Abstract
Cratoxylum cochinchinense (Lour.) Blume is a tropical Asia specie limited distributed from South China to Borneo. The complete chloroplast genome of the species was found to be 157,103 bp in length, with 129 unique genes, including 37 tRNA, eight rRNA, and 84 protein-coding genes. The GC content of C. cochinchinense is 36.2%. As the first complete chloroplast genome in Hypericaceae, it shows the phylogenetic relationship within Malphigiales.
Keywords: Cratoxylum cochinchinense Blume, complete chloroplast genome, phylogeny
The family Hypericaceae consists of ten genera, more than 500 species (Ruhfel et al. 2011; APG 2016), but no complete chloroplast genome has been published yet. Cratoxylum Blume is a native tropical Asia genus including about six species (Gogelein 1967). Cratoxylum cochinchinense (Lour.) Blume is a kind of deciduous flowering shrub or tree, widespread from South China to Borneo. Its stem can be carved with the hard, delicate texture. Young fruit was usually used for cooking spices, and tender leaves can be used as substitutes of tea (Li et al. 1997). The first plastome sequence of C. cochinchinense was reported in this study.
Fresh leaves of C. cochinchinense were collected from Houmiling Provincial Forest Park, Dongfang city, Hainan Province, China. The voucher specimen was deposited in Museum of Beijing Forestry University, BJFC (specimen No.: Z.X. Zhang zzx2017120806). The total genomic DNA was extracted from the silica gel dried leaf by CTAB method (Doyle and Doyle 1987) with minor modification and then sent to Novogene Company (http://www.novogene.com, China) for next-generation sequencing using Illumina Hiseq 4000 platform. About 5 Gb high quality, 2 × 150 bp PE reads were obtained from high-throughput sequencing. Referring to Garcinia mangostana L., we filtered available reads using the Map tool of Geneious R11 (Kearse et al. 2012), these reads were used for de novo assembling. Gaps were bridged by Fine Tuning function. Plann (Huang and Cronk 2015) was employed to annotate the chloroplast genome sequence, and modification was also performed by Geneious R11.
The total length of the chloroplast genome is 157,103 bp. It consists of a pair of inverted repeat regions (IRs) of 26,272 bp, a large single copy region (LSC) of 85,664 bp, and a small single region (SSC) of 18,895 bp. The overall GC-content of the whole pastime genome is 36.2%, while that in LSC, SSC, and IR regions is 34.0%, 29.9%, and 42.1%, respectively. The sequence contained 129 unique genes, including 37 tRNA, eight rRNA, and 84 protein-coding genes.
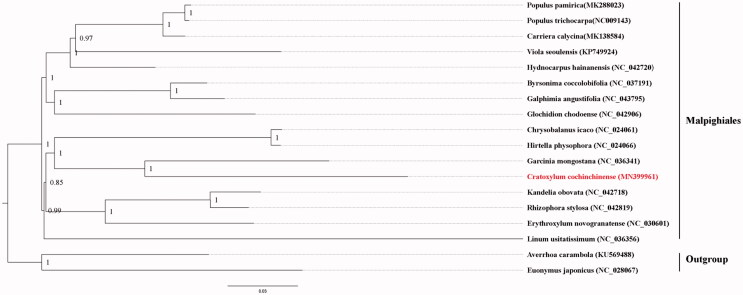
To confirm the phylogenetic relationship of C. cochinchinense in Malphigiales, a Bayesian Inference (BI) tree was constructed using MrBayes 3.2.6 (Ronquist and Huelsenbeck 2003), based on 17 published complete chloroplast genome sequences of Malphigiales and two more sequences that Averrhoa in Oxalidales and Euonymus in Celastrales are used as outgroup. All sequences were aligned using MAFFT v7 (Katoh and Standley 2013). As shown, Cratoxylum in Hypericaceae and Garcinia in Clusiaceae were clustered into a clade perfectly, which is sister to Chrysobalanus and Hirtella in Chrysobalanaceae (Li et al. 2019). It further clarifies the phylogenetic relationship among the generalised Clusiaceae. The plastome sequence of C. cochinchinense will be an important reference for the phylogenetic and evolutionary studies of Hypericaceae and Malphigiales.
Disclosure statement
No potential conflict of interest was reported by the authors.
Figure 1.
Bayesian phylogram of Cratoxylum cochinchinense as well as Malphigiales species inferred from the complete plastome sequences.
Reference
- APG (Angiosperm Phylogeny Group). 2016. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot J Linn Soc. 181:1–20. [Google Scholar]
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf material. Phytochem Bull. 19:11–15. [Google Scholar]
- Gogelein AJF. 1967. A revision of the genus Cratoxylum Bl. (Guttiferae). Blumea. 15:453–475. [Google Scholar]
- Huang DI, Cronk QC. 2015. Plann: a command-line application for annotating plastome sequences. Appl Pl Sci. 3:1500026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H-T, Yi T-S, Gao L-M, Ma P-F, Zhang T, Yang J-B, Gitzendanner MA, Fritsch PW, Cai J, Luo Y, et al. 2019. Origin of angiosperms and the puzzle of the Jurassic gap. Nat Plants. 5:461. [DOI] [PubMed] [Google Scholar]
- Li XW, Li YH, Tong SQ, Tao GD. 1997. Flora of China. Beijing: Science Press. [Google Scholar]
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19:1572–1574. [DOI] [PubMed] [Google Scholar]
- Ruhfel BR, Bittrich V, Bove CP, Gustafsson MHG, Philbrick CT, Rutishauser R, Xi Z, Davis CC. 2011. Phylogeny of the clusioid clade (Malpighiales): evidence from the plastid and mitochondrial genomes. Am J Bot. 98:306–325. [DOI] [PubMed] [Google Scholar]