Abstract
The complete mitochondrial genome of the praying mantises Mekongomantis quinquespinosa was characterized in this study. The circular molecule is 15388 bp in length (GenBank accession no. MN267041), containing 13 protein-coding genes (PCGs), 2 ribosomal RNA (rRNA) genes, 22 transfer RNA (tRNA) genes. The nucleotide composition is asymmetric (39.0% A, 15.8% C, 9.8% G, 35.4% T), with an overall A + T content of 74.4%. The gene arrangement of M. quinquespinosa is identical to that observed in other Mantidae family praying mantises. Ten reading frame overlaps and seven intergenic regions are found in the mitogenome of M. quinquespinosa. The phylogenetic relationships based on 13 PCGs show that M. quinquespinosa clusters closest to the species of Mantidae.
Keywords: Mekongomantis quinquespinosa, praying mantises, mitogenome
Mekongomantis, established and described a new genus of Mantidae by Schwarz et al. (2018), belong to the Mantidae subfamily Hierodulinae. The samples of a female adult were collected at Jinghong, Xishuangbanna, Yunnan, China (101°25′E, 22°36′N), in 2017, and identified to species by morphology. Mekongomantis quinquespinosa captures insect pests as food and is recognized as an important natural enemy for biological control. In this study, samples of M. quinquespinosa were stored in the praying mantises specimen room of College of Plant Medicine, Qingdao Agricultural University with an accession number 17-XSBN.
The complete mitogenome of M. quinquespinosa is a circular molecule of 15388 bp in length (GenBank under accession no. MN267041), and contains 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), 2 ribosomal RNA genes (rrnL and rrnS). The nucleotide composition of M. quinquespinosa mitogenome is asymmetric (39.0% A, 15.8% C, 9.8% G, 35.4% T), with an overall A + T content of 74.4%. The AT-skew and GC-skew of this genome were 0.048 and −0.234, respectively. The gene organization of M. quinquespinosa is similar to that observed in family Mantidae praying mantises (Ye et al. 2016; Rivera and Svenson 2016; Zhang et al. 2018). Twenty-five genes were encoded on the major strand (J-strand), whereas the others were encoded on the minor strand (N-strand).
The M. quinquespinosa mitogenome harbours a total of 53 bp intergenic spacer sequences, which is made up of 7 regions in the range from 7 to 15 bp. Gene overlaps were found at 6 gene junctions and involved a total of 212 bp. The A + T content of the M. quinquespinosa mitogenome was 74.4%. The higher A + T content of M. quinquespinosa was present in all regions, both genes and noncoding regions. Gln, Glu, and Trp are also the most frequently used. Moreover, the A + T content was also reflected further in the codon usage: the relative synonymous codon usages showed that M. quinquespinosa used more NNA and NNT codon. The rrnL was 1322 bp in length with A + T content of 78.7%, and rrnS was 788 bp in length with A + T content of 75.1%.
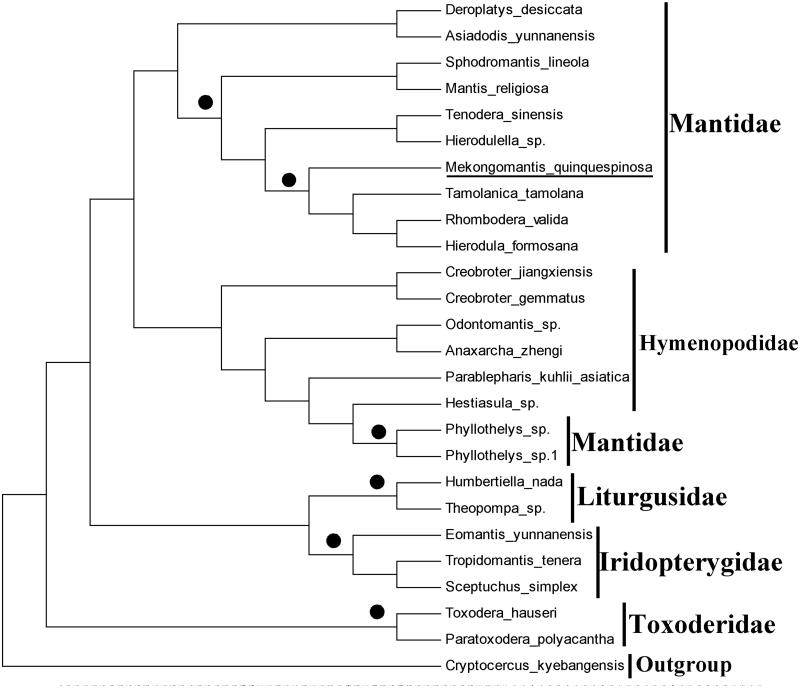
Eight PCGs in M. quinquespinosa mitogenome start with a typical ATN (ATC, ATT, and ATG) codon, nad3 and nad6 begin with TTA, CoI uses CTG, nad1 use GTA and nad4L use CAT as initiation codon. Thirteen PCGs stop with a complete termination codon (TAT, TAA, and TAG). Based on the maximum likelihood analyses, we constructed the phylogenetic relationships of M. quinquespinosa and 24 other praying mantises based on the 13 PCGs amino acids using RAxML. Cryptocercus kyebangensis was used as an outgroup. The results have shown that M. quinquespinosa is closely clustered with other species in family Mantidae (Figure 1), which agree with the morphology.
Figure 1.
Phylogenetic tree showing the relationship between Mekongomantis quinquespinosa (MN267041) and 24 other praying mantises. Cryptocercus kyebangensis (KP872847.1) was used as an outgroup. The black circle indicates popularity >90. GenBank accession numbers used in the study are the following: Rhombodera valida (KX611804.1), Hierodula formosana (KR703238.1), Tamolanica tamolana (DQ241797.1), Sphodromantis lineola (KY689123.1), Tenodera sinensis (KY689132.1), Hierodulella sp. (KY689136.1), Deroplatys desiccate (KY689113.1), Mantis religiosa (KU201317.1), Sceptuchus simplex (KY689133.1), Tropidomantis tenera (KY689127.1), Eomantis yunnanensis (KY689138.1), Mekongomantis quinquespinosa, (MN267041), Theopompa sp. (KU201314.1), Humbertiella nada (KU201315.1), Anaxarcha zhengi (KU201320.1), Odontomantis sp. ( KY689121.1), Creobroter gemmatus (KU201319.1), Creobroter jiangxiensis (KY689134.1), Parablepharis kuhlii asiatica (KY689117.1), Hestiasula sp. (KY689115.1), Phyllothelys sp.1 (KY689119.1), Phyllothelys sp. (KY689129.1), Toxodera hauseri (KX434837.1) and Paratoxodera polyacantha (MG049920.1). Mantis determined in this study was underlined.
Disclosure statement
The authors report no conflicts of interests. The authors alone are responsible for the content and writing of the paper.
References
- Rivera J, Svenson GJ. 2016. The neotropical‘ polymorphic earless praying mantises’ – Part I: molecular phylogeny and revised higher-level systematics (Insecta: mantodea, acanthopoidea). Syst Entomol. 41:607–649. [Google Scholar]
- Schwarz CJ, Ehrmann R, Shcherbakov E. 2018. A new genus and species of praying mantis (Insecta, Mantodea, Mantidae) from Indochina, with a key to Mantidae of South-East Asia[J]. Zootaxa. 4472:581. [DOI] [PubMed] [Google Scholar]
- Ye F, Lan XE, Zhu WB, You P. 2016. Mitochondrial genomes of praying mantises (dictyoptera, mantodea): rearrangement, duplication, and reassignment of trna genes. Sci Rep. 6:25634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang LP, Yu DN, Storey KB. 2018. Higher tRNA gene duplication in mitogenomes of praying mantises (Dictyoptera, Mantodea) and the phylogeny within Mantodea [J]. Int J Biol Macromol. 11:787–795. S014181301733948X. [DOI] [PubMed] [Google Scholar]