Abstract
Phalaenopsis cornu-cervi is a taxonomically and horticulturally important moth orchid. In this study, we report and characterize the complete plastid genome sequence of P. cornu-cervi for the first genomic resources in section Polychilos. Its complete plastome is 147,241 bp in length and contains two inverted repeat (IR) regions of 25,005bp, a large single-copy (LSC) region of 85,714 bp, and a small single-copy (SSC) region of 11,517 bp. The plastome contains 110 genes, consisting of 76 unique protein-coding genes, 30 unique tRNA genes, and 4 unique rRNA genes. It also shows the typical characteristics of Phalaenopsis chloroplast genome, while all ndh genes are non-functional. The complete plastome sequence of P. cornu-cervi will provide a useful resource for future phylogenetic study of Phalaenopsis and its garden utilization.
Keywords: Section Polychilos, Phalaenopsis cornu-cervi, chloroplast genome
Commonly referred to as the ‘moth orchid’, Phalaenopsis is one of the well-known genera in Orchidaceae that occupied a large proportion of ornamental market shares (Van Huylenbroeck 2018). In this genus, subgenus Phalaenopsis has the showy flower and also widely applied as the crossing parents for breeding within moth orchids or between close genera. Subgenus Phalaenopsis was classified into two sections, sect. Phalaenopsis and sect. Polychilos, three chloroplast genomes of section Phalaenopsis have been published (Chang et al. 2006; Jheng et al. 2012; Kim et al. 2016), however, no genetic data of species belonging to sect. Polychilos is obtained yet. Thus, we obtained the plastome of P. cornu-cervi (GenBank accession number: MN395041), the type species of sect. Polychilos, as the sequenced target to provide genetic and genomic information for this section to promote the systematics research and garden utilization of Phalaenopsis.
Phalaenopsis cornu-cervi was sampled from National Orchid Conservation Centre in Guangdong province of China (114°19'01''E, 22°60'34'' N). A voucher specimen (Noccphal052) was deposited in the Herbarium of National Orchid Conservation Centre, Shenzhen, China. DNA was acquired from the young leaves of its plant and the total genome was sequenced using Illumina HiSeq 2000 platform. Then, the plastid source reads were identified by mapping them against the published Phalaenopsis chloroplast genomes by BLAST. Later, we applied PLATANUS to conduct the assembly with the modification by Geneious 2019.0.3. Geneious 2019.0.3 and online pipeline Geseq (Tillich et al. 2017) were used to complete the annotation against the plastome of Phalaenopsis aphrodite var. formosa. We used the BLAST to confirm the IR boundaries for the draft chloroplast genome.
The total chloroplast genome size of P. cornu-cervi is 147,241 bp, including a large single-copy (LSC) region (85,714 bp), a small single-copy region (11,517 bp), and two inverted repeat (IRs) regions (25,005 bp). The overall GC contents of the plastid genome were 36.7%. In total, the plastome contains 110 unique genes, consisting of 76 protein-coding genes, 4 rRNA genes, and 30 tRNA genes. The ndh genes were also non-functional in P. cornu-cervi, and ndhA, ndhF, and ndhH genes were completely absent as previous description by Chang et al. (2006). Moreover, the ycf3, ycf1, rps16, rpoC2 and atpF were also non-functional in P. cornu-cervi, indicating the unique plastome features of sect. Polychilos species.
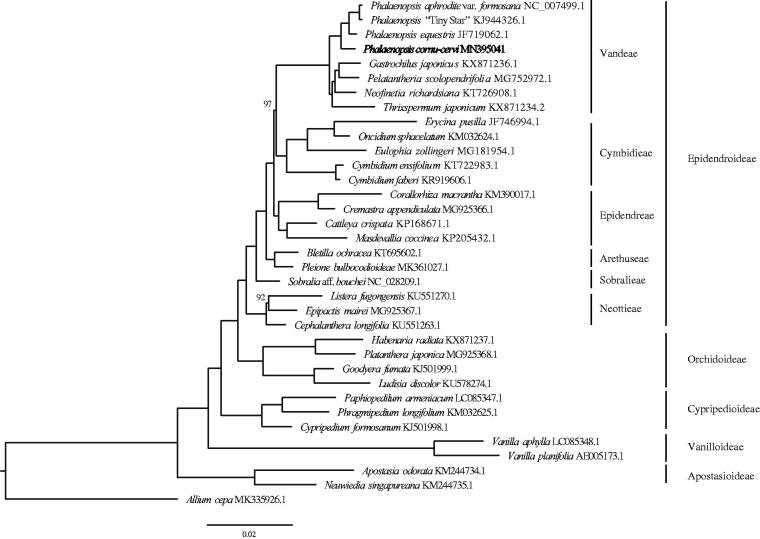
We used RAxML (Stamatakis 2006) with 1000 bootstraps under the GTRGAMMAI substitution model to reconstruct a maximum likelihood (ML) phylogeny of P. cornu-cervi and 13 published complete plastomes of Orchidaceae, using Allium cepa (Amaryllidaceae) as outgroups. The phylogenetic analysis indicated that P. cornu-cervi was situated as the sister of sect. Phalaenopsis clade (Figure 1). The complete plastome sequence of P. cornu-cervi will provide useful information for phylogenetic studies and future breeding in Phalaenopsis even in Orchidaceae.
Figure 1.
Maximum-likelihood phylogenetic tree reconstructed by RAxML based on complete plastome sequences from P. cornu-cervi, and thirty-three other orchids representing different tribes and subfamilies with Allium cepa as outgroup (Amaryllidaceae). Numbers on branches are bootstrap support values below than 100.
Acknowledgements
The authors are grateful to Zhong-Jian Liu for revising the manuscript and Ding-Kun Liu for advice on analysis pipeline.
Authors’ contributions
C.C.P. conceived the study; J.Y.W. and G.Q.Z. obtained the molecular data; J.Y.W. conducted the data analysis; J.Y.W. drafted the manuscript; C.C.P. revised the manuscript. All authors provided comments and final approval.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Chang C-C, Lin H-C, Lin I-P, Chow T-Y, Chen H-H, Chen W-H, Cheng C-H, Lin C-Y, Liu S-M, Chang C-C, et al. 2006. The chloroplast genome of Phalaenopsis aphrodite (Orchidaceae): comparative analysis of evolutionary rate with that of grasses and its phylogenetic implications. Mol Biol Evol. 23(2):279–291. [DOI] [PubMed] [Google Scholar]
- Jheng C-F, Chen T-C, Lin J-Y, Chen T-C, Wu W-L, Chang C-C. 2012. The comparative chloroplast genomic analysis of photosynthetic orchids and developing DNA markers to distinguish Phalaenopsis orchids. Plant Sci. 190:62–73. [DOI] [PubMed] [Google Scholar]
- Kim G-B, Kwon Y, Yu H-J, Lim K-B, Seo J-H, Mun J-H. 2016. The complete chloroplast genome of Phalaenopsis “Tiny Star”. Mitochondrial DNA Part A. 27(2):1300–1302. [DOI] [PubMed] [Google Scholar]
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Huylenbroeck J. 2018. Ornamental crops. New York: Springer. [Google Scholar]