Abstract
Amygdalus ferganensis is a member of the family Rosaceae. In this paper, we report complete chloroplast genome sequences of A. ferganensis (Rosaceae). The results showed that A. ferganensis complete chloroplast genome comprises 158,365 bp, containing a largen single copy (LSC) region of 86,240 bp, a small single copy (SSC) region of 19,012 bp, and a pair of inverted repeats (IRs) region of 26,386 bp. The genome has a GC content of 42.6%. The LSC, SSC, and IR regions represent 54.46, 12.01, and 33.32% of the A. ferganensis chloroplast genome length. We annotated 112 genes, including 78 protein coding genes, 4 rRNAs, and 30 tRNAs. And A. ferganensis is closely the A. persica.
Keywords: Amygdalus ferganensis, complete chloroplast genome, phylogenetic tree
Amygdalus ferganensis is widely distributed throughout Xinjiang, North, and South. In the southern Xinjiang, natural selection and artificial domestication in long-term; it has become one of the most characteristic fruit tree types in the region (Kelimu et al. 2017). As A. ferganensis is an important germplasm resource, many scholars have studied about morphology, cytology, palynology and enzymology, but we are not clear about the positioning of the classification of A. ferganensis still (Chen et al. 2015).
Chloroplast (cp) genome is one of the three sets of genetic systems (cytoblast, chloroplast, and mitochondrion) with different evolutionary histories and origins in higher plants, the chloroplast genomes have independent evolutionary routes and own the characteristics of uniparental inheritance, moderate rates of nucleotide substitutions, haploid status, and no homologous recombination (Hansen et al. 2007; Shaw et al. 2005). However, there is less research on the chloroplast genome characteristics of A. ferganensis; and, in this paper, we report complete chloroplast genome sequences of A. ferganensis (Rosaceae). We hope to provide a theoretical basis for the chloroplast genome characteristics and the phylogenetic relationship of this species.
The fresh leaves of A. ferganensis were provided from Zhengzhou fruit research institute, CAAS (Chinese Academy of Agricultural Sciences) in Henan, China (113°38′E, 34°45′N) in June 2018. The specimens of A. ferganensis (Accession Number: 20180910Yl03) were deposited at the Herbarium of Yulin University, Shaanxi, China. Genomic DNA was extracted from the fresh leaves according to a modified CTAB method (Doyle and Doyle 1987), and the high-throughput sequencing was carried out using the Illumia HiSeq X Ten system. The complete chloroplast genome of A. pedunculata (MG869261) was used as the reference sequence to assembled and annotated. The complete chloroplast genome of A. ferganensis was annotated using the Geneious 8.0.2 (Kearse et al. 2012). The physical map of A. ferganensis was visualised using OGDRAW online tool (Lohse et al. 2013). The complete chloroplast genome sequences were aligned using MAFFT (Kazutaka et al. 2002), and we used the MEGA v7.0 (Kumar et al. 2016) to construct a phylogenetic tree according to the neighbor-joining method, with a bootstrap value of 1000. The complete chloroplast genome sequence of A. ferganensis has been deposited into the GenBank (MK798146).
Our result showed that complete chloroplast genome of A. ferganensis comprises 158,365 bp, containing a large single copy (LSC) region of 86,240 bp, a small single copy (SSC) region of 19,012 bp, and a pair of inverted repeats (IRs) region of 26,386 bp. The genome has a GC content of 42.6%. The LSC, SSC, and IR regions represent 54.46, 12.01, and 33.32% of the A. ferganensis chloroplast genome length. We annotated 112 genes, including 78 protein-coding genes, 4 rRNAs, and 30 tRNAs.
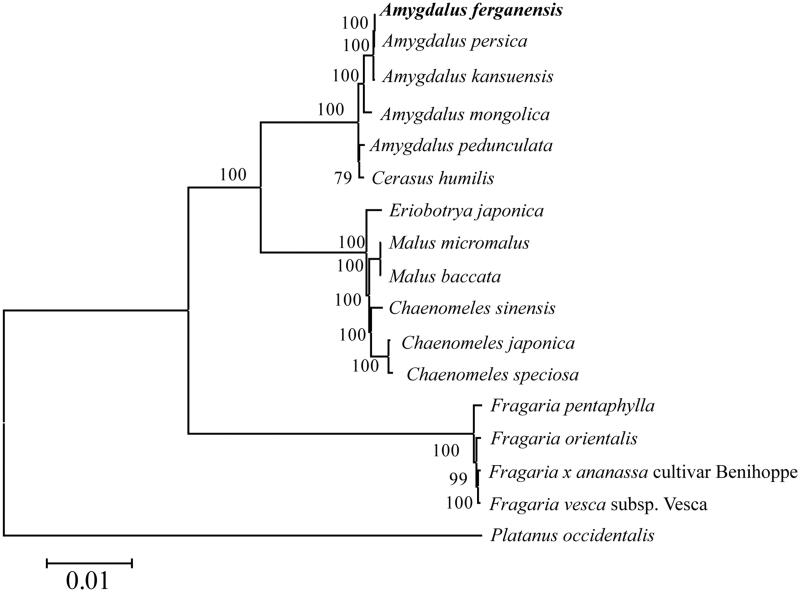
The 17 species of complete chloroplast genomes was constructed using a phylogenetic tree, and the Platanus occidentalis (DQ923116) complete chloroplast genome as an outgroup (Figure 1). The result showed that A. ferganensis is closely the A. persica.
Figure 1.
Phylogenetic tree constructed based on 17 species of chloroplast genomes. Amygdalus mongolica (KY073235); Amygdalus pedunculata (MG869261); Amygdalus persica (HQ336405); Amygdalus kansuensis (NC023956); Amygdalus ferganensis (MK798146); Cerasus humilis (NC035880); Malus baccata (KX499859); Malus × micromalus (NC036368); Chaenomeles japonica (NC035566); Chaenomeles sinensis (KT932967); Chaenomeles speciosa (KT932965); Eriobotrya japonica (NC034639); Fragaria x ananassa cultivar Benihoppe (KY358226); Fragaria vesca subsp. Vesca (JF345175); Fragaria orientalis (NC035501); Fragaria pentaphylla (NC034347); Platanus occidentalis (DQ923116).
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Chen QJ, Meng D, Li W, Gu ZY, Duan XW, Yuan H, Zhang Y, Li TZ. 2015. Cloning and analysis of S-RNase and SFBgenes in Xinjiang peach (Prunus ferganensis Kost.et Riab). Jou of Chi Agr Uni. 20(6):76–83. (In Chinese.) [Google Scholar]
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phyt Bul. 19:11–15. [Google Scholar]
- Hansen DR, Dastidar SG, Cai Z, Penaflor C, Kuehl JV, Boore JL, Jansen RK. 2007. Phylogenetic and evolutionary implications of complete chloroplast genome sequences of four early-diverging angiosperms: Buxus (Buxaceae), Chloranthus (Chloranthaceae), Dioscorea (Dioscoreaceae), and Illicium (Schisandraceae). Mol Phy and Evo. 45(2):547–563. [DOI] [PubMed] [Google Scholar]
- Kazutaka K, Kazuharu M, Kei-Ichi K, Takashi M. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelimu YM, Han LQ, Maerhaba WSM, Aisikaer AHMT, Ghunqam ABDRXT, Abulimiti MS, Wang XW, Ma K, Wang JX. 2017. Investigation of the fruit characters and preliminary evaluation of different Xinjiang peach germplasms. Xinjiang Agr Sci. 54(6):1041–1046. (In Chinese.) [Google Scholar]
- Kumar S, Stecher G, Tamura K. 2016. Mega7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. Organellar Genome DRAW – a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41(W1):W575–W581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw J, Lickey EB, Beck JT, Farmer SB, Liu W, Miller J, Siripun KC, Winder CT, Schilling EE, Small RL. 2005. The tortoise and the hare II: relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. Am J Bot. 92(1):142–166. [DOI] [PubMed] [Google Scholar]