Abstract
Onosma paniculatum is a medicinal plant commonly used in Yunnan and its adjacent regions, China. In the present study, we sequenced the complete chloroplast (cp) genome sequence of O. paniculatum to investigate the phylogenetic relationship in the Tubiflorae. The total length of the chloroplast genome was 151,198 bp, with 37.4% overall GC content and exhibited typical quadripartite structure, a pair of IRs (inverted repeats) of 25,889 bp was separated by a small single copy (SSC) region of 17,274 bp and a large single copy (LSC) region of 82,146 bp. The cp genome was composed of 113 genes, including 79 protein coding genes, 30 tRNA genes, and 4 rRNA genes. The phylogenetic analysis indicated that Boraginaceae was closely related to Convolvulaceae and Solanaceae in Tubiflorae.
Keywords: Onosma paniculatum, Boraginaceae, complete chloroplast genome, Illumina sequencing, phylogenetic analysis
Onosma L. is an important genus of the Boraginaceae family, which includes about 150 species all over the world (Naz et al. 2006). The genus is distributed across Europe to East Asia, especially in Western and Central Asia, the Mediterranean area, Anatolia and Southeast Europe (Kolarčik et al. 2010). In this genus, there are 29 species in China (Zhu et al. 1995), and some species of them, such as Onosma paniculatum Bur. et Franch., possess important medicinal values and are commonly used as the alternative of Arnebiae radix in Yunnan and its adjacent regions, China (Ge et al. 2003; Chinese Pharmacopoeia Commission 2015). Until now, most of the studies on Onosma and its related genus have focused on its chemical compositions, morphological taxonomy and molecular phylogeny (Kumar et al. 2013; Binzet et al. 2018; Nasrollahi et al. 2019); whereas, no complete chloroplast genome sequence has been reported in NCBI and other databases. Therefore, we sequenced the complete chloroplast genome sequence of the important medicinal plant, O. paniculatum, as well as reconstructed its phylogenetic tree with other groups based on cp genomes.
Molecular materials and voucher specimen (No. ZDQ105) of O. paniculatum were collected from Dali county, Yunnan, China (N25.76°, E100.23°), and then deposited at the Herbarium of Medicinal Plants and Crude Drugs of the College of Pharmacy and Chemistry, Dali University (DLUSYX19001). The total genomic DNA was extracted using the improved CTAB method (Doyle 1987; Yang et al. 2014), and sequenced with Illumina Hiseq 2500 (Novogene, Tianjing, China) platform with pair-end (2 × 300 bp) library. About 6.34 Gb of raw reads with 21,147,864 paired-end reads were obtained from high-throughput sequencing. The raw data was filtered using Trimmomatic v.0.32 with default settings (Bolger et al. 2014). Then paired-end reads of clean data were assembled into circular contigs using GetOrganelle.py (Jin et al. 2018). Finally, the cpDNA was annotated by the Dual Organellar Genome Annotator (DOGMA; http://dogma.ccbb.utexas.edu/) (Wyman et al. 2004) and tRNAscan-SE (Lowe and Chan 2016).
The annotated chloroplast genome was submitted to the GenBank with accession number MN175501. Total length of the chloroplast genome was 151,198 bp, with 37.4% overall GC content. With typical quadripartite structure, a pair of IRs (inverted repeats) of 25,889 bp was separated by a small single copy (SSC) region of 17,274 bp and a large single copy (LSC) region of 82,146 bp. The cp genome was composed of 113 genes, including 79 protein coding genes, 30 tRNA genes, and 4 rRNA genes. Among them, 17 genes were duplicated in the inverted repeat regions, 16 genes, and 6 tRNA genes contain one intron, while 2 genes (ycf3 and clpP) have two introns.
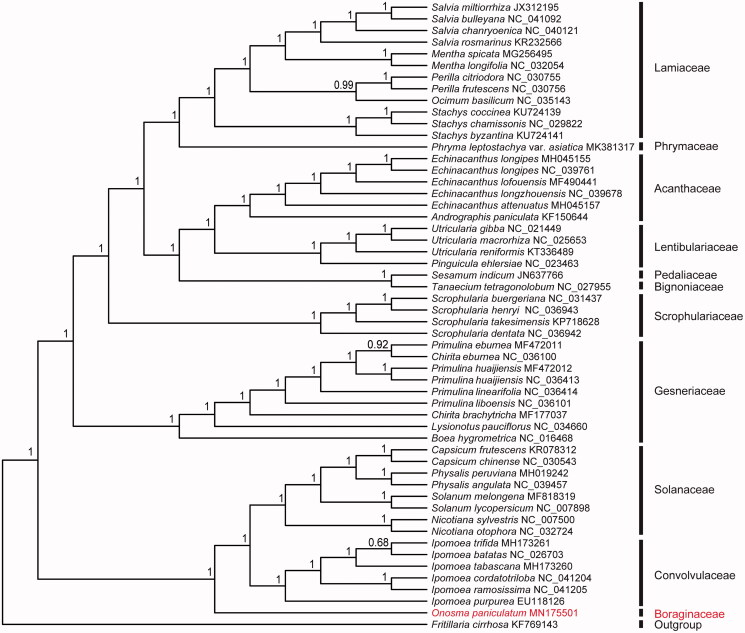
To this day, there is nearly no annotated complete chloroplast genome for species in Onosma and its related genus. To investigate its phylogeny, a total of 52 cp genome sequences of species belonged to the Order Tubiflorae, were downloaded from the NCBI database used for phylogenetic analysis. After using MAFFT V.7.149 for aligning (Katoh and Standley 2013), jModelTest v.2.1.7 (Darriba et al. 2012) was used to determine the best-fitting model for the chloroplast genomes. Then Bayesian inference (BI) was performed by MrBayes v.3.2.6 (Ronquist et al. 2012) with Fritillaria cirrhosa D. Don (No. KF769143) as outgroup. The results showed that Boraginaceae was closely related to Convolvulaceae and Solanaceae, and the three families formed into a monophyletic clade which was divided out in the early evolution of Tubiflorae (Figure 1). Furthermore, complete chloroplast genome of O. paniculatum would be beneficial to phylogeny of Boraginaceae and its related families, as well as developing chloroplast markers for further studies.
Figure 1.
Phylogeny of 53 species of the complete chloroplast genome sequences within the order Tubiflorae based on the Bayesian inference (BI). The GTR + G + I model was employed as the best-fit nucleotide substitution model as suggested using Mr. Bayes 3.2.6 with Fritillaria cirrhosa D. Don (No. KF769143) as an outgroup.
Disclosure statement
The authors are highly grateful to the published genome data in the public database. The authors declare no conflicts of interest and are responsible for the content.
References
- Binzet R, Kandemir I, Kandemir I. 2018. Numerical taxonomic study of the genus onosma L. (boraginaceae) from eastern mediterranean region in turkey. Pak J Bot. 50:561–573. [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinese Pharmacopoeia Commission 2015. Chinese Pharmacopoeia. vol. 1. Beijing (China): China Medical Science and Technology Press. [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9:772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle J. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15. [Google Scholar]
- Ge F, Wang XD, Wang YC. 2003. Advances in studies on medicinal Radix Arnebiae Seu Lithospermi. Chin Tradit Herbal Drugs. 34:app.7–app.9. [Google Scholar]
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. bioRxiv. 1–11. doi: 10.1101/256479 [DOI] [Google Scholar]
- Katoh K, Standley D. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolarčik V, Zozomová-Lihová J, Mártonfi P. 2010. Systematics and evolutionary history of the Asterotricha group of the genus Onosma (Boraginaceae) in central and southern Europe inferred from AFLP and nrDNA ITS data. Plant Syst Evol. 290:21–45. [Google Scholar]
- Kumar N, Kumar R, Kishore K. 2013. Onosma L.: a review of phytochemistry and ethnopharmacology. Pharmacogn Rev. 7:140–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44:W54–W57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasrollahi F, Kazempour-Osaloo S, Saadati N, Mozaffarian V, Zare-Maivan H. 2019. Molecular phylogeny and divergence times of Onosma (Boraginaceae s.s.) based on nrDNA ITS and plastid rpl32-trnL(UAG) and trnH–psbA sequences. Nord J Bot. 37:e02060. [Google Scholar]
- Naz S, Ahmad S, Rasool SA, Sayeed SA, Siddiqi R. 2006. Antibacterial activity directed isolation of compounds from Onosma hispidum. Microbiol Res. 161:43–48. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255. [DOI] [PubMed] [Google Scholar]
- Yang JB, Li DZ, Li HT. 2014. Highly effective sequencing whole chloroplast genomes of angiosperms by nine novel universal primer pairs. Mol Ecol Resour. 14:1024–1031. [DOI] [PubMed] [Google Scholar]
- Zhu GL, Harald R, Rudolf K. 1995. Boraginaceae In: Wu ZY, Raven PH, editors. Flora of China. 16th ed. Beijing (China): Science Press. [Google Scholar]