Abstract
Calycanthaceae is a perennial shrub endemic to China with important ornamental and medicinal values. In this study, Chimonanthus praecox cv. concolor chloroplast (cp) genome was characterized using Illumina paired-end reads data. In total, whole cp genome is 153,254 bp long and contains a small single-copy region of 19,769 bp, a pair of repeat (IRa and IRb) regions of 23,286 bp each, and a large single-copy region of 86,913 bp. This genome contains 129 genes, including 84 protein-genes, 8 rRNA genes, and 37 tRNA genes. Phylogenetic analysis based on 19 cp genomes showed that C. praecox cv. concolor is closely related to Chimonanthus praecox and Chimonanthus nitens.
Keywords: Chimonanthus praecox cv. concolor, chloroplast genome, phylogeny analysis
Calycanthaceae, widely cultivated in many provinces in China, is a Chinese endemic winter-flowering plant which has a long history of cultivation. The chloroplast (cp) DNA has been widely and successfully employed by many plant biologists for the reconstruction of plant evolution (Kellogg 1998, Goremykin et al. 2003). Good knowledge of complete cp genomics is helpful to the establishments of effective management and conservation strategies for species. Studies about Calycanthaceae such as classification, ecological character (Liu et al. 1996) and genetic diversity (Zhou and Ye 2002) have been done. However, genomic information of C. praecox cv. concolor is less well known. Here, in this study, we report the complete cp genome of C. praecox cv. concolor so that the data can provide evidence for Calycanthaceae genomics and phylogenetic research.
Total DNA (Voucher specimen: E118°49′1.19″, N32°4′47.24″, CF201903079, NFU) was stored in the forest breeding laboratory of Nanjing Forestry University. It was extracted from leaf tissue from C. praecox cv. concolor. After filtered and trimmed by fastp (Chen et al. 2018), high-quality paired-end reads were assembled into a complete cp genome using GetOrganelle (Jin et al. 2018) using Calycanthus floridus var. glaucus (NC_004993.1) as reference. The genome annotation was performed with GeSeq (Tillich et al. 2017) and by aligning with the cp genome of relatively related species within the Calycanthaceae, including C. floridus var. glaucus and Calycanthus chinensis (NC_037504.1). The annotation result was inspected using Geneious and adjusted manually as needed. Finally, a physical map of the genome was drawn by using the IQ-Tree program (Nguyen et al., 2015). The annotated cp genome has been submitted to GenBank under the accession number MN172352.
The cp genome was 153,254 bp in size with a typical quadripartite structure, consisting of a large single-copy (LSC, 86,913 bp), a small single-copy (SSC, 19,769 bp), and two inverted repeats (IRs, 23,286 bp each). In total, this cp genome contained 129 genes, including 84 protein-coding genes, 8 rRNA genes, and 37 tRNA genes. Most of the genes occurred as single copy in the LSC or SSC, but 15 genes presented as two copies in the IRs, including all 4 rRNA genes (rrn 4.5, rrn 5, rrn 16, and rrn 23), 7 tRNA genes (trnI-CAU, trnL-CAA, trnV-GAC, trnI-GAU, trnA-UGC, trnR-ACG, and trnN-GUU), and 4 protein-coding genes (rpl23, ycf2, ndhB, and rps7). Among all the genes, 13 genes (rps16, atpF, rpoC1, nahB, ndhA, rpl2, rpl16, petB, petD, trnL-UAA, trnV-UAC, trnI-GAU, and trnA-UGC) contained a single intron, but 3 genes (ycf3, rps12, clpP) contained two introns.
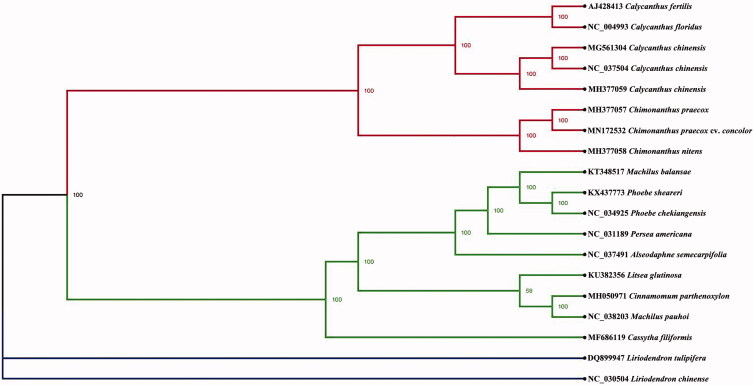
Based on 19 cp genome sequences, the phylogenetic position of C. praecox cv. concolor was inferred using the maximum-likelihood (ML) method. The alignment was achieved by MAFFT (Katoh and Standley 2013), and the phylogenetic tree was constructed using IQ-tree. The best-fitted model was selected by ModelFinder (Kalyaanamoorthy et al. 2017). The result (Figure 1) indicated that C. praecox cv. concolor was closely related to Chimonanthus praecox and Chimonanthus nitens. We believe that the complete cp genome of C. praecox cv. concolor could be subsequently utilized for classification purpose.
Figure 1.
Phylogenetic position of Chimonanthu praecox cv. concolor within Laurales. Phylogenetic tree was constructed using ML method. Numbers in the nodes were the support values.
Disclosure statement
The authors declare no conflict of interest.
Reference
- Chen S, Zhou Y, Chen Y, Gu J. 2018. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 34:i884–i890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goremykin V, Hirsch-Ernst KI, Wölfl S, Hellwig FH. 2003. The chloroplast genome of the “basal” angiosperm Calycanthus fertilis - structural and phylogenetic analyses. Plant Syst Evol. 242:119–135. [Google Scholar]
- Jin J, Yu W, Yang J, Song Y, Yi T, Li D. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. bioRxiv. doi: 10.1101/256479. [DOI] [Google Scholar]
- Kalyaanamoorthy S, Minh B, Wong TKF, Von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14:587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg EA. 1998. Who’s related to whom? Recent results from molecular systematic studies. Curr Opin Plant Biol. 1:149–158. [DOI] [PubMed] [Google Scholar]
- Liu HE, Zhang RH, Huang SF, Zhao ZF. 1996. Study on chromosomes of 8 species of Calycathaceae. J Zhejiang for Coll. 13:28–33. [Google Scholar]
- Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45:W6–W11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou SL, Ye WG. 2002. The genetic diversity and conservation of Sinocalycanthus chinensis. Chin Biodivers. 10:1–6. [Google Scholar]