Abstract
Lipoprotein (a) [Lp(a)] is characterized by an LDL-like composition in terms of lipids and apoB100, and by one copy of a unique glycoprotein, apo(a). The apo(a) structure is mainly based on the repetition of tandem kringle domains with high homology to plasminogen kringles 4 and 5. Among them, kringle IV type 2 (KIV-2) is present in a highly variable number of genetically encoded repeats, whose length is inversely related to Lp(a) plasma concentration and cardiovascular risk. Despite it being the major component of apo(a), the actual function of KIV-2 is still unclear. Here, we describe the first high-resolution crystallographic structure of this domain. It shows a general fold very similar to other KIV domains with high and intermediate affinity for the lysine analog, ε-aminocaproic acid. Interestingly, KIV-2 presents a lysine binding site (LBS) with a unique shape and charge distribution. KIV-2 affinity for predicted small molecule binders was found to be negligible in surface plasmon resonance experiments; and with the LBS being nonfunctional, we propose to rename it “pseudo-LBS”. Further investigation of the protein by computational small-molecule docking allowed us to identify a possible heparin-binding site away from the LBS, which was confirmed by specific reverse charge mutations abolishing heparin binding. This study opens new possibilities to define the pathogenesis of Lp(a)-related diseases and to facilitate the design of specific therapeutic drugs.
Keywords: lipoprotein (a), X-ray crystallography, apolipoproteins, diseases/atherosclerosis, low density lipoprotein, ε-aminocaproic acid
Human lipoprotein (a) [Lp(a)] is a macromolecular complex present in plasma originally described by Kåre Berg (1). Its physiological function is still unknown, but the increase in its concentration can double or even triple cardiovascular risk (2, 3). Lp(a) is an LDL-like lipoprotein, and it shares both lipid composition (4) and apoB100 with LDLs. Unique to Lp(a) is the presence of apo(a) (5–8), a highly polymorphic glycoprotein, which is covalently linked to apoB100 by a disulfide bond.
A characteristic feature of apo(a) is the presence of multiple kringle domains (9, 10), which are identified by their high homology to plasminogen kringle IV (KIV) and kringle V (KV). Particularly, apo(a) is composed of 10 different KIV subtypes (designated from 1 to 10) with high interdomain homology, a KV domain, and a serine protease domain (11). Nine of the 10 kringle IV types (1, 3–10) are present in a single copy, while the number of KIV type 2s (KIV-2s) can vary between 1 and more than 40 (2, 12, 13). This variability is responsible for the great heterogeneity of isoforms present in the general population and is related to CVD risk (2). It has been observed, in fact, that low molecular weight isoforms (less than or equal to 22 total KIV repeats) are associated to 4- to 5-fold higher Lp(a) plasma concentrations than high molecular weight isoforms (higher than 22 KIV repeats) (14), with low concentration variability within families (15, 16).
The molecular structure of the whole apo(a) has not yet been determined. However, the structures of some of its kringle domains are available and the atomic models of KIV-6, KIV-7, KIV-8, KIV-10, and KV have been deposited in the Protein Data Bank (PDB) database [PDB identification numbers (IDs): 1JFN, 1I71, 2FEB, 1KIV, and 4BVV, respectively], with KIV-6 and KIV-8 structures determined by NMR and those of KIV-7, KIV-10, and KV by X-ray crystallography. All these domains show a very high structural homology, with the highest found among the KIV domain subtypes. They all share a common kringle fold, consisting of a single polypeptide chain with a length of approximately 80 amino acids. They exhibit a typical disulfide bond pattern of six cysteine (Cys) residues, which are joined in a 1-6, 2-4, and 3-5 scheme (17). Kringle domains are found in plasminogen (18, 19) and several other plasma proteins (20–25) and membrane receptors (26).
The best characterized function of kringle domains is to mediate protein-protein interactions through their lysine binding site (LBS) pocket. Out of the different apo(a) KIV domains, KIV-10 has the highest homology to plasminogen K4, and it has the most conserved and highest affinity lysine-binding pocket (Table 1). In fact, a dominant role for KIV-10 in the lysine-binding function of apo(a) was first demonstrated by the loss of binding observed in a KIV-10 W70R mutant of Rhesus monkey (27). Further lysine binding capacity was later found localized within the KIV-5 to KIV-9 domains, while the KIV-1 to KIV-4 domains did not show any significant lysine binding capacity (28). Within the KIV-5 to KIV-9 range, domains KIV-6 and KIV-7 have a crucial role in Lp(a) assembly (29–32), and their LBSs show moderate affinity for the lysine analog, ε-aminocaproic acid (EACA; 0.2–0.3 mM, see Table 1). Structurally, the LBS is a surface pocket consisting of a cationic, an anionic, and a central hydrophobic binding center, each defined by specific clusters of residues. The residue types present in each kringle type LBS determine the size of the pocket and the affinity for EACA and for other ligands.
TABLE 1.
Affinities of apo(a) and plasminogen kringles for EACA
While the number of KIV-2 repeats in apo(a) is inversely related to Lp(a) plasma concentration, suggesting a key role for this domain in pathogenesis, its function is not yet completely clear. Its ability to bind EACA has never been experimentally determined and is likely to be affected by the differences observed in the three binding centers when compared with, for example, the high-affinity EACA binding site of KIV-10 (see alignment, Fig. 1). There is experimental evidence based on two-hybrid systems, however, that a single KIV-2 domain can bind fibulin-5 [also known as DANCE (33)] and apoH [also known as β2-glycoprotein I (β2GPI) (34)]. However, for the above reasons, these interactions are unlikely to be based on a classical kringle lysine binding mode.
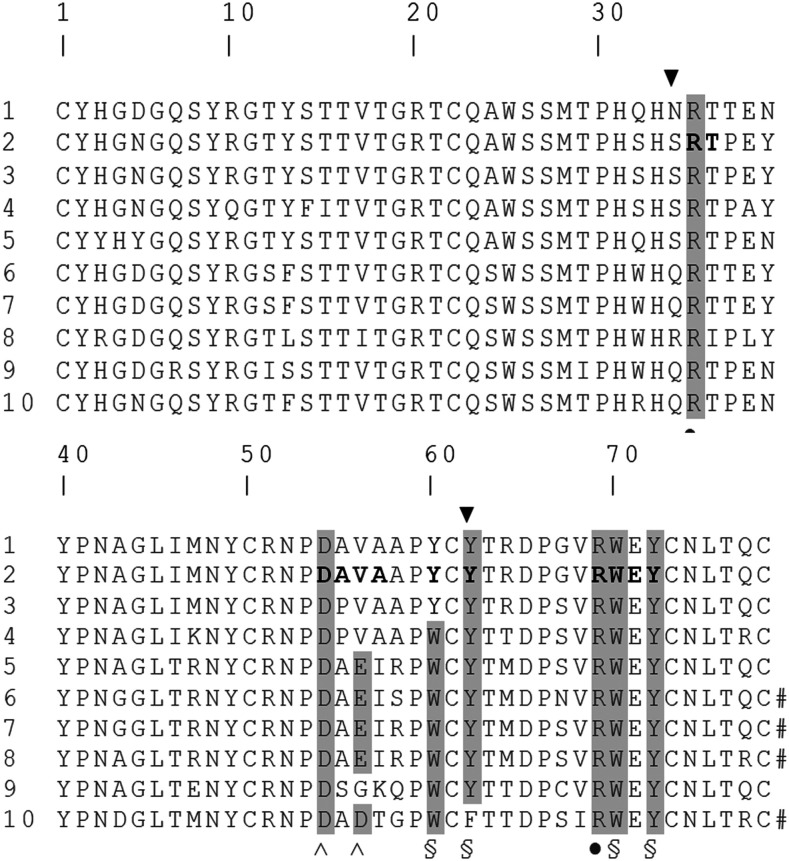
Fig. 1.
Primary sequence alignment of human apo(a) type IV kringles (1 to 10). Residues are numbered starting from the first Cys and ending in the last one. Horizontal light gray highlights the sequence of KIV-2. Vertical dark gray highlights indicate residues belonging to the conserved LBS of KIV. ▼ indicates S34 and Y62, the residues that differ the most in KIV-2 LBS three-dimensional architecture; ● indicates residues belonging to the LBS cationic center; ^ indicates residues belonging to the LBS anionic center; and § indicates residues belonging to the LBS hydrophobic center.
With the number of repeated KIV-2 domains affecting Lp(a) plasma levels, and hence CVD risk, a full understanding of KIV-2 function and molecular structure is essential for the development of new strategies for interfering with Lp(a) pathophysiological function in heart diseases and stroke. Here, we describe the production and purification of the protein in recombinant form and the determination of its high-resolution structure by X-ray crystallography. We have also analyzed its binding to low molecular weight molecules and explored possible interaction modes with relevant proteins by in silico analysis.
MATERIAL AND METHODS
Cloning, expression, and purification
The cDNA sequence encoding for a single KIV-2 domain was derived from the LPA full cDNA sequence deposited in the NCBI Nucleotide database (NM_005577.2) (35). Seven linker amino acids were included at the C terminus of the KIV-2 peptide downstream of the last Cys to facilitate formation of all the correct intra-molecular disulfide bonds. At the 5′-terminus of the KIV-2 encoding sequence, a 6x histidine tag encoding sequence was added, followed by the recognition site for tobacco etch virus (TEV) protease. The KIV-2 gene fragment was obtained by gene synthesis (GeneArt, Thermo Fisher Scientific) and subcloned into the pET-45b(+) expression vector between restriction sites Nco I and Not I. After sequencing of the insert, the construct was transformed into Escherichia coli BL21(DE3). The protein was obtained with the nonclassical inclusion bodies technique using 2xTY medium (16 g/l tryptone, 10 g/l yeast extract, 5 g/l NaCl) supplemented with 100 μg/ml ampicillin for overexpression (36). Induction was performed at an OD600 of 0.85 absorbance units (AU) by adding 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) and incubating the cultures at 18°C, 250 rpm, for 24 h (36).
Cells were collected by centrifugation and resuspended in 50 mM Tris-HCl and 500 mM NaCl (pH 8.5). The cell suspension was subjected to three sequential freeze-thaw cycles (alternating −80°C and room temperature) to disrupt cell walls. Then, sonication was performed at 40% power, on ice, by 10 cycles of 1 min each using an OMNI Ruptor 400 sonicator. The insoluble fraction containing inclusion bodies was precipitated by centrifugation (10,000 g, 4°C, 20 min) and resuspended in 2 M l-arginine, 500 mM NaCl, and 50 mM Tris-HCl (pH 8.5) by homogenization using a glass potter (37). Reduced and oxidized glutathione (5 mM and 0.5 mM final concentration, respectively) were added, prior to incubating the sample for 3–4 days while stirring at 4°C to allow protein solubilization. Then, the remaining insoluble fraction was removed by centrifugation (15,000 g, 4°C, 15 min) and the supernatant diluted 1:50 in 50 mM Tris-HCl and 500 mM NaCl (pH 8.5). The recombinant protein was purified by affinity chromatography using a 5 ml HisTrap HP column (GE Healthcare). The eluted protein was further purified using a Superdex 75 10/300 GL column (GE Healthcare). Pure monomeric KIV-2 protein to be used for crystallization was buffer exchanged versus 20 mM Tris-HCl and 100 mM NaCl (pH 8.0) using centrifugal concentrators (3 kDa cut-off; Millipore).
The Q7E, R10E, R51E triple mutant was obtained by gene synthesis (Twist Biosciences) and subcloned into pET-45b(+) using Nco I and Hind III restriction enzymes. Expression was performed as described above, reducing the pH of the extraction buffer to 7.0 and salt concentration to 200 mM after optimization. The recombinant protein was purified using the same protocol described above for the wild-type, though yields were much lower (typically 0.15 mg/l versus 30 mg/l).
Crystallization and structure determination
Protein was crystallized at 2.2 mg/ml in 20 mM Tris-HCl and 100 mM NaCl (pH 8.0) at 17°C using the sitting drop method. Condition n. 30 of MD1-01 Structure Screen kit [0.1 M HEPES (pH 7.5), 2 M ammonium sulfate, 2% v/v PEG 400] was optimized and the final condition providing diffracting crystals was: 0.1 M HEPES (pH 6.0), 1% v/v PEG 400, and 2 M ammonium sulfate.
Data were collected at the European Synchrotron Radiation Facility (Grenoble, France) at beamline ID30A-1/MASSIF-1 (38). The single KIV-2 protein structure was solved at 1.63 Å by molecular replacement using Phaser 1.11.1 (39) software and PDB IDs 1KIV and 3KIV (KIV-10 M66 variant bound and not bound to EACA, respectively) (40) and 4BVW (KIV-7 structure) (Sandmark, Althage, Andersson, et al., unpublished data) as probes. The obtained model was refined using PHENIX Refine 1.11.1 (41). Hydrogen bonds were calculated with Hbonds (https://cib.cf.ocha.ac.jp/bitool/HBOND/).
Small-molecule and protein binding prediction
If not otherwise stated, KIV-2 chain C (residues 1 to 78) was used as a monomer to predict both small-molecule binding and protein-protein interactions. Small-molecule binding was predicted using 3DLigandSite (42) and ProBis (43); heparin binding prediction was done using ClusPro server (44) and setting heparin as a ligand in the search options. For protein-protein interaction modeling, the Z-DOCK server (45) was used. Protein-protein model refinement and ranking were performed for the top 10 models using the CONSRank server (46) and the best two scoring models analyzed. Modeling of fibulin-5 (Uniprot: Q9UBX5) was obtained by i-Tasser (47). Modeller (48) was used to model KIV-2 self-assembly. Models were visualized with PyMOL (PyMOL Molecular Graphics System, Version 2.0, Schrödinger, LLC). Rotation and translation matrices were calculated by Superpose (49).
Surface plasmon resonance analysis
Affinity of small molecules for the KIV-2 domain was investigated through surface plasmon resonance (SPR), using a Biacore™ T200 (GE Healthcare) apparatus. All experiments were carried out at an operating temperature of 25°C and using PBS-T (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, 0.005% v/v Tween 20) as a running buffer. Purified KIV-2 was immobilized by an affinity capture approach, using a NiHC Nickel 1500M chip (XanTec Bioanalytics, GmbH). One of the four functionalized flow cells of the chip was used as a reference channel. For binding assays, KIV-2 was diluted to 30 μg/ml in running buffer and used to coat the working flow channel, typically obtaining 7,500 RUs of signal. For the determination of the KD value, analysis of a concentration range between 0 and 50 mM was performed for each small molecule with a contact time of 60 s, a dissociation time of 400 s, and a flow rate of 10 μl/min (n = 3).
Heparin binding was investigated on a XanTec heparin chip (XanTec Bioanalytics, GmbH) using 20 mM Tris (pH 7.7) and 100 mM NaCl as a running buffer and injecting different KIV-2 concentrations between 0 and 40 μM. The flow rate was 15 μl/min, the contact time 300 s, and dissociation time 300 s. Regeneration of the surface was obtained by one 60 s pulse of 0.05% w/v SDS in water followed by one 60 s pulse with 1 M NaCl. When needed, preincubation of KIV-2 was performed with heparin (enoxaparin, Clexane) in a 30:1 molar ratio.
KIV-2 triple mutant was always analyzed in parallel with the wild-type in similar running conditions, but the low yield of its purification allowed testing of concentrations only up to 5 μM.
Results were analyzed with Biacore T200 Evaluation Software 3.0 and with Scrubber (https://www.biologic.com.au/scrubber.html). Relative Binding Response Units (RelRUs) were compared by Student’s t-test. RelRUs for heparin binding were obtained for each curve using the Biacore T200 Evaluation Software 3.0. Data points were then used for nonlinear regression fitting using the one-site specific binding equation by GraphPad Prism.
RESULTS
Overall structure
KIV-2 crystallized in space group C 1 2 1 with three molecules per asymmetric unit. Details of data processing and structure refinement are given in Table 2. In the PDB file (PDB ID: 6RX7), residue numbering starts with 1 at the first Cys residue to facilitate comparison to other previously described kringles (Fig. 1). The Glu located immediately before residue C1 is indicated with zero and further N-terminal residues with negative numbers. Residues 0 to −10 belong to the apo(a) interdomain linker and from −11 to −13 represent part of the tobacco etch virus (TEV) recognition site.
TABLE 2.
Crystallography data collection and refinement statistics
| Data collection | |
| Space group | C 1 2 1 |
| Unit cell dimensions | |
| a, b, c (Å) | 120.25, 42.84, 61.17 |
| α, β, γ (°) | 90.00, 110.23, 90.00 |
| Resolution (Å) | 34.92–1.63 |
| R merge (%) | 0.05 |
| I/σI | 2.24 (at 1.63Å) |
| Completeness (%) | 100.0 (34.92–1.63) |
| Redundancy | 3 |
| CC 1/2 | 0.976 (0.556) |
| Refinement | |
| Resolution (Å) | 34.92–1.63 |
| Number of unique reflections | 36,781 |
| Rwork (Rfree) | 0.232 (0.269) |
| Number of atoms | 2,350 |
| Wilson B (Å2) | 17.420 |
| Average refined B factors (Å2) | 2.823 |
| r.m.s. deviations | |
| Bond lengths (Å) | 0.0067 |
| Bond angles (°) | 0.93 |
| Ramachandran analysis | |
| Favored (%) | 96 |
| Allowed (%) | 3 |
| Outliers (%) | 1 |
Numbers in parenthesis refer to the outer resolution shell.
The KIV-2 core structure was well defined in all of the three copies present in the asymmetric unit. Residues −10 to 78 could be modeled for chain A, 0 to 78 for chain B, and −13 to 79 for chain C. The relatively large part of the structure that could not be rebuilt was the main reason for the relatively high Rwork and Rfree values observed. Superposition of the Cα traces of the three chains showed no significant differences (root mean square deviation values were 0.37 Å for chain A vs. chain B, 0.36 Å for chain B vs. chain C, and 0.34 Å for chain C vs. chain A) and the Ramachandran plot revealed that 1% of the residues (Val56 from all the three molecules: electron density in supplemental Fig. S10) did not lie in the allowed regions (Table 2). The LBS of chain A contained a glycerol molecule, and the one of chain B a sulfate ion deriving from the cryoprotective and crystallization solutions, respectively. Superposition of the same region of chain C, which had nothing bound, showed no relevant changes in side chain orientation in all local residues, except for R35 in chain A, whose side chain was slightly turned and faced the LBS pocket, compared with the other two chains (supplemental Fig. S1).
The overall structure was lentil-shaped and very similar to that of other apo(a) kringles previously determined, which include KIV-6, KIV-7, KIV-8, KIV-10, and KV (supplemental Fig. S2 illustrates the superposition of α-traces). KIV-8 showed a significant divergence, but its structure was determined by NMR and only one member of the ensemble had been randomly chosen for this superposition.
The secondary structure of KIV-2 was mainly composed of turns and coils (supplemental Fig. S3), with two short stretches of anti-parallel β-sheets comprising residues 60-62 and 70-72, and several isolated β-bridges. Similarly to KIV-10 and plasminogen K4, KIV-2 did not contain α-helices in contrast to other kringles (50–53).
The LBS
KIV-10 has the highest affinity among apo(a) kringles for the lysine mimic EACA. Its sequence homology to KIV-2 is 79% (Fig. 1). Seven out of the 16 residues that are different are located along the LBS border, which, in both kringles, is made up of residues S32-R35, D54-V56, Y60-Y62, and R69-Y72 (Fig. 2A, light blue).
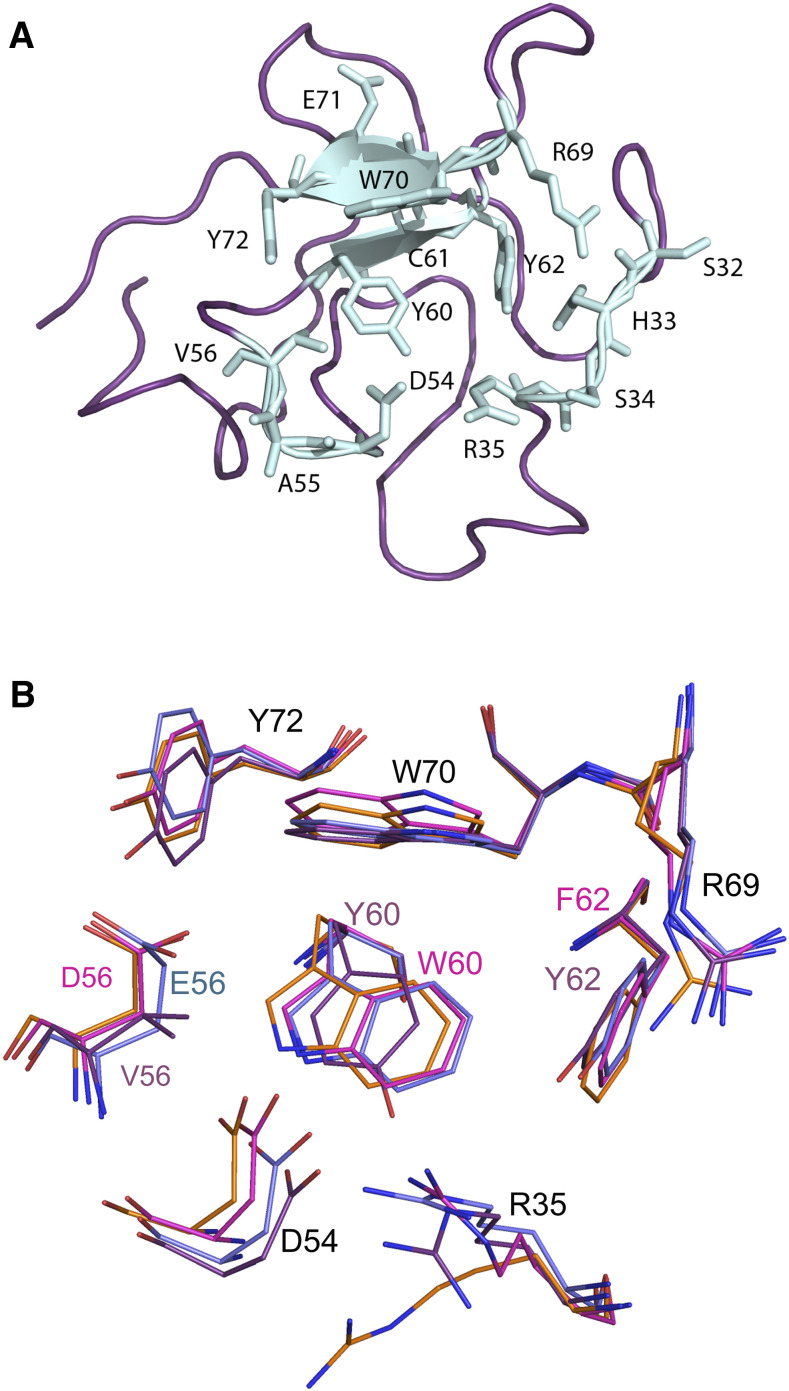
Fig. 2.
A: KIV-2 LBS residues (chain B was used to generate the figure). B: KIV-2, KIV-7, and KIV-10 LBS residues superimposition. Residues are represented as sticks colored in purple (KIV-2; PDB ID: 6RX7), cyan (KIV-7; PDB ID: 1I71), orange (KIV-10; PDB ID: 1KIV), or magenta (KIV-10 EACA complex; PDB ID: 3KIV). Residue numbers are from KIV-2 sequence (Fig. 1).
Three out of the four residues forming hydrogen bonds with EACA in KIV-10 (R35, D54, and R69) were conserved in KIV-2, while D56 was replaced by a Val residue (Fig. 2B). In the LBS cationic center (formed by residues R35 and R69), the side chain of residue R35, which forms a hydrogen bond with the O2 atom of EACA in the bound form of KIV-10, was oriented outwards in the unbound form, and it established a hydrogen bond with P53 instead. In KIV-2 (chain C), R35 pointed outwards and formed hydrogen bonds with D54, Y40, S34, and H33. P53 could not be reached in KIV-2 because of the displacement of Y40 compared with its position in KIV-10. The R69 orientation was well conserved in both KIV-2 and KIV-10 (Fig. 2B).
Regarding the anionic center (D54 and D56), the orientation of residue D54 was nearly identical in all the three chains of KIV-2 and in both the bound and unbound form of KIV-10. Its interactions with R35 were maintained in all the structures and it also established contacts with the residue at the base of the LBS, which is W60 in KIV-10 and Y60 in KIV-2. The effect of the presence of a Val residue in position 56 of KIV-2 in place of the Asp of KIV-10 was quite disruptive. In KIV-10, D56 provides bonds to EACA, to Y72, and to T57. All these bonds were not present in KIV-2 and, moreover, the D to V side-chain change modified the charge from negative to neutral, generating a hydrophobic patch in the otherwise extensive negative surface of the anionic center (supplemental Fig. S4).
Considering the hydrophobic component of the LBS, residues W60, F62, W70, and Y72 interact with the methylene chain of EACA in KIV-10. In KIV-2, W60 and F62 were replaced by Y60 and Y62, respectively. Thus, the bottom surface of the KIV-2 pocket carried two oxygen atoms that could hamper interaction with the hydrophobic part of EACA because of the charge and steric hindrance they created (Fig. 3A). Moreover, S34 and Y62 contributed hydroxyl groups in the anionic center, further reducing EACA binding affinity (Fig. 3B).
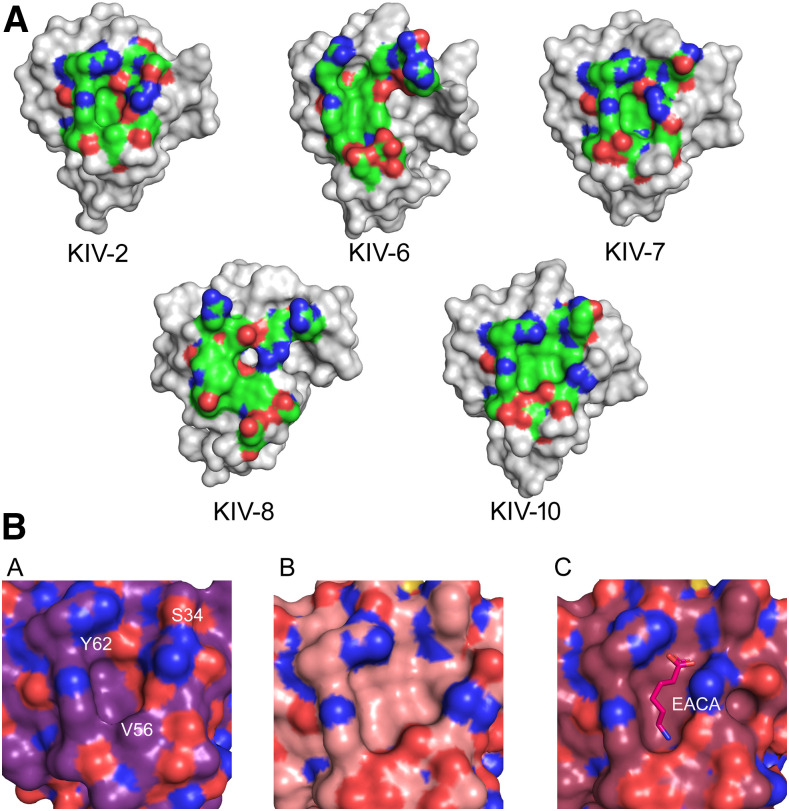
Fig. 3.
A: LBS of Lp(a) KIV-2, -6, -7, -8, and -10. Residues belonging to the LBS are represented as surface and colored by atom type (green C atoms, red O atoms, blue N atoms). KIV-6 and KIV-8 structures are derived from NMR data. PDB IDs: KIV-2, 6RX7; KIV-6, 1JFN; KIV-7, 1I71; KIV-8, 2FEB; KIV-10, 1KIV. B: Close-up of the LBS pocket (surface representation) of KIV-2 (panel a), KIV-10 apo (panel b), and EACA-bound (panel c). In panel a, residues that most vary in KIV-2 LBS are labeled. In panel c, EACA is represented as sticks.
Comparison of the KIV-2 structure to apo(a) kringles with intermediate affinity for EACA (KIV-6, KIV-7, and KIV-8) was also useful. For example, the side-chains involved in the LBS of KIV-2 superposed very well to those of KIV-7 (Fig. 2B). However, the KIV-7 side chains of the 62-64 stretch, forming the floor of the binding pocket, showed an intermediate situation compared with KIV-10 and KIV-2. Position 60 in KIV-7 carried a Trp, like in KIV-10, and position 62 a Tyr, like in KIV-2. The orientation of the R35 residue, which contributed to the cationic center, is optimal in KIV-7 to form a hydrogen bond with Y62 and D54. This was not the case in KIV-2, where the R35 side chain did not contact either Y62 or D54 in any of the three chains. The anionic center was well conserved in KIV-7, though the side chain of E64 could hamper binding of EACA compared with the one of KIV-10 D64. All this explains the affinity of KIV-7 for EACA, which sits between that of KIV-2 and KIV-10 (Table 1).
A detailed comparison of the crystallographic structure of KIV-2 with the NMR structures of KIV-6 and KIV-8 was not trivial because of the variability of the side chain positions in the different models of the ensemble. Both kringles, however, show a preserved cationic center (R35 and R60) and, in the anionic center, KIV-6 and KIV-8 show a sequence (DAE) similar to that present in KIV-7 (Fig. 1). Their hydrophobic center is perfectly conserved, while the floor of the LBS pocket in KIV-6 and KIV-8 preserves the configuration observed in KIV-7.
Binding of small molecules to KIV-2
The partial conservation of the LBS residues present in the high-affinity EACA binding site of KIV-10 leaves open the question of the real capability of KIV-2 to bind EACA and, hence, a Lys residue of another protein. To investigate the interaction of EACA and KIV-2, SPR analysis was performed, testing concentrations of EACA up to 25 mM (supplemental Fig. S5). The resulting KD was extrapolated to be greater than 87 ± 7.5 mM, indicating a very low affinity.
In order to investigate whether other small molecules could be candidates for KIV-2 binding, its complete structure was submitted to the 3DLigandSite server. The prediction of interactions of KIV-2 with small molecules was done by a homology search, obtaining positive hits with human plasminogen K1 (PDB ID: 1CEB, chain B) (51) and K2 (PDB ID: 1B2I, chain A) (52) and bovine prothrombin fragment 1 (PDB IDs: 2SPT, chain A; 1NL2, chain A; and 1NL1, chain A) (54, 55). Two molecules were identified as potential binders: trans-4-aminomethylcyclohexane-1-carboxylic acid (tranexamic acid, t-AMCHA) within the LBS and N-acetyl-D-glucosamine (NAG), binding at a location away from the LBS. However, no significant binding was found by SPR analysis, which showed an average KD of 62.5 ± 33.6 mM for t-AMCHA and of 445.8 ± 192.8 mM for NAG (supplemental Fig. S5).
Binding of heparin to KIV-2
Heparan sulfate, structurally related to heparin, binds a wide range of proteins of different functionality, some of which through an interaction with a kringle domain, and is involved in various physiological and pathological processes. KIV-2 heparin binding sites were predicted by homology to the hepatocyte growth factor/scatter factor kringle structure in complex with heparin (PDB ID: 3SP8) (Recacha, R., Mulloy, B., Gherardi, E., unpublished data). The hit was obtained using the ProBis server and using KIV-2 chain C as a search model. The interaction with heparin was also investigated by ClusPro. Nine out of the 10 simulation results clustered into two main regions (supplemental Fig. S6A–C), neither involving the LBS. Particularly, binding was predicted to occur through the highly conserved residues Q7, R10, P41, N42, R51, and P53 (Fig. 1, supplemental Fig. S6D). Subsequent SPR experiments allowed the calculation of an average KD for heparin binding of 77.61 ± 21.80 μM (supplemental Fig. S7a). Sensorgrams suggested a complex kinetics, with a very stable residual interaction with the chip after an initially fast dissociation phase, especially at higher concentrations (supplemental Fig. S7b). Preincubation of 20 μM KIV-2 with a 30 times molar excess of heparin reduced binding to the heparin-coated chip by approximately 10 times (stability at 10 s after injection end: 695.9 ± 131.4 RUs vs. 71.9 ± 11.3 RUs, P = 0.001). The introduction of three reverse charge mutations, Q7E, R10E, and R51E, completely abolished binding to the heparin-coated chip (i.e., resulted in flattened SPR sensorgrams) (supplemental Fig. S7b).
KIV-2 self assembly
Crystal packing can often provide relevant information about biological interactions. In the case of the KIV-2 domain, the asymmetric unit included three monomers (A, B, and C) assembled in a horse-shoe shape. The A and C chains were related by a complex rigid transformation, which included both a 164° rotation around an axis centered within the B-domain and a translation. Interdomain interactions occurred through LBS-independent binding interfaces. For chain B some residues of the LBS were partially involved, but not in a classical lysine binding mechanism, as the binding pocket was completely interaction free (supplemental Fig. S8a).
Interfaces involved in trimer formation were of similar sizes (255.5 Å and 240.5 Å for interfaces AB and BC, respectively). Chain B, which was sandwiched between chains A and C, interacted with them through two independent surface patches comprising residues 3-8, 55-57, and 77 (for the interaction with chain A) and residues 32-35 and 67-70 (for the interaction with chain C), respectively, whose sequences are unrelated. Interestingly, the same group of residues (38-42 and 53-55) on both chains A and C interacted with chain B. Further interactions provided by residues S34 and R35 of chain A justified the higher interface surface area provided by the latter. Each KIV-2 domain could therefore provide at least three different interaction surfaces for self-assembly, none directly involving the LBS pocket.
Crystal contacts also generated a linear chain of KIV-2 tandem repeats (supplemental Fig. S8b). In fact, a model including three KIV-2 monomers and generated in silico by using Modeller showed a configuration highly similar to that experimentally determined for the asymmetric unit, suggesting that the type of crystal contacts observed in this kind of interaction might really be physiologically favored, and not only a crystal feature. Interestingly, the structure of angiostatin kringles 1–3 (PDB ID:1KI0) (56) closely resembles the one observed in the KIV-2 asymmetric unit.
Prediction of KIV-2 binding modes to selected protein interactors
The interaction of KIV-2 with known protein ligands was explored considering its monomeric state and selecting predicted complexes according to CONSRANK score (supplemental Table S2).
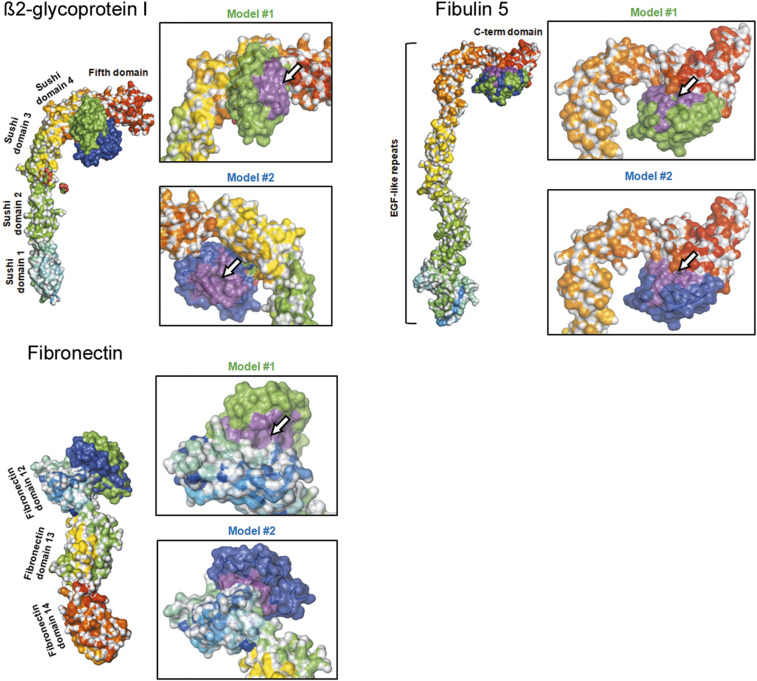
β2GPI (PDB ID: 1C1Z) (57) is a single chain polypeptide with a molecular mass of about 38 kDa, comprising 326 amino acids (58). It is organized into five sushi domains (Fig. 4) and a C-terminal domain named “fifth domain” oriented perpendicularly to the sushi domain repeats. Binding of KIV-2 to β2GPI occurred in the first two top-ranking models in the hinge region between sushi domain 4 and the fifth domain. Consensus contacts involved the two NAG molecules associated to β2GPI, while interface analysis showed that the KIV-2 LBS was not involved (Fig. 4).
Fig. 4.
Predicted KIV-2 protein-protein interactions. Proteins are shown as surface colored by domain. KIV-2 is represented as a surface, colored either in green (top ranking model) or dark blue (second ranking model). The white arrow indicates the KIV-2 LBS, where visible.
In order to study the interaction of KIV-2 with fibulin-5 (33), a model of the latter was generated by homology modeling using I-Tasser (supplemental Table S1). Though a direct comparison with the original data was not possible, the general architecture of the fibulin-5 model looked similar to the one obtained by SAXS (59) with the C-terminus domain 90° bend with respect to the stalk formed by the EGF-like tandem repeats. This structure is also similar to the one of β2GPI (Fig. 4).
According to experimental data (33), KIV-2 binding to fibulin-5 occurred in a very specific region comprising 98 C-terminal amino acids (59). Interface consensus analysis of the two top-ranking KIV-2-fibulin-5 complexes showed the partial involvement of the region comprising the LBS of KIV-2 (Fig. 4).
According to the literature, even fibronectin could interact with full-length apo(a) through a very short peptide belonging to type III fragment 12 (FN12) in a non-lysine-dependent way (60). Therefore, interaction models of KIV-2 and FN12 were built that showed good consensus. Their interface surfaces comprised the LBS of KIV-2, but the pocket was free and most of the interactions were hydrophobic (Fig. 4).
DISCUSSION
In this work, we report the first high-resolution crystallographic structure of the so far elusive KIV-2 domain of apo(a). This domain showed a high conservation of the general fold compared with other known KIV domains, but, as detailed above, the fine structure of the KIV-2 LBS was significantly different from that of apo(a) kringles with high and medium affinity for EACA (Table 1). The main differences affected the LBS pocket shape and the surface charges typically involved in EACA binding, leading to the negligible affinity found for EACA and t-AMCHA. These data indicate that the main functional role of KIV-2 cannot be related to a classical lysine binding mechanism mediated by an optimized LBS and is in line with previous sequence-based hypotheses and experimental findings (28). In fact, structural differences generate an “affinity gradient” for EACA going from the high-affinity C-terminal KIV-10 to the low affinity N-terminal KIV-2 of apo(a). Interestingly, from the alignment of KIV-2 to the other apo(a) KIV-type kringles (Fig. 1), it is possible to observe that the anionic center of the LBS comprises a DAD motif in KIV-10 (with high affinity for EACA), which becomes a DAE motif in KIV-5, KIV-6, KIV-7, and KIV-8 (domains with intermediate affinity for EACA), a DPV motif in KIV-3 and KIV-4 and a DAV motif in KIV-2 (the latter with low affinity for EACA). The absolute conservation of the D residue of the motif indicates its essential structural role, while the presence of a second negative charge (either D or E) in the third position seems to be important to optimize binding affinity to EACA.
For the hydrophobic region, the best combination of residues for EACA binding in apo(a) is the WCF motif found in KIV-10. The WCY variant (from KIV-4 to KIV-8) introduces the terminal hydroxyl group of Tyr. Its charge and size can hamper EACA binding and even more so, when the motif becomes YCY, as in KIV-2.
The cationic center is totally conserved in R35 and R69 throughout all apo(a) kringles. Notably, in KIV-2 the R35 side chain was oriented in a conformation more similar to KIV-10 with bound EACA than to the KIV-10 unbound structure. A similar orientation was found in all three molecules in the asymmetric unit, with minor changes in glycerol and sulfate bound chains A and B, respectively. In the three-dimensional structure of KIV-2, the presence of the S34 OH group in the proximity of the LBS binding site contributed an impairment to EACA binding. According to the primary structure of apo(a) KIVs, S34 is conserved in kringles with low affinity for EACA (KIV-2 to KIV-5) and is replaced by a charged Gln in higher affinity kringles (KIV-6, KIV-7, KIV-9, and KIV-10) and by an Arg in KIV-8, suggesting a major hindrance effect deriving from S34.
The fact that KIV-2-mediated interactions can occur without involvement of the LBS is very well illustrated by the molecular arrangement present in the crystal, where the three different KIV-2-KIV-2 interactions systematically occurred out of the LBS. KIV-2 is the only KIV kringle crystallizing in a trimeric asymmetric unit. Although trimers were never observed in solution in SEC, considering the unique feature of KIV-2 to be present in tandem repeats in apo(a), we speculate that similar interdomain interactions might also be present in the full-length protein.
Another example of KIV-2 interaction is the one with heparin. Our investigation indicated that KIV-2 could potentially interact with heparin and indeed KIV-2 bound to a heparin-coated SPR chip with an affinity of 78 μM, on the low side of the spectrum described in (61). It is worthwhile pointing out that the affinity derived by SPR describes the behavior of a single KIV-2 domain, but, considering the presence in apo(a) of repeated KIV-2 heparin binding units, it is plausible to expect a cumulative effect that might result in high heparin avidity. Moreover, the high level of conservation of presumed heparin binding residues throughout apo(a) KIV domains suggests that binding might participate into intermolecular or intramolecular domain bridging. Heparin binding sites are typically composed of clusters of positively charged residues that interact with the negatively charged groups of the glycosaminoglycan. In fact, the main binding site was predicted and experimentally confirmed to be located in a positively charged crevice, involving residues Q7, R10, and R51, close to the LBS, a situation similar to the one observed for the hepatocyte growth factor NK1 fragment (61, 62), suggesting a potential role of heparin in in vivo interactions. According to the results derived from the triple mutant, charge reversal is the main mechanism expected to be involved in the loss of heparin binding. However, as residues R10 and R51 establish critical hydrogen bonds with neighboring residues through their side chains, we consider alteration of the surface of this specific region, which modifies complementarity, an alternative possibility. apoB and apoE are also known to have heparin binding motifs and to bind to heparin and other glycosaminoglycans contributing to lipoprotein lipolysis and to receptor-mediated uptake of the remnants (63). Heparin binding of KIV-2 therefore deserves particular attention, as it might contribute to both processes. In this respect, its specificity and complex kinetics, which resemble those observed for the 10 kDa fragment of apoE3 (64), require further dissection.
Structural features of KIV-2 protein-protein interactions were explored for fibulin-5 and β2GPI (33, 34). Protein docking showed that interactions between them and KIV-2 do not involve the kringle LBS. Interestingly, the models suggest an interaction pose in which β2GPI and fibulin-5 might be “hooked” onto Lp(a) KIV-2 tandem repeats. Fibronectin type III interaction with apo(a) was described as well, but no definitive proof of its direct interaction with the KIV-2 component of apo(a). The interaction between fibronectin and apo(a) was described as being independent from a lysine-binding mechanism. We therefore speculated a possible main role of the lysine-binding-deficient KIV-2 in the interaction with apo(a). The obtained models were quite similar and showed a recurrent mode of interaction between the KIV-2 and FN12. The modeled information must still be confirmed by experimental data, but it is an indication of a possible further involvement of the repetitive module of apo(a) in binding with a protein which is part of the extracellular matrix involved in atherogenesis.
The availability of KIV-2 atomic structure also allows a detailed mapping of the SNPs very recently described through a systematic population analysis (65). Only 25 positions out of the 78 residues of the KIV-2 were found to be absolutely conserved (light blue in supplemental Fig. S9). They are either single or paired residues along the primary sequence. The only exception is the 68-72 stretch, which includes the five consecutive residue motif (VRWEY) forming one of the two KIV-2 anti-parallel β-sheets, suggesting that this strand represents a structural core element. For all the other positions, one or more nonsynonymous SNPs were described (purple in supplemental Fig. S9), indicating a strong structural tolerance to mutations and raising questions about the role of these regions in KIV-2 function.
In conclusion, our work reveals the first available high-resolution crystal structure of KIV-2 and provides a structural basis for very low affinity toward small molecules like EACA and t-AMCHA of the novel LBS that was experimentally determined. As the structure confirms that the KIV-2 LBS is, in fact, not structurally suitable for lysine and EACA binding, we propose to solve the derived semantic issue by renaming the KIV-2 LBS as pseudo-LBS (pLBS). For the first time, we also contribute in silico and in vitro experimental evidence for the interactions of KIV-2 with heparin, which we proved not to be mediated by the pLBS. This study has clarified that KIV-2 pLBS has, if any, a nonclassical role in apo(a) function and has identified possible alternative sites for protein-protein and protein-heparin interactions, opening possibilities for the design of new therapeutic drugs for Lp(a)-related diseases.
Data availability
All data are available in this article, except for the structural atomic coordinates of the KIV-2 domain, which are accessible in the PDB repository through PDB ID 6RX7. Modeling and docking raw data are available upon request to the corresponding author (Claudia Scotti, Department of Molecular Medicine, Unit of Immunology and General Pathology, University of Pavia, Via Ferrata, 9, 27100 Pavia, Italy, e-mail: claudia.scotti@unipv.it).
Supplementary Material
Acknowledgments
The authors thank Paolo Giordano (ABS Advanced Biological Systems) and Federico Pontigia (Neomed S.r.l.) for partnering the projects, and the Ardis S.r.l. staff for help with SPR measurements. The authors here remember Vittorio Molina for his continuous support and his sharp scientific insights into Lp(a) and other subjects.
Footnotes
This article contains supplemental data.
Author contributions—A.S. KIV2 experiments; M.M. data collection; M.M. structure solution and PDB deposition; M.M. mutant production and purification; M.M. modelling and docking; M.M. SPR data analysis; M.M. figure preparation; M.M. and H.DJ. writing-review and editing; C.S. conceptualization; C.S. supervision; C.S. SPR experiments and analysis; C.S. mutant subcloning; C.S. writing-original draft.
Author ORCIDs—Alice Santonastaso https://orcid.org/0000-0002-4492-0760; Maristella Maggi https://orcid.org/0000-0002-9075-8143; Hugo De Jonge https://orcid.org/0000-0003-2777-4084; Claudia Scotti https://orcid.org/0000-0002-5790-1833
Funding and additional information—This work was supported by Bando MISE-ICE-CRUI 2010, Project 166, and by Bando congiunto Regione Lombardia-Fondazione Cariplo Project 42657763 (2013-0380). This research was also supported by a grant from the Italian Ministry of Education, University and Research (MIUR) to the Department of Molecular Medicine of the University of Pavia under the initiative “Dipartimenti di Eccellenza (2018–2022)”. This publication is distributed under the terms of open access policies implemented by the Italian Ministry of Education, University and Research (MIUR).
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
Abbreviations—
- Cys
- cysteine
- EACA
- ε-aminocaproic acid
- KD
- affinity dissociation constant
- KIV
- kringle IV
- KIV-X
- kringle IV type X
- KV
- kringle V
- LBS
- lysine binding site
- Lp(a)
- lipoprotein (a)
- NAG
- N-acetyl-D-glucosamine
- PDB
- Protein Data Bank
- PDB ID
- Protein Data Bank identification number
- pLBS
- pseudo-lysine binding site
- SPR
- surface plasmon resonance
- t-AMCHA
- trans-4-aminomethylcyclohexane-1-carboxylic acid (tranexamic acid)
Manuscript received July 10, 2020, and in revised form September 1, 2020. Published, JLR Papers in Press, September 9, 2020, DOI 10.1194/jlr.RA120001023.
REFERENCES
- 1.Berg K. 1963. A new serum type system in man–the Lp system. Acta Pathol. Microbiol. Scand. 59: 369–382. [DOI] [PubMed] [Google Scholar]
- 2.Kronenberg F., and Utermann G.. 2013. Lipoprotein(a): resurrected by genetics. J. Intern. Med. 273: 6–30. [DOI] [PubMed] [Google Scholar]
- 3.Kamstrup P. R., Tybjaerg-Hansen A., Steffensen R., and Nordestgaard B. G.. 2009. Genetically elevated lipoprotein(a) and increased risk of myocardial infarction. JAMA. 301: 2331–2339. [DOI] [PubMed] [Google Scholar]
- 4.Sattler W., Kostner G. M., Waeg G., and Esterbauer H.. 1991. Oxidation of lipoprotein Lp(a). A comparison with low-density lipoproteins. Biochim. Biophys. Acta. 1081: 65–74. [DOI] [PubMed] [Google Scholar]
- 5.Ehnholm C., Garoff H., Renkonen O., and Simons K.. 1972. Protein and carbohydrate composition of Lp(a)lipoprotein from human plasma. Biochemistry. 11: 3229–3232. [DOI] [PubMed] [Google Scholar]
- 6.Gaubatz J. W., Heideman C., Gotto A. M. Jr., Morrisett J. D., and Dahlen G. H.. 1983. Human plasma lipoprotein [a]. Structural properties. J. Biol. Chem. 258: 4582–4589. [PubMed] [Google Scholar]
- 7.Utermann G., and Weber W.. 1983. Protein composition of Lp(a) lipoprotein from human plasma. FEBS Lett. 154: 357–361. [DOI] [PubMed] [Google Scholar]
- 8.Utermann G. 1989. The mysteries of lipoprotein(a). Science. 246: 904–910. [DOI] [PubMed] [Google Scholar]
- 9.Eaton D. L., Fless G. M., Kohr W. J., McLean J. W., Xu Q. T., Miller C. G., Lawn R. M., and Scanu A. M.. 1987. Partial amino acid sequence of apolipoprotein(a) shows that it is homologous to plasminogen. Proc. Natl. Acad. Sci. USA. 84: 3224–3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kratzin H., Armstrong V. W., Niehaus M., Hilschmann N., and Seidel D.. 1987. Structural relationship of an apolipoprotein (a) phenotype (570 kDa) to plasminogen: homologous kringle domains are linked by carbohydrate-rich regions. Biol. Chem. Hoppe Seyler. 368: 1533–1544. [DOI] [PubMed] [Google Scholar]
- 11.Schmidt K., Noureen A., Kronenberg F., and Utermann G.. 2016. Structure, function, and genetics of lipoprotein (a). J. Lipid Res. 57: 1339–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kraft H. G., Kochl S., Menzel H. J., Sandholzer C., and Utermann G.. 1992. The apolipoprotein (a) gene: a transcribed hypervariable locus controlling plasma lipoprotein (a) concentration. Hum. Genet. 90: 220–230. [DOI] [PubMed] [Google Scholar]
- 13.Lackner C., Boerwinkle E., Leffert C. C., Rahmig T., and Hobbs H. H.. 1991. Molecular basis of apolipoprotein (a) isoform size heterogeneity as revealed by pulsed-field gel electrophoresis. J. Clin. Invest. 87: 2153–2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kronenberg F. 2016. Human genetics and the causal role of lipoprotein(a) for various diseases. Cardiovasc. Drugs Ther. 30: 87–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perombelon Y. F., Soutar A. K., and Knight B. L.. 1994. Variation in lipoprotein(a) concentration associated with different apolipoprotein(a) alleles. J. Clin. Invest. 93: 1481–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Puckey L. H., Lawn R. M., and Knight B. L.. 1997. Polymorphisms in the apolipoprotein(a) gene and their relationship to allele size and plasma lipoprotein(a) concentration. Hum. Mol. Genet. 6: 1099–1107. [DOI] [PubMed] [Google Scholar]
- 17.Trexler M., and Patthy L.. 1983. Folding autonomy of the kringle 4 fragment of human plasminogen. Proc. Natl. Acad. Sci. USA. 80: 2457–2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ponting C. P., Marshall J. M., and Cederholm-Williams S. A.. 1992. Plasminogen: a structural review. Blood Coagul. Fibrinolysis. 3: 605–614. [PubMed] [Google Scholar]
- 19.Soff G. A. 2000. Angiostatin and angiostatin-related proteins. Cancer Metastasis Rev. 19: 97–107. [DOI] [PubMed] [Google Scholar]
- 20.Chinnaraj M., Planer W., and Pozzi N.. 2018. Structure of coagulation factor II: molecular mechanism of thrombin generation and development of next-generation anticoagulants. Front. Med. (Lausanne). 5: 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McMullen B. A., and Fujikawa K.. 1985. Amino acid sequence of the heavy chain of human alpha-factor XIIa (activated Hageman factor). J. Biol. Chem. 260: 5328–5341. [PubMed] [Google Scholar]
- 22.van Zonneveld A. J., Veerman H., MacDonald M. E., van Mourik J. A., and Pannekoek H.. 1986. Structure and function of human tissue-type plasminogen activator (t-PA). J. Cell. Biochem. 32: 169–178. [DOI] [PubMed] [Google Scholar]
- 23.Günzler W. A., Steffens G. J., Otting F., Kim S. M., Frankus E., and Flohe L.. 1982. The primary structure of high molecular mass urokinase from human urine. The complete amino acid sequence of the A chain. Hoppe Seylers Z. Physiol. Chem. 363: 1155–1165. [DOI] [PubMed] [Google Scholar]
- 24.Holmes O., Pillozzi S., Deakin J. A., Carafoli F., Kemp L., Butler P. J., Lyon M., and Gherardi E.. 2007. Insights into the structure/function of hepatocyte growth factor/scatter factor from studies with individual domains. J. Mol. Biol. 367: 395–408. [DOI] [PubMed] [Google Scholar]
- 25.Sigurdardottir A. G., Winter A., Sobkowicz A., Fragai M., Chirgadze D., Ascher D. B., Blundell T. L., and Gherardi E.. 2015. Exploring the chemical space of the lysine-binding pocket of the first kringle domain of hepatocyte growth factor/scatter factor (HGF/SF) yields a new class of inhibitors of HGF/SF-MET binding. Chem. Sci. 6: 6147–6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma X., Zhang Y., Liu B., Yang J., and Hu K.. 2018. Backbone and side-chain chemical shift assignments of the kringle domain of human receptor tyrosine kinase-like orphan receptor 1 (ROR1). Biomol. NMR Assign. 12: 145–148. [DOI] [PubMed] [Google Scholar]
- 27.Tomlinson J. E., McLean J. W., and Lawn R. M.. 1989. Rhesus monkey apolipoprotein(a). Sequence, evolution, and sites of synthesis. J. Biol. Chem. 264: 5957–5965. [PubMed] [Google Scholar]
- 28.Ernst A., Helmhold M., Brunner C., Petho-Schramm A., Armstrong V. W., and Muller H. J.. 1995. Identification of two functionally distinct lysine-binding sites in kringle 37 and in kringles 32-36 of human apolipoprotein(a). J. Biol. Chem. 270: 6227–6234. [DOI] [PubMed] [Google Scholar]
- 29.Gabel B. R., and Koschinsky M. L.. 1998. Sequences within apolipoprotein(a) kringle IV types 6-8 bind directly to low-density lipoprotein and mediate noncovalent association of apolipoprotein(a) with apolipoprotein B-100. Biochemistry. 37: 7892–7898. [DOI] [PubMed] [Google Scholar]
- 30.Frank S., and Kostner G. M.. 1997. The role of apo-(a) kringle-IVs in the assembly of lipoprotein-(a). Protein Eng. 10: 291–298. [DOI] [PubMed] [Google Scholar]
- 31.Frank S., Durovic S., and Kostner G. M.. 1994. Structural requirements of apo-a for the lipoprotein-a assembly. Biochem. J. 304: 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keesler G. A., Li Y., Skiba P. J., Fless G. M., and Tabas I.. 1994. Macrophage foam cell lipoprotein(a)/apoprotein(a) receptor. Cell-surface localization, dependence of induction on new protein synthesis, and ligand specificity. Arterioscler. Thromb. 14: 1337–1345. [DOI] [PubMed] [Google Scholar]
- 33.Kapetanopoulos A., Fresser F., Millonig G., Shaul Y., Baier G., and Utermann G.. 2002. Direct interaction of the extracellular matrix protein DANCE with apolipoprotein(a) mediated by the kringle IV-type 2 domain. Mol. Genet. Genomics. 267: 440–446. [DOI] [PubMed] [Google Scholar]
- 34.Köchl S., Fresser F., Lobentanz E., Baier G., and Utermann G.. 1997. Novel interaction of apolipoprotein(a) with beta-2 glycoprotein I mediated by the kringle IV domain. Blood. 90: 1482–1489. [PubMed] [Google Scholar]
- 35.Tolbus A., Mortensen M. B., Nielsen S. F., Kamstrup P. R., Bojesen S. E., and Nordestgaard B. G.. 2017. Kringle IV type 2, not low lipoprotein(a), as a cause of diabetes: a novel genetic approach using SNPs associated selectively with lipoprotein(a) concentrations or with kringle IV type 2 repeats. Clin. Chem. 63: 1866–1876. [DOI] [PubMed] [Google Scholar]
- 36.Jevsevar S., Gaberc-Porekar V., Fonda I., Podobnik B., Grdadolnik J., and Menart V.. 2005. Production of nonclassical inclusion bodies from which correctly folded protein can be extracted. Biotechnol. Prog. 21: 632–639. [DOI] [PubMed] [Google Scholar]
- 37.Tsumoto K., Abe R., Ejima D., and Arakawa T.. 2010. Non-denaturing solubilization of inclusion bodies. Curr. Pharm. Biotechnol. 11: 309–312. [DOI] [PubMed] [Google Scholar]
- 38.Svensson O., Gilski M., Nurizzo D., and Bowler M. W.. 2018. Multi-position data collection and dynamic beam sizing: recent improvements to the automatic data-collection algorithms on MASSIF-1. Acta Crystallogr. D Struct. Biol. 74: 433–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J.. 2007. Phaser crystallographic software. J. Appl. Crystallogr. 40: 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mochalkin I., Cheng B., Klezovitch O., Scanu A. M., and Tulinsky A.. 1999. Recombinant kringle IV-10 modules of human apolipoprotein(a): structure, ligand binding modes, and biological relevance. Biochemistry. 38: 1990–1998. [DOI] [PubMed] [Google Scholar]
- 41.Afonine P. V., Grosse-Kunstleve R. W., Echols N., Headd J. J., Moriarty N. W., Mustyakimov M., Terwilliger T. C., Urzhumtsev A., Zwart P. H., and Adams P. D.. 2012. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68: 352–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wass M. N., Kelley L. A., and Sternberg M. J.. 2010. 3DLigandSite: predicting ligand-binding sites using similar structures. Nucleic Acids Res. 38: W469–W473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Konc J., and Janezic D.. 2014. ProBiS-ligands: a web server for prediction of ligands by examination of protein binding sites. Nucleic Acids Res. 42: W215–W220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vajda S., Yueh C., Beglov D., Bohnuud T., Mottarella S. E., Xia B., Hall D. R., and Kozakov D.. 2017. New additions to the ClusPro server motivated by CAPRI. Proteins. 85: 435–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pierce B. G., Wiehe K., Hwang H., Kim B. H., Vreven T., and Weng Z.. 2014. ZDOCK server: interactive docking prediction of protein-protein complexes and symmetric multimers. Bioinformatics. 30: 1771–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chermak E., Petta A., Serra L., Vangone A., Scarano V., Cavallo L., and Oliva R.. 2015. CONSRANK: a server for the analysis, comparison and ranking of docking models based on inter-residue contacts. Bioinformatics. 31: 1481–1483. [DOI] [PubMed] [Google Scholar]
- 47.Zhang Y. 2008. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 9: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Webb B., and Sali A.. 2017. Protein structure modeling with MODELLER. Methods Mol. Biol. 1654: 39–54. [DOI] [PubMed] [Google Scholar]
- 49.Krissinel E., and Henrick K.. 2004. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60: 2256–2268. [DOI] [PubMed] [Google Scholar]
- 50.Hansen A. P., Petros A. M., Meadows R. P., Nettesheim D. G., Mazar A. P., Olejniczak E. T., Xu R. X., Pederson T. M., Henkin J., and Fesik S. W.. 1994. Solution structure of the amino-terminal fragment of urokinase-type plasminogen activator. Biochemistry. 33: 4847–4864. [DOI] [PubMed] [Google Scholar]
- 51.Mathews I. I., Vanderhoff-Hanaver P., Castellino F. J., and Tulinsky A.. 1996. Crystal structures of the recombinant kringle 1 domain of human plasminogen in complexes with the ligands epsilon-aminocaproic acid and trans-4-(aminomethyl)cyclohexane-1-carboxylic Acid. Biochemistry. 35: 2567–2576. [DOI] [PubMed] [Google Scholar]
- 52.Marti D. N., Schaller J., and Llinas M.. 1999. Solution structure and dynamics of the plasminogen kringle 2-AMCHA complex: 3(1)-helix in homologous domains. Biochemistry. 38: 15741–15755. [DOI] [PubMed] [Google Scholar]
- 53.de Vos A. M., Ultsch M. H., Kelley R. F., Padmanabhan K., Tulinsky A., Westbrook M. L., and Kossiakoff A. A.. 1992. Crystal structure of the kringle 2 domain of tissue plasminogen activator at 2.4-A resolution. Biochemistry. 31: 270–279. [DOI] [PubMed] [Google Scholar]
- 54.Seshadri T. P., Skrzypczak-Jankun E., Yin M., and Tulinsky A.. 1994. Differences in the metal ion structure between Sr- and Ca-prothrombin fragment 1. Biochemistry. 33: 1087–1092. [DOI] [PubMed] [Google Scholar]
- 55.Huang M., Rigby A. C., Morelli X., Grant M. A., Huang G., Furie B., Seaton B., and Furie B. C.. 2003. Structural basis of membrane binding by Gla domains of vitamin K-dependent proteins. Nat. Struct. Biol. 10: 751–756. [DOI] [PubMed] [Google Scholar]
- 56.Abad M. C., Arni R. K., Grella D. K., Castellino F. J., Tulinsky A., and Geiger J. H.. 2002. The X-ray crystallographic structure of the angiogenesis inhibitor angiostatin. J. Mol. Biol. 318: 1009–1017. [DOI] [PubMed] [Google Scholar]
- 57.Schwarzenbacher R., Zeth K., Diederichs K., Gries A., Kostner G. M., Laggner P., and Prassl R.. 1999. Crystal structure of human beta2-glycoprotein I: implications for phospholipid binding and the antiphospholipid syndrome. EMBO J. 18: 6228–6239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lozier J., Takahashi N., and Putnam F. W.. 1984. Complete amino acid sequence of human plasma beta 2-glycoprotein I. Proc. Natl. Acad. Sci. USA. 81: 3640–3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jones R. P., Wang M. C., Jowitt T. A., Ridley C., Mellody K. T., Howard M., Wang T., Bishop P. N., Lotery A. J., Kielty C. M., et al. 2009. Fibulin 5 forms a compact dimer in physiological solutions. J. Biol. Chem. 284: 25938–25943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van der Hoek Y. Y., Sangrar W., Cote G. P., Kastelein J. J., and Koschinsky M. L.. 1994. Binding of recombinant apolipoprotein(a) to extracellular matrix proteins. Arterioscler. Thromb. 14: 1792–1798. [DOI] [PubMed] [Google Scholar]
- 61.Peysselon F., and Ricard-Blum S.. 2014. Heparin-protein interactions: from affinity and kinetics to biological roles. Application to an interaction network regulating angiogenesis. Matrix Biol. 35: 73–81. [DOI] [PubMed] [Google Scholar]
- 62.Lietha D., Chirgadze D. Y., Mulloy B., Blundell T. L., and Gherardi E.. 2001. Crystal structures of NK1-heparin complexes reveal the basis for NK1 activity and enable engineering of potent agonists of the MET receptor. EMBO J. 20: 5543–5555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lookene A., Savonen R., and Olivecrona G.. 1997. Interaction of lipoproteins with heparan sulfate proteoglycans and with lipoprotein lipase. Studies by surface plasmon resonance technique. Biochemistry. 36: 5267–5275. [DOI] [PubMed] [Google Scholar]
- 64.Futamura M., Dhanasekaran P., Handa T., Phillips M. C., Lund-Katz S., and Saito H.. 2005. Two-step mechanism of binding of apolipoprotein E to heparin: implications for the kinetics of apolipoprotein E-heparan sulfate proteoglycan complex formation on cell surfaces. J. Biol. Chem. 280: 5414–5422. [DOI] [PubMed] [Google Scholar]
- 65.Coassin S., Schonherr S., Weissensteiner H., Erhart G., Forer L., Losso J. L., Lamina C., Haun M., Utermann G., Paulweber B., et al. 2019. A comprehensive map of single-base polymorphisms in the hypervariable LPA kringle IV type 2 copy number variation region. J. Lipid Res. 60: 186–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Maderegger B., Bermel W., Hrzenjak A., Kostner G. M., and Sterk H.. 2002. Solution structure of human apolipoprotein(a) kringle IV type 6. Biochemistry. 41: 660–668. [DOI] [PubMed] [Google Scholar]
- 67.Becker L., Cook P. M., Wright T. G., and Koschinsky M. L.. 2004. Quantitative evaluation of the contribution of weak lysine-binding sites present within apolipoprotein(a) kringle IV types 6–8 to lipoprotein(a) assembly. J. Biol. Chem. 279: 2679–2688. [DOI] [PubMed] [Google Scholar]
- 68.Rahman M. N., Becker L., Petrounevitch V., Hill B. C., Jia Z., and Koschinsky M. L.. 2002. Comparative analyses of the lysine binding site properties of apolipoprotein(a) kringle IV types 7 and 10. Biochemistry. 41: 1149–1155. [DOI] [PubMed] [Google Scholar]
- 69.LoGrasso P. V., Cornell-Kennon S., and Boettcher B. R.. 1994. Cloning, expression, and characterization of human apolipoprotein(a) kringle IV37. J. Biol. Chem. 269: 21820–21827. [PubMed] [Google Scholar]
- 70.Marti D. N., Hu C. K., An S. S., von Haller P., Schaller J., and Llinas M.. 1997. Ligand preferences of kringle 2 and homologous domains of human plasminogen: canvassing weak, intermediate, and high-affinity binding sites by 1H-NMR. Biochemistry. 36: 11591–11604. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in this article, except for the structural atomic coordinates of the KIV-2 domain, which are accessible in the PDB repository through PDB ID 6RX7. Modeling and docking raw data are available upon request to the corresponding author (Claudia Scotti, Department of Molecular Medicine, Unit of Immunology and General Pathology, University of Pavia, Via Ferrata, 9, 27100 Pavia, Italy, e-mail: claudia.scotti@unipv.it).