Abstract
Cynanchum auriculatum is a Chinese herbal medicine species in the family Apocynaceae. The mitochondrial genome of C. auriculatum has heteroplasmy and consists of two chromosomes (chromosomes I and II), the lengths of which are 614,836 and 426,495 nucleotides. The multipartite mitochondrial genome encodes 57 genes, including 37 protein-coding genes, 17 transfer RNA genes, and three ribosomal RNA genes. Including 44 overlapping genes, we identified 57 genes on chromosome I and 44 genes on chromosome II. A phylogenetic tree revealed that C. auriculatum is most closely related to Asclepias syriaca.
Keywords: Apocynaceae family, Cynanchum auriculatum, mitochondrial genome
Introduction
Cynanchum auriculatum is a perennial herb that is used to treat digestive disorders in Asia. Cynanchum auriculatum contains many bioactive components with antioxidant, anticancer, and antidepressant activities (Liu et al. 2018). A sample of C. auriculatum was obtained from the National Institute of Horticultural and Herbal Science (voucher number: NIHHS2014-3, geographic coordinate: N 36°94′38ʺ, E 127°75′33ʺ) in Eumseong, Korea. Whole-genome sequencing was performed using the Illumina Miseq platform (Illumina, CA, USA). A total of 6.6 Gb of raw reads were generated. Low-quality bases (<Q20) were trimmed using the NGS QC Toolkit (Patel and Jain 2012). The screened contigs were subjected to gap-filling and error-correction procedures using the CLC Assembly Cell package (QIAGEN Bioinformatics, CA, USA). In addition, chloroplast-derived sequences of C. auriculatum (Jang et al. 2016) were used as a reference.
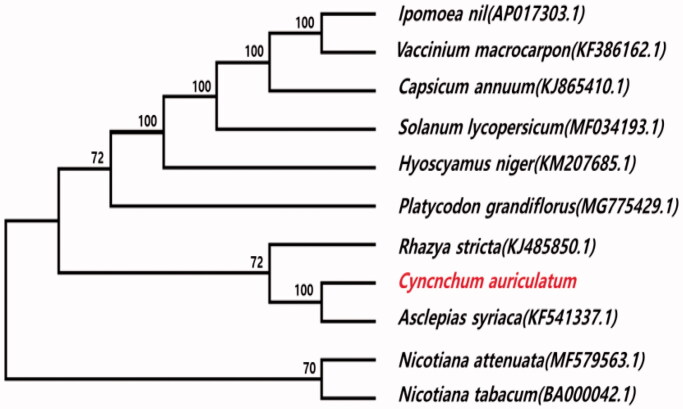
The multipartite mitochondrial genome of C. auriculatum consists of the linear chromosome I (GenBank accession number: MH410146) and the circular chromosome II (MH410147). The linear nature of chromosome I was verified by an alignment of Illumina reads that showed the presence of both proximal sequences and no bridge reads with which to make a circular chromosome. We assumed that C. auriculatum has heteroplasmy (Woloszynska 2010) resulting from homologous recombination between the linear and circular mitochondrial chromosomes (Wei et al. 2012). The C. auriculatum mitochondrial chromosomes were numbered on the basis of their size. Chromosomes I and II contain 614,836 and 426,495 base pairs, respectively. The mitochondrial genome encodes 57 genes, including 37 protein-coding genes, 17 transfer RNA genes, and 3 ribosomal RNA genes. Including 44 overlapping genes, we identified 57 genes on chromosome I and 44 genes on chromosome II. In addition, we identified 17 open reading frames (ORFs) and splicing variants of the nad1, nad2, nad4, nad5, and nad7 genes. The nad5 splicing gene includes five exons in chromosome I, whereas only exons 1 and 2 exist in chromosome II. A phylogeny of C. auriculatum and the genomes of 10 reported Asterid species was constructed using MEGA7.0 (Kumar et al. 2016). The phylogenetic relationships were analyzed using six common protein-coding genes (cox2, cox3, nad2, nad7, nad9, and rps13). The phylogenetic tree indicated that C. auriculatum is closely related to Asclepias syriaca of the Asterids (Figure 1).
Figure 1.
Phylogeny of C. auriculatum and 10 related species based on their mitochondrial genome sequences. The phylogenetic tree was constructed using the neighbor-joining method and a bootstrap test with 1000 iterations based on five common protein-coding genes from the mitochondrial genomes of 11 species.
Geolocation
The genomic DNA sample used for sequencing was obtained from the National Institute of Horticultural and Herbal Science (voucher number: NIHHS2014-3, geographic coordinate: N 36°94′38ʺ, E 127°75′33ʺ).
Disclosure statement
The authors report no conflicts of interest.
References
- Jang WJ, Kim KY, Kim KH, Lee SC, Park HS, Lee JK, Rack Seon S, Shim YH, Sung SH, Yang TJ. 2016. The complete chloroplast genome sequence of Cynanchum auriculatum Royle ex Wight (Apocynaceae). Mitochondrial DNA A. 27:4549–4550. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Wang X, Yong H, Kan J, Zhang N, Jin C. 2018. Preparation, characterization, digestibility and antioxidant activity of quercetin grafted Cynanchum auriculatum starch. Int J Biol Macromol. 114:130–136. [DOI] [PubMed] [Google Scholar]
- Patel RK, Jain M. 2012. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 7:e30619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei DD, Shao R, Yuan ML, Dou W, Barker SC, Wang JJ. 2012. The multipartite mitochondrial genome of Liposcelis bostrychophila: insights into the evolution of mitochondrial genomes in bilateral animals. PLoS One. 7:e33973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woloszynska M. 2010. Heteroplasmy and stoichiometric complexity of plant mitochondrial genomes-though this be madness, yet there's method in't. J Exp Bot. 61:657–671. [DOI] [PubMed] [Google Scholar]