Abstract
Bougainvillea glabra is an ornamental plant that is domesticated in many tropical and sub-tropical countries. The focus on its breeding programmes has overshadowed genetic studies of this important species. In this study, we reported on the complete chloroplast (cp) genome of B. glabra. The cp genome is 154,763 bp in size, comprised of a large single copy (LSC) region of 85,921 bp, a small single copy (SSC) region of 18,018 bp, and a pair of inverted repeat (IR) regions of 25,412 bp each. A total of 131 genes were predicted, including 37 tRNA, 8 rRNA, and 86 protein-coding genes. Phylogenetic analysis revealed that B. glabra is placed under Nyctaginaceae and sister to B. spectabilis under a strong bootstrap support.
Keywords: Bougainvillea, genetic resources, landscaping, illumina sequencing, plant breeding
Main text
Bougainvillea glabra Choisy is an evergreen, flowering plant native to the South America and is domesticated in many places with warm climates due to its ornamental values (Salam et al. 2017). The plant produces flowers that are attached with thin and papery bracts which come in different attractive bright colours. Its fast-growing characteristic and all-year flowering period make it suitable as a landscaping and gardening plant species in parks, office places and road dividers (Kumar et al. 2014). Due to its wide application in landscaping purposes, various breeding programmes on this species has produced hundreds of varieties and cultivars which display variations in leaf (shape, colour, and arrangement), and flower (colour, size, and arrangement) (Wu and Tang 2010; He et al. 2011). Although several genetic fingerprinting studies have been conducted to characterise the species, there is no report on the complete chloroplast (cp) genome of this commercially important ornamental species. Therefore, in this study, we characterised the complete cp genome sequence of B. glabra using next-generation sequencing technique.
Fresh leaf samples were collected from the B. glabra planted in the Germplasm Resource Nursery of Ornamental Plants, Guangzhou Institute of Forestry and Landscape Architecture, Guangdong province of China (N113°20′25″, E23°13′47′) with voucher specimen (voucher record number: GIFLA-Bogl-2019-07-20) deposited at the voucher collection room of Guangzhou Institute of Forestry and Landscape Architecture, Guangzhou, China. Upon total DNA extraction, a 300-bp insert size genomic library was constructed using TruSeq DNA Sample Prep Kit (Illumina, USA) and sequencing was conducted on an Illumina Novaseq platform. Approximately 8 Gb of raw data of 150-bp paired-end reads were generated and assembled using NOVOPlasty (Dierckxsens et al. 2017), using the rbcL gene sequence of B. glabra (GenBank accession number: MG833637) as the seed sequence. Genome annotation was conducted using DOGMA (Wyman et al. 2004) and manually corrected.
The complete cp genome sequence of B. glabra (GenBank accession number: MN449976) was 154,263 bp in length, comprised of a large single copy (LSC) region of 85,921 bp, a small single copy (SSC) region of 18,018 bp, and a pair of inverted repeat (IR) regions of 25,412 bp each. A total of 131 genes were predicted, consisting of 86 protein-coding genes, 37 tRNA, and 8 rRNA. The overall GC content was 36.4%.
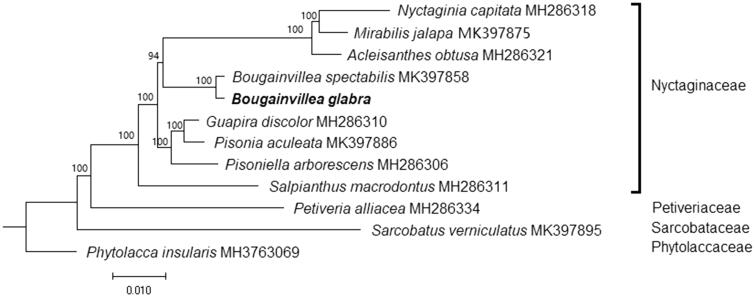
A maximum-likelihood (ML) tree was constructed using RAxML software (Stamatakis 2014) with 1000 bootstrap replicates, based on cp genome sequences of nine species from the family Nyctaginaceae and three species, Petiveria alliacea (Petiveriaceae; MH286334), Sarcobatus vermiculatus (Sarcobataceae; MK397895), and Phytolacca insularis (Phytolaccaceae; MH376309), were included as outgroup. Phylogenetic analysis showed that B. glabra is clustered together with other species from the family Nyctaginaceae, and is sister to B. spectabilis under a strong bootstrap support (100%) (Figure 1). The finding could provide valuable genomic information toward the development of breeding and cultivation of B. glabra.
Figure 1.
Maximum likelihood tree based on the chloroplast genome sequences of 9 species from the family Nyctaginaceae, with Petiveria alliacea (Petiveriaceae), Sarcobatus vermiculatus (Sarcobataceae), and Phytolacca insularis (Phytolaccaceae) as outgroup. Shown next to the nodes are bootstrap support values based on 1000 replicates.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, He Y, Li B, Yang Y. 2011. Review of the studies on Bougainvillea's introduction and cultivation in China. Hubei Agric Sci. 50:1519–1521. [Google Scholar]
- Kumar PP, Janakiram T, Bhatt KV, Jain R, Prasad KV, Prabhu KV. 2014. Molecular characterization and cultivar identification in Bougainvillea spp. using SSR markers. Indian J Agr Sci. 84:1024–1030. [Google Scholar]
- Salam P, Bhargav V, Gupta YC, Nimbolkar PK. 2017. Evolution in Bougainvillea (Bougainvillea Commers). A review. JANS. 9:1489–1494. [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Tang Y. 2010. Research advances in the germplasm resources and their applications of landscape architecture and horticulture of Bougainvillea. South Chin Agric. 4:40–43. [Google Scholar]
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255. [DOI] [PubMed] [Google Scholar]