Abstract
Aristolochia manshuriensis is a medicinal plant belonging to the family Aristolichiaceae. In this study, complete chloroplast (cp) genome sequence of A. manshuriensis was characterized through de novo assembly with next-generation sequencing data. The cp genome is 160,182 bp long and has a typical quadripartite organization consisting of a large single-copy (LSC), a small single-copy (SSC), and a pair of inverted repeats (IRs). The cp genome harboured 79 protein-coding genes, 30 tRNA genes, and 4 rRNA genes. Phylogenetic analysis revealed that A. manshuriensis has close relationship with Aristolochia macrophylla.
Keywords: Aristolochia manshuriensis, medicinal plant, chloroplast genome, Aristolochiaceae
The genus Aristolochia L. sensu lato (Aristolochiaceae), which consists of ∼400 species, is distributed from temperate to tropical regions around the world (Ohi-Toma et al. 2006). In Korea, only two species are distributed; Aristolochia contorta and Aristolochia manshuriensis (Oh 2007). For thousands of years, Aristolochia species have been used as medicinal plants in East Asia including China, Korea, and Japan (Chinese Company of Medicinal Materials 1995; Hu et al. 2004). In particular, aristolochic acid (AA) from root, rootstock or stem of Aristolochia species is used to treat gout, rheumatoid arthritis, wound festering, or to reduce inflammation (Negi et al. 2003; Heinrich et al. 2009; Nie et al. 2015). However, AA is also reported as mutagenic, nephrotoxic, and carcinogenic to animals and humans (Arlt et al. 2002, 2007; International Agency for Research on Cancer 2002; Cheng et al. 2006; Huang et al. 2007; Nie et al. 2015). Recently, it has been banned to utilize AA for medicinal purposes in many countries (International Agency for Research on Cancer 2002; Cheng et al. 2006; Arlt et al. 2007; Martena et al. 2007; Lai et al. 2010).
In this study, we determined the chloroplast (cp) genome of A. manshuriensis to contribute to the classification and development of DNA markers for the authentication of Aristolochia species. The specimen was collected from Samil-ri, Sanae-myeon, Hwacheon-gun, Gangwon-do, South Korea (38°0′23.5″N, 127°31′22.2″E) and deposited at NIBR herbarium (KB) with the accession number NIBR-VP0000575956. Total genomic DNA was prepared and sequenced by the Illumina MiSeq platform (Illumina Inc., San Diego, CA) and obtained high-quality paired-end reads of ca. 2.5 Gb. The complete cp genome of A. manshuriensis was revealed to GenBank (Accession no. MN132862), as described previously (Kim et al. 2015).
The cp genome was 160,182 bp in length with 38.7% overall GC content. The cp genome structure of A. manshuriensis has the typical quadripartite organization featuring two copies (IRa and IRb) of inverted repeat (IR) regions (25,691 bp) that are separated by a large single-copy (LSC) region (89,503 bp), and a small single-copy (SSC) region (19,297 bp). The total number of identified encoded genes is 113 with 79 protein-coding genes, 30 tRNA genes, and 4 rRNA genes.
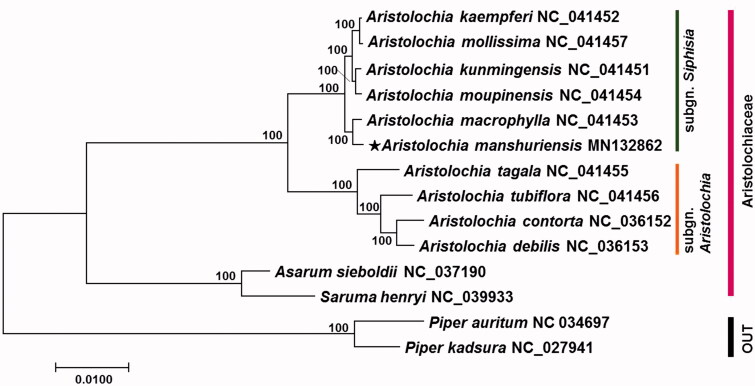
To understand the phylogenetic relationship of A. manshuriensis with relative taxa, a maximum-likelihood (ML) tree was constructed using 75 common protein-coding genes of A. manshuriensis and 11 taxa in Aristolochiaceae. Outgroup includes two species of Piper (Piper autrittum and Piper kadsura) in Piperaceae, which is the most likely sister group of Aristolochiaceae (Figure 1). The Aristolochia clade comprised well-supported monophyletic group (pp = 100), and the two major subclades were circumscribed; these include (1) subgn. Aristolochia, and (2) subgn. Siphisia (Murata et al. 2001) (Figure 1). Aristolochia manshuriensis was contained in subgn. Siphisia clade with Aristolochia kaempferi, Aristolochia kunmingensis, Aristolochia macrophylla, Aristolochia mollissima, and Aristolochia moupinensis. Among those species, A. manshuriensis has more close relationship with A. macrophylla (Figure 1).
Figure 1.
Maximum-likelihood (ML) tree based on the 75 chloroplast protein-coding genes of 14 taxa including A. manshuriensis. Sequences of 75 chloroplast protein-coding gene from 14 taxa were aligned using MAFFT (http://mafft.cbrc.jp/alignment/server/index.html) and used to generate ML phylogenetic tree by MEGA 7.0 (Kumar et al. 2016).
Disclosure statement
None of the authors report any conflict of interest. The authors alone are responsible for the content and writing of the paper.
References
- Arlt VM, Stiborová M, Schmeiser HH. 2002. Aristolochic acid as a probable human cancer hazard in herbal remedies: a review. Mutagenesis. 17(4):265–277. [DOI] [PubMed] [Google Scholar]
- Arlt VM, Stiborová M, Vom Brocke J, Simões ML, Lord GM, Nortier JL, Hollstein M, Phillips DH, Schmeiser HH. 2007. Aristolochic acid mutagenesis: molecular clues to the aetiology of Balkan endemic nephropathy-associated urothelial cancer. Carcinogenesis. 28(11):2253–2261. [DOI] [PubMed] [Google Scholar]
- Cheng CL, Chen KJ, Shih PH, Lu LY, Hung CF, Lin WC, Yesong GJ. 2006. Chronic renal failure rats are highly sensitive to aristolochic acids, which are nephrotoxic and carcinogenic agents. Cancer Lett. 232(2):236–242. [DOI] [PubMed] [Google Scholar]
- Chinese Company of Medicinal Materials. 1995. China medicinal herbs most in use. Beijing (China): Science Publishing House; p. 838–839. [Google Scholar]
- Heinrich M, Chan J, Wanke S, Neinhuis C, Simmonds M. 2009. Local uses of Aristolochia species and content of nephrotoxic aristolochic acid 1 and 2–A global assessment based on bibliographic sources. J Ethnopharmacol. 125(1):108–144. [DOI] [PubMed] [Google Scholar]
- Hu SL, Zhang HQ, Chan K, Mei QX. 2004. Studies on the toxicity of Aristolochia manshuriensis (Guanmuton). Toxicology. 198(1–3):195–201. [DOI] [PubMed] [Google Scholar]
- Huang CC, Chen PC, Huang CW, Yu J. 2007. Aristolochic acid induces heart failure in zebrafish embryos that is mediated by inflammation. Toxicol Sci. 100(2):486–494. [DOI] [PubMed] [Google Scholar]
- International Agency for Research on Cancer 2002. Some traditional herbal medicines, some mycotoxins, naphthalene and styrene. IARC Monogr Eval Carcinog Risks Hum. 82:1–556. [PMC free article] [PubMed] [Google Scholar]
- Kim K, Lee SC, Lee J, Yu Y, Yang K, Choi BS, Koh HJ, Waminal NE, Choi HI, Kim NH, et al. 2015. Complete chloroplast and ribosomal sequences for 30 accessions elucidate evolution of Oryza AA genome species. Sci Rep. 5(1):15655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai MN, Wang SM, Chen PC, Chen YY, Wang JD. 2010. Population-based case-control study of Chinese herbal products containing aristolochic acid and urinary tract cancer risk. J Natl Cancer Inst. 102(3):179–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martena MJ, van der Wielen JCA, van de Laak LFJ, Konings EJM, de Groot HN, Rietjens I. 2007. Enforcement of the ban on aristolochic acids in Chinese traditional herbal preparations on the Dutch market. Anal Bioanal Chem. 389(1):263–275. [DOI] [PubMed] [Google Scholar]
- Murata J, Ohi T, Wu S, Darnaedi D, Sugawara T, Nakanishi T, Murata H. 2001. Molecular phylogeny of Aristolochia (Aristolochiaceae) inferred from matK sequences. Acta Phytotax Geobot. 52(1):75–83. [Google Scholar]
- Negi PS, Anandharamakrishnan C, Jayaprakasha GK. 2003. Antibacterial activity of Aristolochia bracteata root extracts. J Med Food. 6(4):401–403. [DOI] [PubMed] [Google Scholar]
- Nie W, Lv Y, Yan L, Chen X, Lv H. 2015. Prediction and characterization of the system effects of aristolochic acid: a novel joint network analysis towards therapeutic and toxicological mechanisms. PLoS One. 5:17646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh BU. 2007. Aristolochia In: Park CW, editor. The genera of vascular plants of Korea. Seoul (Korea): Academy Publishing Company; p. 153–154. [Google Scholar]
- Ohi-Toma T, Sugawara T, Murata H, Wanke S, Neinhuis C, Murata J. 2006. Molecular phylogeny of Aristolochia sensu lato (Aristolochiaceae) based on sequences of rbcL, matK, and phyA genes, with special reference to differentiation of chromosome number. Synth Bot. 31(3):481–492. [Google Scholar]