Abstract
Eremurus robustus Regel (Eremurus, Asphodelaceae) is an ornamental plant and endemic to the Tien Shan and Pamir Alai mountains. In this study, the complete chloroplast genome of E. robustus was determined. The cp genome of E. robustus is 155,647 bp in length consisting of a large single-copy (LSC) region of 84,776 bp, a small single-copy (SSC) region of 17,786 bp, and a pair of identical inverted repeat regions (IRs) of 26,490 bp. The cp genome encodes 114 unique genes, including 80 protein-coding genes, 4 ribosomal RNAs and 30 transfer RNAs. Among them, ten coding genes contained one intron each and two genes (clpP and ycf3) contained two introns each. The phylogenetic analysis demonstrates a close relationship between genus Aloe with genus Eremurus.
Keywords: Chloroplast genome, phylogenetic analysis, Eremurus robustus
The genus Eremurus M.Bieb is naturally distributed in Central and West Asia, extending East to China and West to Turkey and Ukraine (Xinqi et al. 2000). It can be easily distinguished from other genera of Asphodelaceae by having flowers more than 50, leafless flowering stem and rhizomatous rootstock (Fedchenko 1935) Eremurus robustus Regel is an ornamental plant endemic to the Tien Shan and Pamir Alai mountains. The roots of E. robustus are used for glue extraction and food. Here, we characterise the complete cp genome of E. robustus in an effort to provide genomic and genetic sources useful for further research on ornamental important species of the family Asphodelaceae.
Fresh leaves of E. robustus were collected from Uzbekistan, Kashkadarya province, Langar village (E66° 40′ 580″, N39° 24′ 990″). Voucher specimen of E. robustus was deposited in the National Herbarium of Uzbekistan (TASH, DM0012), Institute of Botany, Uzbekistan Academy of Sciences. Total DNA was extracted according to the modified CTAB method (Doyle and Doyle, 1987), then fragmented into 200 bp for library construction and sequenced using an Illumina HiSeq 2500 system at BGI (Shenzhen, Guangdong, China). We assembled the cp genome based on the methods of Jin et al. (2018), and the plastome of Aloe maculata was used as a reference genome (Genbank accession: NC_035505). We annotated the cp genome of E. robustus using Geneious v11.1.5 (Kearse et al. 2012), then start and stop codons and intron/exon boundaries were manual edited. Phylogenetic analysis was performed using RAxML-HPC BlackBox v8.1.24 software (Stamatakis 2006) with the GTRGAMMAI model. The cp genome sequence of E. robustus was submitted to GenBank with the accession number of MN315570.
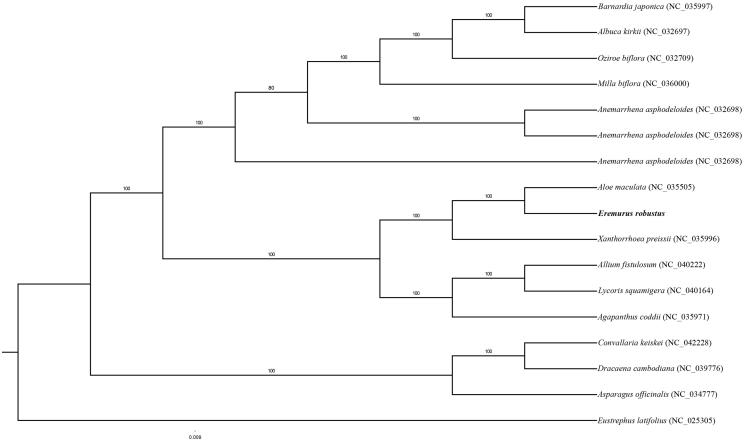
The cp genome of E. robustus was 155,647 bp in length, containing a large single copy region (LSC) of 84,881 bp (GC-35.1%), a small single copy region (SSC) of 17,786 bp (GC-31.1%), and a pair of inverted repeats (IR) regions of 26,490 bp (GC-42.8%). A total of 114 genes were identified including 80 protein-coding genes, 30 tRNA genes, four ribosomal RNA genes. Among these, twelve coding genes contained introns. The phylogenetic analysis of 16 chloroplast genomes showed that E. robustus is closely clustered with A. maculata (Figure 1). The cp genome sequence of E. robustus will be useful for further population genomic studies, phylogenetic analyses, and genetic engineering studies of family Asphodelaceae.
Figure 1.
Phylogenetic analysis of E. robustus with 16 related species. Numbers in the nodes are the bootstrap values from 1000.
Acknowledgement
The authors are grateful to the opened raw genome data from public database.
Disclosure statement
The authors declare no conflicts of interest and are responsible for the content.
References
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15. [Google Scholar]
- Fedchenko BA. 1935. Eremurus In: Komarov VL, editor. Flora of the USSR. Vol. 4 Leningrad: USSR Acad. Sci; p. 37–52. [Google Scholar]
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. bioRxiv. 256479. [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analysis with thousands of taxa and mixed models. Bioinformatics. 22(21):2688–2690. [DOI] [PubMed] [Google Scholar]
- Xinqi C, Songyun L, Jiemei X, Tamura MN. 2000. Liliaceae. Flora of China In: Wu ZY, Raven PH, editors. Flora of China. Vol 24 Beijing and St. Louis (MO): Science Press and Missouri Botanical Garden Press; p. 73–263. [Google Scholar]