Abstract
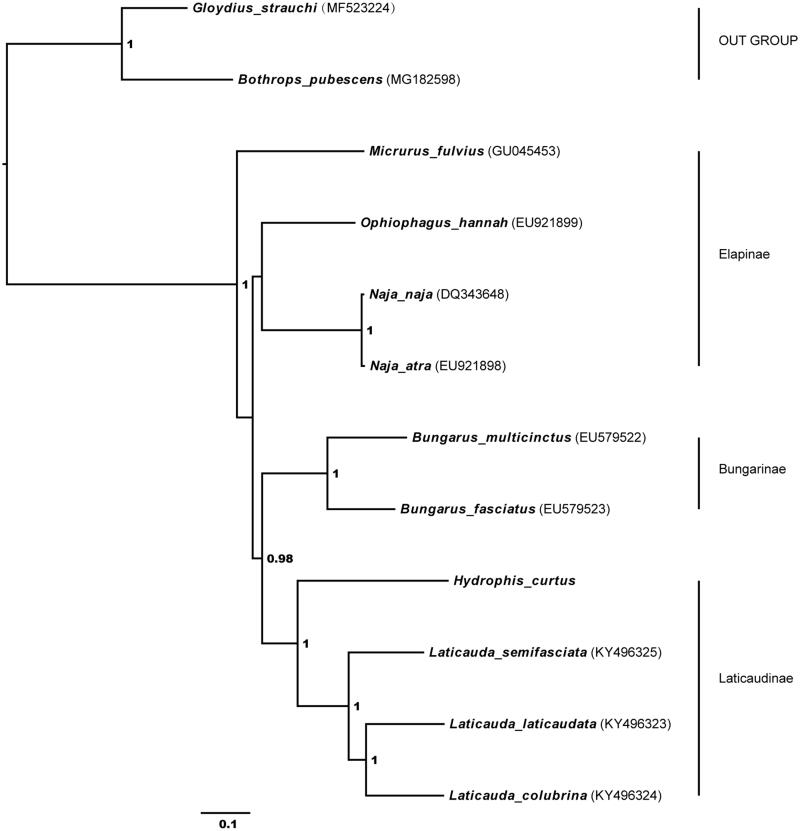
We report the complete mitogenome of Hydrophis curtus, which is 17,702 bp in size and includes 13 protein-coding (PCGs), two rRNA genes, 22 tRNA genes, and two control regions. PCGs, with 13 genes, are 11,261 bp in length. All PCGs use as the start codon ATN except ND1 (CTA) and COX1 (GTG); ATP8, ATP6, ND4L, ND5, and Cytb use the typical stop codon TAA; but COX2 and ND4 with a single T-. Phylogeny recon-structed using the Bayesian inference (BI) method with 13 PCGs indicates that H. curtus at the root of Laticaudinae.
Keywords: Complete mitochondrial genome, phylogeny, Elapidae, Hydrophis curtus
Shaw’s sea-snake (Hydrophis curtus, HYCU), a viviparous species, belongs to the genus Hydrophis, and inhabits shallow coastal waters around western Pacific Ocean and Indian Ocean (Zhao and Adler 1993). The genus Hydrophis belongs to the family Elapidae and including 48 species (Uetz et al. 2019). The taxonomy of the genus Hydrophis has been questioned due to minor morphological variations among species (Zhao and Adler 1993). In order to obtain more basic genetic information for phylogeny on this group of snakes, we determined the complete mitogenome of H. curtus.
The sample (HKHR‒HYCU‒20180303) was donated by the local fishermen from the coastal of Hainan island, China, and stored in a freezer (WUF‒300, DAIHAN® Digital Ultra-Low Temp. Freezer, DAIHAN Scientific Co., Ltd., Korea) at ‒80 °C in the Museum of Hainan Key Laboratory of Herpetological Research (HKHR) at Hainan Tropical Ocean University in Sanya, Hainan, China. We chose the muscle tissue to extract the total genomic DNA using EasyPure@ Genomic DNA Kit according to the manufacturer's instructions (TransGen Biotech Co, Beijing, China). The mitogenomes of H. curtus were sequenced by next-generation sequencing (Illumina HiSeq X ten), and clean data without sequencing adapters were de novo assembled using the NOVO Plasty 2.7.2 software (Dierckxsens et al. 2017). The complete and circular mitochondrial genome is 17,430 bp in size, with an AT bias of 60.0%, and includes 13 PCGs (11,261 bp, ND1‒6, ND4L, CO1‒3, Cytb, ATP6 and ATP8), two rRNA genes (rrnS and rrnL), and 22 tRNA genes and two non-coding regions (CR or D-loop). All PCGs use as the start codon ATN except ND1 (CTA) and COX1 (GTG); ATP8, ATP6, ND4L, ND5, Cytb use the typical stop codon TAA; but COX2 and ND4 with a single T. Twenty-eight gens are encoded on the H-strand and eight genes (ND6 gene and 7 tRNAs) are located on the L-strand. The 22 tRNA genes varied in size from 57 to 73 bp. There are 14 overlapping regions in the mitogenome with 1–38 bp size. The mitogenome of H. curtus has been deposited in Genbank under accession number MK953549.
Using two Viperidae species as outgroups, the phylogeny of 10 Elapidae species including H. curtus was reconstructed based on 13 PCGs by the Bayesian inference (BI) methods through MrBayes (version 3.2.2). The resultant BI tree distinctly indicated that H. curtus belongs to a monophyletic group including Elapinae, Laticaudinae and Bungarinae, and H. curtus is located at the root of the Laticaudinae (Figure 1).
Figure 1.
Bayesian inference (BI) phylogenetic tree inferred from the nucleotide sequence data of mitogenomic 13 PCGs.
Disclosure statement
The authors report no conflict of interest. All the authors approved the final version of the manuscript for publication and agreed to be held accountable for the content of this manuscript.
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P, Freed P, Hošek J. 2019. The Reptile Database. [accessed 2019 May 11]. http://www.reptile-database.org.
- Zhao EM, Adler K. 1993. Herpetology of China. Oxford, Ohio: SSAR. [Google Scholar]