genotyping of HE and HO mice for the LDLRW483STOP mutation was validated by Western blot (Fig. 1B). A total of 10 LDLR HO mice (seven males and three females) and 10 LDLR HE mice (five males and five females) were obtained. There were eight (four males and four females) and five (three males, two females) of WT and APOE KO mice, respectively. The body weights of LDLR HO mice fed HFD increased progressively during the feeding period and reached statistical significance after 2 weeks. LDLR HE and APOE KO mice fed HFD showed an increasing trend but nonsignificance on body weight compared with those fed ND (Fig. 1C). As shown in Fig. 1D and E, LDL-C levels in LDLR HO mice fed HFD were significantly higher than those of APOE KO mice fed HFD. Moreover, TG levels in LDLR HO mice fed HFD were significantly increased, showing a distinctive lipid profile compared with WT or APOE KO mice (Fig. 1F). LDLR HE mice fed ND or HFD showed no changes in TC, LDL-C, and TG levels over 12 weeks.
As shown on oil red O-staining images, APOE KO mice had more plaques in the aortic roots than WT animals (n = 4) (Fig. 1G, H); this pattern was also evident with hematoxylin and eosin staining (Fig. 1I, J). Similarly, LDLR HO mice, but not LDLR HE mice, also presented plaques in the aortic roots (Fig. 1G–J). Although plaque areas in the aortic roots of LDLR HO mice were comparable to those of APOE KO mice (Fig. 1I, J), the percentage of plaque area of the whole aortic surface was significantly higher in LDLR HO mice (Fig. 1G, H) compared with APOE KO mice. No plaques were found in WT or LDLR HE mice (Fig. 1G–J).
oxLDL-induced inflammation and apoptosis in HAECs
As the initial event in the process of atherogenesis, vascular endothelial cells are activated to recruit leukocytes, leading to lipid accumulation and foam cell formation (5, 6). oxLDL has been reported to contribute to plaque generation by inducing endothelial cell apoptosis (4, 6, 38). To screen for atherosclerosis-related miRNAs, we developed an in vitro model based on oxLDL-treated HAECs. Different concentrations of oxLDL (25, 50, 100, or 200 μg/ml, respectively) were incubated with HAECs for 24 and 48 h; cell apoptosis was then analyzed by flow cytometry. As shown in Fig. 2A and B, oxLDL induced cell apoptosis in HAECs in a dose- and time-dependent manner, which reached statistical significance at every dose at both time points (24 and 48 h) compared with untreated cells. Moreover, the inflammatory cytokines, E-selectin and ICAM-1, were also significantly increased under 50 μg/ml oxLDL treatment for 24 and 48 h, respectively, compared with untreated cells (Fig. 2C, D). These results suggested that oxLDL activated HAECs and ultimately initiated the inflammatory process that led to apoptosis. Considering the high degree of cell death at 200 μg/ml oxLDL and similar effects at 50 and 100 μg/ml oxLDL on apoptotic induction, we selected an oxLDL treatment of 50 μg/ml for 48 h for subsequent experiments to investigate its pro-inflammatory and apoptotic effects.
Bioinformatic analysis of atherosclerosis-related miRNAs
In both in vivo and in vitro models, a total of 4,430 miRNAs (2,549 and 1881 for the human and mouse, respectively) and 53,205 mRNAs (26,083 and 27,122 for the human and mouse, respectively) were detected by microarray analysis. Among them, 302 human miRNAs (123 downregulated and 179 upregulated) and 108 mouse miRNAs (69 downregulated and 39 upregulated) were differentially expressed in the in vitro and in vivo models of atherosclerosis, respectively. Significantly altered expression levels were found in 9,757 human mRNAs (4,574 upregulated and 5,183 downregulated) and 5,020 mouse mRNAs (2,868 upregulated and 2,152 downregulated). To identify novel miRNAs related to atherosclerosis, a reverse expression-based screening flow was designed and illustrated in Fig. 3A. A total of 14 DE miRNAs were identified in both models, including 3 upregulated and 11 downregulated compared with controls (Fig. 3B, C). With the exception of three previously reported miRNAs, including miR-23b-3p (39), miR-30a-5p (40), and miR-30c-5p (41, 42), the other 11 identified miRNAs not previously linked to the pathogenesis of atherosclerosis were retained as candidates for further selection. Meanwhile, there were 242 upregulated DE mRNAs and 246 downregulated DE mRNAs overlapped between the in vivo and in vitro models (supplemental Table S3). As listed in supplemental Table S3, 29 common DE mRNAs (including APOE and LDLR) were already known to be involved in the development of atherosclerosis. Through miRNA target gene prediction by TargetScan (36), the candidate miRNAs targeting 29 common mRNAs with inverse changes in expression levels were selected as novel hits. As a result, a novel atherosclerosis-related miRNA, namely, miR-23a-3p, was identified with significantly decreased expression levels in both in vivo and in vitro models (Fig. 3D, E).
MiR-23a-3p promotes atherogenic inflammation and apoptosis in HAECs
To further explore the functional roles of miR-23a-3p in oxLDL-induced inflammatory response, endothelial cell apoptosis was evaluated by adding miR-23a-3p mimic and its inhibitor under oxLDL stress. As shown in Fig. 4A and B, overexpression of miR-23a-3p worsened oxLDL-induced cell apoptosis, which was reversed by co-application of miR-23a antisense inhibitor. The pro-inflammatory cytokines, E-selectin and ICAM-1, were also detected extracellularly. Figure 4C indicated that E-selectin protein levels were elevated after oxLDL stimulation, and miR-23a-3p strengthened this upregulation, which was also blocked by miR-23a-3p inhibitor. ICAM-1 showed no obvious changes at the protein level after miR-23a-3p addition, probably due to the different response time (Fig. 4D) (43). We further examined the effect of miR-23a-3p on endothelial cell adhesion. In accordance with endothelial apoptosis results, miR-23a-3p overexpression promoted THP-1 cell attachment to HAECs, which was blocked by miR-23a-3p inhibitor (Fig. 4E). These data jointly illustrated that miR-23a-3p promoted atherogenesis by enhancing endothelial inflammation and apoptosis.
TNFAIP3 is the key target gene of miR-23a-3p in atherogenesis
TNFAIP3, encoding the inflammation regulatory protein, TNFAIP3, was upregulated in both in vivo and in vitro models in microarray analysis and confirmed by qPCR (supplemental Fig. S2). Although target prediction algorithms indicated that TNFAIP3 is a potential conserved target gene of miR-23a-3p (Fig. 5A), we sought to further verify the regulatory effect of miR-23a-3p on TNFAIP3. To this end, luciferase reporters containing a WT or mutant variant of TNFAIP3’s 3′UTR in the miR-23a-3p binding site (Fig. 5A) were cotransfected into 293T cells with miR-23a-3p to determine whether the 3′UTR of TNFAIP3 is the binding site of miR-23a-3p. In this study, miR-23a-3p significantly reduced luciferase activity in the WT group, with no effects in the mutant and vector control groups (Fig. 5B), confirming TNFAIP3 as a target of miR-23a-3p. TNFAIP3 mRNA expression was also examined in HAECs after transfection with miR-23a-3p. miR-23a-3p significantly suppressed TNFAIP3 expression in a dose-dependent manner (Fig. 5C). Further, miR-23a-3p inhibitor blocked the suppression of TNFAIP3 by miR-23a-3p at both mRNA and protein levels (Fig. 5D, E). These findings suggested that TNFAIP3 acted as a biological effector of miR-23a-3p in human endothelial cells.
miR-23a-3p regulates atherogenesis through the NF-κB and p38/MAPK pathways
The miR-23a-3p-mediated signaling pathways in the cell model of atherosclerosis were investigated next. As previously reported, both NF-κB and p38/MAPK signaling pathways play critical roles in inflammatory responses and atherogenesis (44, 45). As an anti-inflammatory signaling molecule, TNFAIP3 inhibits the activation of NF-κB by multiple mechanisms, such as modulating ubiquitin-dependent signaling cascades (46), or suppresses the p38/MAPK pathway by inactivating transforming growth factor β-activated kinase 1 (TAK1) (47). To explore whether miR-23a-3p regulates endothelial inflammation through these pathways, we measured the phosphorylation levels of p38/MAPK and IκBα. As shown in Fig. 6A and B, oxLDL treatment of HAECs induced TNFAIP3 expression and increased the phosphorylation levels of both p38/MAPK and IκBα. In addition, miR-23a-3p treatment prior to oxLDL administration reduced TNFAIP3 expression and enhanced both IκBα and p38/MAPK phosphorylation; these effects were all blocked by miR-23a-3p inhibitor (Fig. 6). Interestingly, TNFAIP3-specific siRNA showed a slightly different effect on both pathways, with a stronger increase in the phosphorylation of p38/MAPK than IκBα, compared with miR-23a-3p treatment (Fig. 6C, D), suggesting that miR-23a-3p and TNFAIP3 siRNA may have a stronger effect on one pathway than the other.
DISCUSSION
In this study, we developed a novel mouse model based on human FH genetic characteristics, to identify novel atherosclerosis-related miRNAs. Ultimately, this approach allowed us to further our understanding of miRNA regulation in atherogenesis under disease conditions. FH is an autosomal dominant disorder characterized by elevated plasma levels of LDL-C (48), and individuals with FH present a dramatically increased risk of atherosclerotic cardiovascular diseases (49). The novel model showed phenotypic differences from previous commonly used mouse models of atherosclerosis such as APOE or LDLR KO mice (14, 19, 24, 30). More specifically, we were interested in generating a mouse model that carried a human LDLR gene presenting a mutation previously identified in Chinese FH individuals. As reported, both HE and HO FH with the W483X mutation in LDLR were found in humans. However, individuals with the HO phenotype were younger with a more severe phenotype and higher lipid values compared with HE individuals (34). According to the structure-based functional impact prediction for mutation identification (SFIP-MutID) model, a structure-based computational prediction model for the pathogenicity of the LDLR single missense mutation, a variant of LDLR W483X was predicted to induce very high pathogenicity, as LDLR folding and recycling are known to be defective (50). Based on genetic information, we generated a mouse model containing the FH-causing LDLR W483X mutation, which may share a greater degree of similarity with human atherogenesis compared with other commonly used models. Consistent with HE and HO individuals with the LDLR W483X mutation, HO mice fed HFD developed plaques in the aorta and showed much higher lipid values than HE animals in 12 weeks. Compared with a complete KO of the LDLR gene, individuals with the LDLR W483X mutation demonstrate measurable levels of LDLR expression but truncated extracellular structure of the LDLR protein, whose function is impaired (49). Although APOE deficiency is considered an inherited familial disease correlated with the risk of cardiovascular events, few subjects have been identified with APOE deficiency (51, 52), which is much rarer than LDLR variants.
As a well-developed in vitro model, oxLDL-induced endothelial cells have been used in numerous miRNA studies (20, 21, 42, 53–55). As previously reported (22–24, 27, 56, 57), the HAEC model validated oxLDL-induced cell apoptosis and elevated E-selectin and ICAM-1 expression levels, providing an in vitro tool and human-relevant evidence for the identification and validation of novel atherosclerosis-related miRNAs. With both in vitro and in vivo atherosclerosis models, we identified 14 miRNAs with expression changes in the same variation trend by screening 2,549 human mature miRNAs in the in vitro model and 1,881 mouse miRNAs in the in vivo model. Parallel analysis of miRNA and mRNA profiles has been widely used in disease studies to identify novel diagnostic and prognostic biomarkers, to explore miRNA-mRNA coregulation in pathologic processes, and to identify candidate therapeutic targets (58–60). Fourteen miRNAs were identified as atherosclerosis related in both models, including three previously reported miRNAs that contribute to atherogenesis in different mechanisms of action. For example, miR-23b-3p has been shown to inhibit the synthesis of apo(a), one of the most important components of Lp(a), by targeting Ets1, increasing the risk of cardiovascular diseases (39). miR-30a-5p is correlated with calcification extension by targeting RUNX family transcription factor 2, and significantly upregulated in the arterial calcified culprit plaques of patients with acute coronary syndromes (40). Lastly, miR-30c-5p has been shown to be reversely correlated with total and LDL cholesterol, and promoted caspase-3 expression, leading to apoptosis (41, 42). Among the overlapping miRNAs, only miR-23a-3p was identified as a novel candidate linked to atherosclerosis.
Hsa-miR-23a-3p has been previously reported to be upregulated and is known to have a promoting role in several cancer types, including colorectal, bladder, and gastric cancers, exerting its effects by targeting various tumor suppressor genes (61–64). For example, miR-23a-3p regulates tumorigenesis by targeting proline-rich nuclear receptor coactivator 2 (PNRC2) in renal cell carcinoma (65). Another line of evidence suggests that miR-23a-3p may exert cardioprotective effects through induction of angiogenesis in ischemic heart after myocardial infarction (66). Other studies have revealed that miR-23a may promote cardiac hypertrophy by regulating forkhead box O3 (FOXO3a) and nuclear factor of activated T cells 3 (NFATc3) (67, 68). In this study, miR-23a-3p levels were significantly decreased in both in vitro and in vivo models, suggesting that it plays an important role in atherosclerosis. By pairing with predicted target genes screened in both models, the atherosclerosis-related gene, TNFAIP3, was identified and experimentally validated as a target of miR-23a-3p. Among the three literature-reported targets of miR-23a, significant overexpression of PNRC2 (2.073-fold, P = 0.005) and FOXO3 (3.275-fold, P = 0.002) were found in the in vitro model only. Conversely, Nfatc3 was downregulated (1.459-fold, P = 0.002) in the in vivo model but not in the in vitro model. In humans, miR-23a is also found in a cluster with miR-27a and miR-24-2 (69). In the above microarray database, miR-27a was downregulated significantly in the cell model only (data not shown). The expression of miR-24-2 was decreased in the cell model but upregulated in the mouse model (data not shown). Both of the miRNAs did not meet our screening criteria for further validation.
TNFAIP3, also known as A20, is a well-known anti-inflammatory molecule that regulates multiple intracellular signaling pathways (46). It was first described as a primary response gene following stimulation of human umbilical vein endothelial cells with stimulators such as TNFα, interleukin (IL)-1, and lipopolysaccharide (LPS) (70). The level of TNFAIP3 expression in specific cell types influences the type of inflammation and susceptibility to inflammatory diseases (46). Atherosclerosis is a chronic inflammatory disease, which is attenuated by TNFAIP3 through NF-κB signaling pathway inhibition in arterial endothelial cells (71–74). A recent study reported that TNFAIP3-mediated TAK1 inactivation suppresses the p38/MAPK pathway, which also contributes to decreased inflammation and subsequent endothelial cell survival (47). In this study, downregulation of TNFAIP3 by miR-23a-3p in HAECs enhanced NF-κB activation upstream of IκBα degradation and promoted the activation of p38/MAPK signaling. It subsequently activated NF-κB and p38/MAPK-dependent pro-inflammatory and proatherogenic proteins such as E-selectin, leading to endothelial dysfunction. In addition to inflammatory regulation, inhibition of TNFAIP3, an antiapoptotic protein in endothelial cells, by miR-23a-3p aggravated apoptosis in oxLDL-treated HAECs. Both endothelial inflammation and apoptosis play critical roles in endothelial dysfunction and atherosclerotic lesion formation. Therefore, miR-23a-3p promotes the atherogenic process through TNFAIP3-regulated NF-κB and p38/MAPK pathways.
In summary, a new mouse model of atherosclerosis in conjunction with human endothelial cells was used to identify a new pro-inflammatory role for miR-23a-3p in atherosclerosis. These findings may help better understand the pathogenesis of atherosclerotic diseases and accelerate the search for potential targets for therapeutic intervention. The potential role of this novel miRNA as an early diagnostic or prognostic biomarker also remains unknown and requires further investigation.
Data availability
Data have been deposited in the Gene Expression Omnibus under accession numbers GSE137578, GSE137580, GSE137581, and GSE137582.
Supplementary Material
Footnotes
This article contains supplemental data.
Author contributions—J.G., M.M.V., and H.Y. study concept; J.G., M.M.V., and H.Y. design; H.M. in vitro model development; H.M. cell functional experiments; Z.S. bioinformatic analysis. Q.M. in vivo model atherosclerotic plaque analysis; J.G., H.M., Z.S., Q.M., M.M.V., and H.Y. data collection; Q.M., M.M.V., and H.Y. data interpretation; Q.M., M.M.V., and H.Y. writing. All authors reviewed and approved the final manuscript.
Funding and additional information—This work was funded by Amgen Inc.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
Abbreviations—
- DE mRNA
- differentially expressed mRNA
- DE miRNA
- differentially expressed miRNA
- FH
- familial hypercholesterolemia
- FOXO3a
- forkhead box O3a
- gRNA
- guide RNA
- HAEC
- human aorta endothelial cell
- HE
- heterozygous
- HO
- homozygous
- ICAM
- intercellular adhesion molecules
- IL
- interleukin
- KI
- knockin
- KO
- knock-out
- LOX1
- lectin-like oxLDL receptor 1
- LPS
- Lipopolysaccharide
- MAPK
- mitogen-activated protein kinase
- ND
- normal diet
- NFATc3
- nuclear factor of activated T cells 3
- oxLDL
- oxidized low density lipoprotein
- PNRC2
- proline rich nuclear receptor coactivator 2
- RUNX-2
- RUNX family transcription factor 2
- SFIP-MutID
- structure-based factional impact prediction for mutation identification
- TAK1
- transforming growth factor β-activated kinase 1
- TC
- total cholesterol
- TNFAIP3
- tumor necrosis factor α-induced protein 3
- UTR
- untranslated region
Manuscript received September 8, 2020. Published, JLR Papers in Press, October 2, 2020, DOI 10.1194/jlr.RA120001121.
REFERENCES
- 1.Hansson G. K., Libby P., and Tabas I.. 2015. Inflammation and plaque vulnerability. J. Intern. Med. 278: 483–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sakao S., Taraseviciene-Stewart L., Lee J. D., Wood K., Cool C. D., and Voelkel N. F.. 2005. Initial apoptosis is followed by increased proliferation of apoptosis-resistant endothelial cells. FASEB J. 19: 1178–1180. [DOI] [PubMed] [Google Scholar]
- 3.Tardy Y., Resnick N., Nagel T., Gimbrone M. A. Jr., and Dewey C. F. Jr.. 1997. Shear stress gradients remodel endothelial monolayers in vitro via a cell proliferation-migration-loss cycle. Arterioscler. Thromb. Vasc. Biol. 17: 3102–3106. [DOI] [PubMed] [Google Scholar]
- 4.Cancel L. M., and Tarbell J. M.. 2010. The role of apoptosis in LDL transport through cultured endothelial cell monolayers. Atherosclerosis. 208: 335–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li D., Chen H., Romeo F., Sawamura T., Saldeen T., and Mehta J. L.. 2002. Statins modulate oxidized low-density lipoprotein-mediated adhesion molecule expression in human coronary artery endothelial cells: role of LOX-1. J. Pharmacol. Exp. Ther. 302: 601–605. [DOI] [PubMed] [Google Scholar]
- 6.Pirillo A., Norata G. D., and Catapano A. L.. 2013. LOX-1, OxLDL, and atherosclerosis. Mediators Inflamm. 2013: 152786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esau C., Davis S., Murray S. F., Yu X. X., Pandey S. K., Pear M., Watts L., Booten S. L., Graham M., McKay R., et al. . 2006. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 3: 87–98. [DOI] [PubMed] [Google Scholar]
- 8.Vickers K. C., Shoucri B. M., Levin M. G., Wu H., Pearson D. S., Osei-Hwedieh D., Collins F. S., Remaley A. T., and Sethupathy P.. 2013. MicroRNA-27b is a regulatory hub in lipid metabolism and is altered in dyslipidemia. Hepatology. 57: 533–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vickers K. C., Landstreet S. R., Levin M. G., Shoucri B. M., Toth C. L., Taylor R. C., Palmisano B. T., Tabet F., Cui H. L., Rye K. A., et al. . 2014. MicroRNA-223 coordinates cholesterol homeostasis. Proc. Natl. Acad. Sci. USA. 111: 14518–14523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goedeke L., Rotllan N., Canfran-Duque A., Aranda J. F., Ramirez C. M., Araldi E., Lin C. S., Anderson N. N., Wagschal A., de Cabo R., et al. . 2015. MicroRNA-148a regulates LDL receptor and ABCA1 expression to control circulating lipoprotein levels. Nat. Med. 21: 1280–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zernecke A., Bidzhekov K., Noels H., Shagdarsuren E., Gan L., Denecke B., Hristov M., Koppel T., Jahantigh M. N., Lutgens E., et al. . 2009. Delivery of microRNA-126 by apoptotic bodies induces CXCL12-dependent vascular protection. Sci. Signal. 2: ra81. [DOI] [PubMed] [Google Scholar]
- 12.Suárez Y., Wang C., Manes T. D., and Pober J. S.. 2010. Cutting edge: TNF-induced microRNAs regulate TNF-induced expression of E-selectin and intercellular adhesion molecule-1 on human endothelial cells: feedback control of inflammation. J. Immunol. 184: 21–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun D., Zhang J., Xie J., Wei W., Chen M., and Zhao X.. 2012. MiR-26 controls LXR-dependent cholesterol efflux by targeting ABCA1 and ARL7. FEBS Lett. 586: 1472–1479. [DOI] [PubMed] [Google Scholar]
- 14.Loyer X., Potteaux S., Vion A. C., Guerin C. L., Boulkroun S., Rautou P. E., Ramkhelawon B., Esposito B., Dalloz M., Paul J. L., et al. . 2014. Inhibition of microRNA-92a prevents endothelial dysfunction and atherosclerosis in mice. Circ. Res. 114: 434–443. [DOI] [PubMed] [Google Scholar]
- 15.Chen Z., Wen L., Martin M., Hsu C. Y., Fang L., Lin F. M., Lin T. Y., Geary M. J., Geary G. G., Zhao Y., et al. . 2015. Oxidative stress activates endothelial innate immunity via sterol regulatory element binding protein 2 (SREBP2) transactivation of microRNA-92a. Circulation. 131: 805–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun X., Icli B., Wara A. K., Belkin N., He S., Kobzik L., Hunninghake G. M., Vera M. P., Registry M., Blackwell T. S., et al. . 2012. MicroRNA-181b regulates NF-kappaB-mediated vascular inflammation. J. Clin. Invest. 122: 1973–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Aguiar Vallim T. Q., Tarling E. J., Kim T., Civelek M., Baldan A., Esau C., and Edwards P. A.. 2013. MicroRNA-144 regulates hepatic ATP binding cassette transporter A1 and plasma high-density lipoprotein after activation of the nuclear receptor farnesoid X receptor. Circ. Res. 112: 1602–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kozomara A., Birgaoanu M., and Griffiths-Jones S.. 2019. miRBase: from microRNA sequences to function. Nucleic Acids Res. 47: D155–D162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soh J., Iqbal J., Queiroz J., Fernandez-Hernando C., and Hussain M. M.. 2013. MicroRNA-30c reduces hyperlipidemia and atherosclerosis in mice by decreasing lipid synthesis and lipoprotein secretion. Nat. Med. 19: 892–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bao M. H., Li J. M., Zhou Q. L., Li G. Y., Zeng J., Zhao J., and Zhang Y. W.. 2016. Effects of miR590 on oxLDLinduced endothelial cell apoptosis: Roles of p53 and NFkappaB. Mol. Med. Rep. 13: 867–873. [DOI] [PubMed] [Google Scholar]
- 21.Zhang H., Zheng J., Lin J., Chen J., Yu Z., Chen C., and Liu T.. 2018. miR-758 mediates oxLDL-dependent vascular endothelial cell damage by suppressing the succinate receptor SUCNR1. Gene. 663: 1–8. [DOI] [PubMed] [Google Scholar]
- 22.Glass C. K., and Witztum J. L.. 2001. Atherosclerosis. the road ahead. Cell. 104: 503–516. [DOI] [PubMed] [Google Scholar]
- 23.DeGraba T. J. 1997. Expression of inflammatory mediators and adhesion molecules in human atherosclerotic plaque. Neurology. 49: S15–S19. [DOI] [PubMed] [Google Scholar]
- 24.Kitagawa K., Matsumoto M., Sasaki T., Hashimoto H., Kuwabara K., Ohtsuki T., and Hori M.. 2002. Involvement of ICAM-1 in the progression of atherosclerosis in APOE-knockout mice. Atherosclerosis. 160: 305–310. [DOI] [PubMed] [Google Scholar]
- 25.Takei A., Huang Y., and Lopes-Virella M. F.. 2001. Expression of adhesion molecules by human endothelial cells exposed to oxidized low density lipoprotein. Influences of degree of oxidation and location of oxidized LDL. Atherosclerosis. 154: 79–86. [DOI] [PubMed] [Google Scholar]
- 26.Di Santo S., Diehm N., Ortmann J., Volzmann J., Yang Z., Keo H. H., Baumgartner I., and Kalka C.. 2008. Oxidized low density lipoprotein impairs endothelial progenitor cell function by downregulation of E-selectin and integrin alpha(v)beta5. Biochem. Biophys. Res. Commun. 373: 528–532. [DOI] [PubMed] [Google Scholar]
- 27.Chen J., Mehta J. L., Haider N., Zhang X., Narula J., and Li D.. 2004. Role of caspases in Ox-LDL-induced apoptotic cascade in human coronary artery endothelial cells. Circ. Res. 94: 370–376. [DOI] [PubMed] [Google Scholar]
- 28.White S. J., Sala-Newby G. B., and Newby A. C.. 2011. Overexpression of scavenger receptor LOX-1 in endothelial cells promotes atherogenesis in the ApoE(-/-) mouse model. Cardiovasc. Pathol. 20: 369–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kataoka H., Kume N., Miyamoto S., Minami M., Moriwaki H., Murase T., Sawamura T., Masaki T., Hashimoto N., and Kita T.. 1999. Expression of lectinlike oxidized low-density lipoprotein receptor-1 in human atherosclerotic lesions. Circulation. 99: 3110–3117. [DOI] [PubMed] [Google Scholar]
- 30.Zhang T., Tian F., Wang J., Jing J., Zhou S. S., and Chen Y. D.. 2015. Atherosclerosis-associated endothelial cell apoptosis by miR-429-mediated down regulation of Bcl-2. Cell. Physiol. Biochem. 37: 1421–1430. [DOI] [PubMed] [Google Scholar]
- 31.Hayek T., Oiknine J., Brook J. G., and Aviram M.. 1994. Role of HDL apolipoprotein E in cellular cholesterol efflux: studies in apo E knockout transgenic mice. Biochem. Biophys. Res. Commun. 205: 1072–1078. [DOI] [PubMed] [Google Scholar]
- 32.Ishibashi S., Brown M. S., Goldstein J. L., Gerard R. D., Hammer R. E., and Herz J.. 1993. Hypercholesterolemia in low density lipoprotein receptor knockout mice and its reversal by adenovirus-mediated gene delivery. J. Clin. Invest. 92: 883–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chora J. R., Medeiros A. M., Alves A. C., and Bourbon M.. 2018. Analysis of publicly available LDLR, APOB, and PCSK9 variants associated with familial hypercholesterolemia: application of ACMG guidelines and implications for familial hypercholesterolemia diagnosis. Genet. Med. 20: 591–598. [DOI] [PubMed] [Google Scholar]
- 34.Jiang L., Sun L. Y., Pan X. D., Chen P. P., Tang L., Wang W., Zhao L. M., Yang S. W., and Wang L. Y.. 2016. Characterization of the unique Chinese W483X mutation in the low-density lipoprotein-receptor gene in young patients with homozygous familial hypercholesterolemia. J. Clin. Lipidol. 10: 538–546.e5. [DOI] [PubMed] [Google Scholar]
- 35.Kanai N., Fujii T., Saito K., and Tokoyama T.. 1994. Rapid and simple method for preparation of genomic DNA from easily obtainable clotted blood. J. Clin. Pathol. 47: 1043–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Agarwal V., Bell G. W., Nam J. W., and Bartel D. P.. 2015. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 4: e05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Waterhouse A. M., Procter J. B., Martin D. M., Clamp M., and Barton G. J.. 2009. Jalview version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics. 25: 1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sata M., and Walsh K.. 1998. Oxidized LDL activates fas-mediated endothelial cell apoptosis. J. Clin. Invest. 102: 1682–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zeng J. F., Zeng Z. L., Zhang K., Zhao Y., Liu Y. M., Chen J. J., Tong H., Wei D. H., Jiang Z. S., and Wang Z.. 2018. miR-23b-3p and miR-125b-5p downregulate apo(a) expression by targeting Ets1 in HepG2 cells. Cell Biol. Int. 42: 313–323. [DOI] [PubMed] [Google Scholar]
- 40.Vasuri F., Ciavarella C., Fittipaldi S., Pini R., Vacirca A., Gargiulo M., Faggioli G., and Pasquinelli G.. 2020. Different histological types of active intraplaque calcification underlie alternative miRNA-mRNA axes in carotid atherosclerotic disease. Virchows Arch. 476: 307–316. [DOI] [PubMed] [Google Scholar]
- 41.Ceolotto G., Giannella A., Albiero M., Kuppusamy M., Radu C., Simioni P., Garlaschelli K., Baragetti A., Catapano A. L., Iori E., et al. . 2017. miR-30c-5p regulates macrophage-mediated inflammation and pro-atherosclerosis pathways. Cardiovasc. Res. 113: 1627–1638. [DOI] [PubMed] [Google Scholar]
- 42.Li P., Zhong X., Li J., Liu H., Ma X., He R., and Zhao Y.. 2018. MicroRNA-30c-5p inhibits NLRP3 inflammasome-mediated endothelial cell pyroptosis through FOXO3 down-regulation in atherosclerosis. Biochem. Biophys. Res. Commun. 503: 2833–2840. [DOI] [PubMed] [Google Scholar]
- 43.Haraldsen G., Kvale D., Lien B., Farstad I. N., and Brandtzaeg P.. 1996. Cytokine-regulated expression of E-selectin, intercellular adhesion molecule-1 (ICAM-1), and vascular cell adhesion molecule-1 (VCAM-1) in human microvascular endothelial cells. J. Immunol. 156: 2558–2565. [PubMed] [Google Scholar]
- 44.Kumar S., Boehm J., and Lee J. C.. 2003. p38 MAP kinases: key signalling molecules as therapeutic targets for inflammatory diseases. Nat. Rev. Drug Discov. 2: 717–726. [DOI] [PubMed] [Google Scholar]
- 45.Pasparakis M. 2009. Regulation of tissue homeostasis by NF-kappaB signalling: implications for inflammatory diseases. Nat. Rev. Immunol. 9: 778–788. [DOI] [PubMed] [Google Scholar]
- 46.Catrysse L., Vereecke L., Beyaert R., and van Loo G.. 2014. A20 in inflammation and autoimmunity. Trends Immunol. 35: 22–31. [DOI] [PubMed] [Google Scholar]
- 47.Li L., Huang B., Song S., Sohun H., Rao Z., Tao L., Jin Q., Zeng J., Wu R., Ji K., et al. . 2017. A20 functions as mediator in TNFalpha-induced injury of human umbilical vein endothelial cells through TAK1-dependent MAPK/eNOS pathway. Oncotarget. 8: 65230–65239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hovingh G. K., Davidson M. H., Kastelein J. J., and O’Connor A. M.. 2013. Diagnosis and treatment of familial hypercholesterolaemia. Eur. Heart J. 34: 962–971. [DOI] [PubMed] [Google Scholar]
- 49.Sniderman A. D., Tsimikas S., and Fazio S.. 2014. The severe hypercholesterolemia phenotype: clinical diagnosis, management, and emerging therapies. J. Am. Coll. Cardiol. 63: 1935–1947. [DOI] [PubMed] [Google Scholar]
- 50.Guo J., Gao Y., Li X., He Y., Zheng X., Bi J., Hou L., Sa Y., Zhang M., Yin H., et al. . 2019. Systematic prediction of familial hypercholesterolemia caused by low-density lipoprotein receptor missense mutations. Atherosclerosis. 281: 1–8. [DOI] [PubMed] [Google Scholar]
- 51.Ghiselli G., Schaefer E. J., Gascon P., and Breser H. B. Jr.. 1981. Type III hyperlipoproteinemia associated with apolipoprotein E deficiency. Science. 214: 1239–1241. [DOI] [PubMed] [Google Scholar]
- 52.Gabelli C., Gregg R. E., Zech L. A., Manzato E., and Brewer H. B. Jr.. 1986. Abnormal low density lipoprotein metabolism in apolipoprotein E deficiency. J. Lipid Res. 27: 326–333. [PubMed] [Google Scholar]
- 53.Chen M., Li W., Zhang Y., and Yang J.. 2018. MicroRNA-20a protects human aortic endothelial cells from Ox-LDL-induced inflammation through targeting TLR4 and TXNIP signaling. Biomed. Pharmacother. 103: 191–197. [DOI] [PubMed] [Google Scholar]
- 54.Yang L., and Gao C.. 2019. MiR-590 inhibits endothelial cell apoptosis by inactivating the TLR4/NF-kappaB pathway in atherosclerosis. Yonsei Med. J. 60: 298–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yin J., Hou X., and Yang S.. 2019. microRNA-338-3p promotes ox-LDL-induced endothelial cell injury through targeting BAMBI and activating TGF-beta/Smad pathway. J. Cell. Physiol. 234: 11577–11586. [DOI] [PubMed] [Google Scholar]
- 56.Ross R. 1999. Atherosclerosis–an inflammatory disease. N. Engl. J. Med. 340: 115–126. [DOI] [PubMed] [Google Scholar]
- 57.Laberge M. A., Moore K. J., and Freeman M. W.. 2005. Atherosclerosis and innate immune signaling. Ann. Med. 37: 130–140. [DOI] [PubMed] [Google Scholar]
- 58.Dweep H., Sticht C., Kharkar A., Pandey P., and Gretz N.. 2013. Parallel analysis of mRNA and microRNA microarray profiles to explore functional regulatory patterns in polycystic kidney disease: using PKD/Mhm rat model. PLoS One. 8: e53780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu F., Zhu Y., He Q., Wu L. Y., Zhang Z., Shi W. H., Liu L., Chang C. K., and Li X.. 2016. Identification of microRNA-regulated pathways using an integration of microRNA-mRNA microarray and bioinformatics analysis in CD34+ cells of myelodysplastic syndromes. Sci. Rep. 6: 32232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu B., Yang H., Taher L., Denz A., Grutzmann R., Pilarsky C., and Weber G. F.. 2018. Identification of prognostic biomarkers by combined mRNA and miRNA expression microarray analysis in pancreatic cancer. Transl. Oncol. 11: 700–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wen Y. C., Lee W. J., Tan P., Yang S. F., Hsiao M., Lee L. M., and Chien M. H.. 2015. By inhibiting snail signaling and miR-23a-3p, osthole suppresses the EMT-mediated metastatic ability in prostate cancer. Oncotarget. 6: 21120–21136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen F., Qi S., Zhang X., Wu J., Yang X., and Wang R.. 2019. miR-23a-3p suppresses cell proliferation in oral squamous cell carcinomas by targeting FGF2 and correlates with a better prognosis: miR-23a-3p inhibits OSCC growth by targeting FGF2. Pathol. Res. Pract. 215: 660–667. [DOI] [PubMed] [Google Scholar]
- 63.Liu J., Fan L., Yu H., Zhang J., He Y., Feng D., Wang F., Li X., Liu Q., Li Y., et al. 2019. Endoplasmic reticulum stress causes liver cancer cells to release exosomal miR-23a-3p and up-regulate programmed death ligand 1 expression in macrophages. Hepatology. 70: 241–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ma M., Dai J., Tang H., Xu T., Yu S., Si L., Cui C., Sheng X., Chi Z., Mao L., et al. . 2019. MicroRNA-23a-3p inhibits mucosal melanoma growth and progression through targeting adenylate cyclase 1 and attenuating cAMP and MAPK pathways. Theranostics. 9: 945–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Quan J., Pan X., Li Y., Hu Y., Tao L., Li Z., Zhao L., Wang J., Li H., Lai Y., et al. . 2019. MiR-23a-3p acts as an oncogene and potential prognostic biomarker by targeting PNRC2 in RCC. Biomed. Pharmacother. 110: 656–666. [DOI] [PubMed] [Google Scholar]
- 66.Moghaddam A. S., Afshari J. T., Esmaeili S. A., Saburi E., Joneidi Z., and Momtazi-Borojeni A. A.. 2019. Cardioprotective microRNAs: Lessons from stem cell-derived exosomal microRNAs to treat cardiovascular disease. Atherosclerosis. 285: 1–9. [DOI] [PubMed] [Google Scholar]
- 67.Lin Z., Murtaza I., Wang K., Jiao J., Gao J., and Li P. F.. 2009. miR-23a functions downstream of NFATc3 to regulate cardiac hypertrophy. Proc. Natl. Acad. Sci. USA. 106: 12103–12108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang K., Lin Z. Q., Long B., Li J. H., Zhou J., and Li P. F.. 2012. Cardiac hypertrophy is positively regulated by MicroRNA miR-23a. J. Biol. Chem. 287: 589–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chhabra R., Dubey R., and Saini N.. 2010. Cooperative and individualistic functions of the microRNAs in the miR-23a∼27a∼24–2 cluster and its implication in human diseases. Mol. Cancer. 9: 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Verstrepen L., Verhelst K., van Loo G., Carpentier I., Ley S. C., and Beyaert R.. 2010. Expression, biological activities and mechanisms of action of A20 (TNFAIP3). Biochem. Pharmacol. 80: 2009–2020. [DOI] [PubMed] [Google Scholar]
- 71.Wolfrum S., Teupser D., Tan M., Chen K. Y., and Breslow J. L.. 2007. The protective effect of A20 on atherosclerosis in apolipoprotein E-deficient mice is associated with reduced expression of NF-kappaB target genes. Proc. Natl. Acad. Sci. USA. 104: 18601–18606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shrikhande G. V., Scali S. T., da Silva C. G., Damrauer S. M., Csizmadia E., Putheti P., Matthey M., Arjoon R., Patel R., Siracuse J. J., et al. . 2010. O-glycosylation regulates ubiquitination and degradation of the anti-inflammatory protein A20 to accelerate atherosclerosis in diabetic ApoE-null mice. PLoS One. 5: e14240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Prasad A. S., Bao B., Beck F. W., and Sarkar F. H.. 2011. Zinc-suppressed inflammatory cytokines by induction of A20-mediated inhibition of nuclear factor-kappaB. Nutrition. 27: 816–823. [DOI] [PubMed] [Google Scholar]
- 74.Qin M., Luo Y., Lu S., Sun J., Yang K., Sun G., and Sun X.. 2017. Ginsenoside F1 Ameliorates Endothelial Cell Inflammatory Injury and Prevents Atherosclerosis in Mice through A20-Mediated Suppression of NF-kB Signaling. Front. Pharmacol. 8: 953. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data have been deposited in the Gene Expression Omnibus under accession numbers GSE137578, GSE137580, GSE137581, and GSE137582.