Abstract
Autophagy is an evolutionarily conserved mechanism that maintains nutrient homeostasis by degrading protein aggregates and damaged organelles. Autophagy is reduced in aging, which is implicated in the pathogenesis of aging-related diseases, including cancers, obesity, type 2 diabetes, cardiovascular diseases, and neurodegenerative diseases. Mitochondria-derived phospholipids cardiolipin, phosphatidylethanolamine, and phosphatidylglycerol are critical throughout the autophagic process, from initiation and phagophore formation to elongation and fusion with endolysosomal vesicles. Cardiolipin is also required for mitochondrial fusion and fission, an important step in isolating dysfunctional mitochondria for mitophagy. Furthermore, genetic screen in yeast has identified a surprising role for cardiolipin in regulating lysosomal function. Phosphatidylethanolamine plays a pivotal role in supporting the autophagic process, including autophagosome elongation as part of lipidated Atg8/LC3. An emerging role for phosphatidylglycerol in AMPK and mTORC1 signaling as well as mitochondrial fission may provide the first glimpse into the function of phosphatidylglycerol apart from being a precursor for cardiolipin. This review examines the effects of manipulating phospholipids on autophagy and mitophagy in health and diseases, as well as current limitations in the field.
Keywords: autophagy, cardiolipin, mitophagy, phosphatidylethanolamine, phosphatidylglycerol
1. Introduction
Autophagy, or “self-eating”, is an evolutionarily conserved mechanism that degrades bulk cytosolic contents and organelles through the lysosomal pathway in response to stressors and damage, recycling amino acids, lipids, and carbohydrates during periods of nutrient starvation. There are three recognized forms of autophagy: micro-autophagy, where cargo is engulfed via lysosomal or vacuolar invaginations; chaperone-mediated autophagy, in which proteins are shuttled through the lysosomal membrane; and macro-autophagy, characterized by the formation of double-membrane autophagosomes that deliver cargo to lysosomes. Cells have a basal level of autophagy to maintain protein homeostasis that degrade excess proteins and toxic aggregates that may form through physiological processes. Cellular organelles also utilize macro-autophagic machinery to degrade damaged or exhausted components, such as the endoplasmic reticulum (ER) (reticulophagy), ribosomes (ribophagy)[1], peroxisomes (pexophagy)[2], intracellular pathogens (xenophagy)[3], lipid droplets (lipophagy)[4], and mitochondria (mitophagy)[5, 6]. Macro-autophagy (hereafter referred to as “autophagy” ) is the most extensively studied of the three autophagic processes. The hallmark of autophagy is the formation of an autophagosome, a double-layered structure that transports its contents to the endo-lysosomal system for degradation. Basal levels of autophagy are required for normal physiology, but its reduction is implicated in aging-related diseases [7–9].
The origins of the autophagosome, the lipid sources for the developing autophagosome, and mechanisms for recognizing of specific cargo and organelles for degradation have been under extensive scrutinization in recent years. Steady progress has been made uncovering the signaling cascades and protein scaffolds involved in autophagosome formation[10]. The specific roles of mitochondrial lipids in membrane biogenesis and maturation of autophagosomes have been inferred indirectly via enzymes of lipid metabolism with few exceptions. The difficulty in visualizing lipid traffic and protein interactions in vivo and in vitro stymies attempts at more profound mechanistic understanding of the autophagosome[11]. The current understanding of the roles of mitochondrial lipids, specifically cardiolipin (CL), phosphatidylethanolamine (PE), and phosphatidylglycerol (PG), in autophagy and mitophagy will be the subject of this review.
2. Phospholipids and autophagosome biogenesis
The life cycle of the double-membrane autophagosome requires the organized accretion of lipid vesicles from multiple sources at the nucleation site, elongation, engulfment of cytoplasmic cargo or organelles, and resolution through fusion with the endolysosomal system. The process is dependent on the coordination of autophagy-related genes (Atg) (thoroughly reviewed elsewhere[12, 13]) and phospholipids to form autophagosomes de novo. Of particular interest to a discussion on phospholipids involved in regulating autophagy are the Beclin1/vacuolar protein sorting 34 (VPS34) complex, the Atg5 complex, and Atg8/microtubule-associated protein 1A/1B-light chain 3 (LC3). The Beclin1/VPS34 is a PI3K class III (PI3KC3) complex that recruits other autophagy-related protein complexes at the beginning of autophagosome biogenesis. The Atg5 complex-mediates Atg8/LC3 lipidation with PE, which is required for elongation and cargo sequestration. The phospholipids most directly involved in this process are phosphatidylinositol (PI) and PE as depicted in Figure 1.
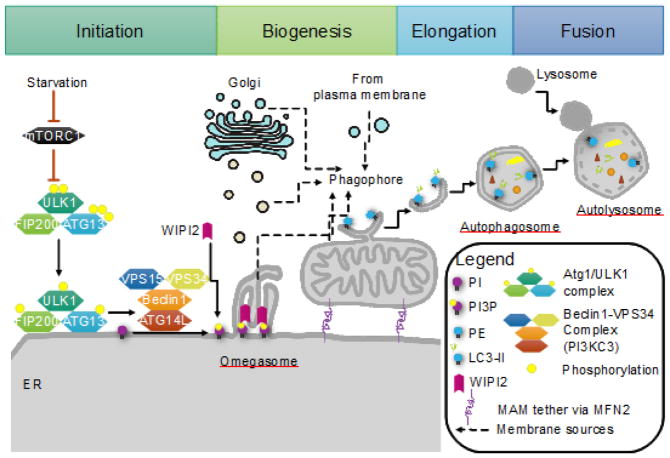
FIGURE 1. The initiation of autophagy.
Initiation of autophagy requires the coordination of the Atg1/ULK1 complex with Beclin1/VPS34 complex (PI3KC3) to form a platform for the nucleation of the autophagosome. Upon starvation, mTORC1 inhibition on the Atg1/ULK1 complex is released. This complex is translocated onto the ER. Atg1/ULK1 then recruits the PI3KC3 complex and phosphorylates Beclin1, activating the local synthesis of PI(3)P. This pool of PI(3)P marks the site for autophagosome formation, allowing WD repeat domain, phospoinositide interacting 2 (WIPI2) to bind and recruit other protein complexes to the nascent phagophore. The phagophore forms within the omegasome at the ER, and is enlarged through membrane contributions from direct ER and mitochondria contact as well as vesicles from the ER, Golgi, and the plasma membrane. The mitochondria-associated membrane (MAM) or ER membrane adjacent to mitochondria is purported to be the site of autophagosome formation, as disruption of the MAM by MFN2 knockout impairs autophagy. The autophagosome elongates and engulfs bulk cytoplasm as well as targeted cargo for trafficking to and fusion with lysosomes for degradation.
PI plays an important role in regulating autophagy through its multiple phosphorylated forms. A comprehensive review on PI in regulating autophagy can be found elsewhere [14, 15], though we would be remiss to not discuss the role of phosphatidylinositol-3-phosphate (PI(3)P) in autophagosome biogenesis. In mammals, canonical autophagy activates the PI3KC3 complex on the ER surface[16], the process of which is illustrated in Figure 1. The ER surface is typically devoid of PI(3)P so the local production of PI(3)P provides a unique signal for autophagy. The PI(3)P-rich structure forms an extrusion on the ER membrane called the omegasome [17], which is believed to be the nucleation site from which the isolation membrane/phagophore arises. PI(3)P allows proteins to bind and stably associate with the ER membrane. WD repeat domain phosphoinositide-interacting protein 2 (WIPI2) is one such protein that localizes onto the PI(3)P-enriched platform in response to autophagic stimulation [18, 19]. On the ER membrane, WIPI2 is critical for the recruitment of the Atg5 complex onto the phagophore, thus linking PI(3)P generation with PE lipidation of Atg8/LC3 and elongation of the autophagosome[20].
Autophagosome maturation proceeds with the covalent lipidation of LC3. LC3 is a member of the Atg8 family in mammals, and primarily consists of two forms: LC3-I, which is cytosolic, and LC3-II is a mature and membrane-bound form[21]. The importance of Atg8/LC3 in autophagy is underscored by defective autophagosome in Atg8 mutant yeast in response to starvation [22]. As previously mentioned, the Atg5 and Atg8/LC3 complexes are involved in the PE-dependent maturation of the nascent autophagosomes. The process occurs in an ubiquitination-like manner where Atg7 (E1), Atg3 (E2), and Atg5-Atg12-Atg16L1 complex (E3-like) covalently link PE onto the C-terminal glycine. The conjugated Atg8/LC3 can act as an adaptor linking autophagic cargo to the membrane, and is essential for selective autophagy as LC3 G120A mutants fail to localize to autophagic puncta[21, 23–25]. In addition, blocking Atg8/LC3-lipidation inhibits the elongation or closure steps of nascent autophagosomes[26]. These findings underscore the importance of LC3 lipidation in autophagosome biogenesis, which will be discussed in details in section 6.
The composition of autophagic membrane is not well defined, but is likely to include membrane from multiple sources (Figure 1). A handful of studies show vesicle contributions from the ER[27, 28], mitochondria[29], plasma membrane[30], Golgi and endosomes[31, 32]. Recently, the mitochondria-associated membrane (MAM), the contact site between the ER and mitochondria has been identified as the putative origin of mammalian autophagosome formation[17, 18]. This is a critical location given our understanding of the MAM is a major site for lipid synthesis, remodeling, and transport. As we will discuss later, the autophagosome requires mitochondria-derived PE, whose synthesis relies on an intact MAM. After it is formed, the autophagosome encloses cytoplasm for bulk degradation of cellular content as well specific degradation of cargo. This cargo recognition relies on autophagy receptors which link aggregated proteins or ubiquitinated organelles to autophagosomes (reviewed here [33]).
3. MAM as the signaling hub for phospholipids remodeling and autophagic initiation
The importance of mitochondrial lipids in autophagy is tightly linked with the functional coupling of the ER and mitochondrial network. The MAM (known as ER-mitochondria encounter structure (ERMES) in yeast) is a suborganellar region of closely juxtaposed ER and mitochondria, and serves as a metabolic signaling hub, since it is enriched in enzymes involved in lipid synthesis and remodeling. The full scope of MAM functions include ER-mitochondria calcium dynamics, innate immune signaling, lipid droplet biogenesis, and insulin and mTOR signaling (reviewed here [34–36]). As a key site for ER lipid biosynthesis and remodeling, the MAM provides a pathway for inter-organelle lipid transfer between mitochondria and ER. Kornmann et al. found that ERMES is critical for PE and CL synthesis[34]. This interface ranges from approximately 10–25 nm across, and permits shuttling of phosphatidylserine (PS) and phosphatidylcholine (PC) to mitochondria for mitochondria-specific synthesis of PE, PG, and CL. The precise mechanisms or mediators of this lipid trafficking is unknown, though it appears to shuttle in a non-vesicular manner[11]. As pointed out by Voelker, interorganelle transfer of lipids, not just between mitochondria and ER, remains largely opaque, and may involve mechanisms evolutionarily ancient compared to vesicular transport[11, 34]. Disrupting this interaction by knocking out components of the tethering complex significantly reduced cell viability[34]. Genetic alteration of MAM structures has a profoundly negative effect on viability through defects in mitochondrial function and autophagosome biogenesis[35, 36]. Thus, the MAM is emerging as a major mediator of autophagy.
The MAM is held together by protein complexes involved mitochondrial quality control. As the best studied MAM tether, mitofusin 2 (MFN2) is localized on the outer mitochondrial membrane (OMM) and the ER, and is required for mitochondrial fusion. MFN2 belongs to a family of GTPases that regulate mitochondrial dynamics and mitochondrial quality control. MFN2 on the ER forms heteromers with mitofusin 1 (MFN1) and homodimers on the OMM[37]. Mutations in MFN2 result in defects of MAM morphology and calcium mobilization[38]. Interestingly, a study in cardiomyocytes also identified a defect at the step of autophagosome-lysosome fusion in cardiac-specific MFN2 knockout mice[39, 40]. Reduction of MFN2 has been shown not only to impair mitochondrial fusion, but also to reduce autophagy[39, 41]. Knockdown of MFN2 by shRNA in cardiomyocytes, HeLa cells, and smooth muscle cells disrupted autophagosome-lysosome fusion, leading to an accumulation of autophagosomes[39, 40]. Zhao et al. found a reduction in autophagic flux in MFN2 knockout hearts[40]. This defect was also localized at the autophagosome-lysosome fusion step. Finally, MAM also regulates insulin signaling, as dilation of MAM in obesity is associated with insulin resistance and defective autophagy [41–43]. Tethering of MAM space with molecular linkers significantly improves insulin signal transduction [44].
4. The origin of phospholipids in autophagosomes
The inability to pinpoint a precise source(s) of membranes for autophagosome biogenesis arises from both the difficulty in imaging organelle-specific phospholipids in live cells and the apparent selective exclusion of transmembrane proteins, with the exception of Atg9[45, 46]. Genetic manipulations have revealed that autophagosome formation requires membranes derived from multiple sources including the ER[27, 28], Golgi, [28], plasma membrane [30], and recycling endosomes [47] (Figure 1).
The strongest evidence for organelle-specific lipid contribution to date arises from direct visualization of NBD-tagged PS in autophagosomes. The fluorescence work done by Hailey et al. showed that mitochondria provide a source of membrane for autophagosomes[29]. The authors demonstrated continuity between the OMM and autophagosome, and showed that MAM integrity was necessary for starvation-induced autophagy[29]. Using NBD-tagged PS they were able to directly visualize lipids from the ER to mitochondria to autophagosome[29]. To further support the importance of MAM integrity in autophagy, they also found that knocking out MFN2 significantly blocked formation of autophagic puncta[29]. This may have been a result of a loss of mitochondrial PE and CL (see sections 5 and 6) or a disruption of autophagic signaling cascades.
5. CL as the master regulator of mitophagy
Mitochondrial quality control is critical for cellular health by clearing dysfunctional mitochondria, which requires the mitochondrial fusion and fission process to isolate damaged portion of mitochondria. Mitophagy is a critical part of the mitochondrial quality control process to prevent the accumulation of dysfunctional mitochondria that have less efficient oxidative phosphorylation and generate more oxidative stress [48, 49]. Mitochondrial DNA (mtDNA) encodes for 13 subunits of the ETC, 22 tRNAs, and 2 rRNAs, and is more vulnerable to oxidative stress compared to nuclear DNA[50]. Importantly, oxidative stress and hydrolytic damage can cause polymerase blocks or mtDNA mutations, resulting in defective ETC enzymes in individual mitochondria. In response to this, mitochondria frequently undergo fusion, permitting exchange of contents and restoring protein synthesis- though notably restoration of mtDNA is infrequent[51]. Mitochondrial fission serves the dual purpose of segregating damaged components and reducing the organelle size to be engulfed by autophagosomes[52], whereas mitochondrial fusion can protect mitochondria from degradation[53].
The mitochondria-specific CL is a complex phospholipid located primarily in the inner mitochondrial membrane (IMM) that is critical for oxidative phosphorylation and mitophagy[54]. The prevailing view is that CL is exclusively localized in the IMM, however this has been challenged by a recent study that demonstrated 25% of total CL localized at the outer membrane in yeast [55]. Additionally, there is evidence for a role for CL outside of mitochondria[56, 57]. Work done by Sorice’s group has shown that CL translocates to the plasma membrane during apoptosis[57–59]. Moreover, studies in yeast have further demonstrated a role for CL in lysosomes (see 5.3 and 5.4). As a non-bilayer forming phospholipid, CL interacts with mitochondrial membrane proteins, and is required for cristae formation and membrane fusion[60].
CL is synthesized by CL synthase (CRD1) with limited specificity on acyl chain composition by condensing CDP-diacylglycerol (CDP-DAG) and PG[61, 62]. After de novo synthesis, CL is subjected to both physiological and pathological remodeling by a family of enzymes, which include Tafazzin (TAZ), monolysocardiolipin acyltransferase (MLCL AT), and acyl-CoA:lysoCL acyltransferase 1 (ALCAT1) (Figure 2). The composition of the remodeled, mature CL adopts a highly symmetric acyl composition, varies between species and between different cell types[63]. In mammals, tetralinoleoyl CL (TLCL) is the predominant CL species in metabolic tissues with high energy demand, such as heart, liver, and skeletal muscles, while yeast CL contain palmitoleoyl (C16:1) and oleoyl (C:18:1) chains[63]. The physiological remodeling of CL requires TAZ, a mitochondrial transacylase whose activity and substrate specificity is dependent on membrane curvature[64]. In mammalian heart mitochondria, TAZ demonstrates an exclusive preference for linoleic acid (C18:2) in vivo using other phospholipids as the acyl donors[65, 66], leading to the production of TLCL. An alternative pathway for remodeling CL involves a monolysocardiolipin (MLCL) intermediate generated by phospholipase A2 (PLA2)[67, 68], followed by subsequent reacylation by MLCL AT or ALCAT1. Although a cardiolipin-specific PLA2 has recently identified in yeast, known as cardiolipin-specific deacylase 1, or CLD1,[69], the PLA2 specific for cardiolipin in mammals remains unknown. MLCL AT is a mitochondrial acyl-CoA dependent acyltransferease initially identified in the liver. The enzyme demonstrates exclusive preference for linoleoyl-CoA and monolysocardiolipin as substrates, leading to the production of TLCL[70, 71]. In contrast, ALCAT1 is localized in MAM where it catalyzes the pathological remodeling of CL using both monolysocardiolipin and dilysocardiolipin and acyl-CoAs dominated with long-chain highly unsaturated fatty acids, including arachidonic acid (C20:4) and docosahexaneonic acid (C22:6)[42]. Changes in CL species, specifically a decrease in TLCL in mammalian heart, have been shown to disrupt mitophagy[5, 6, 72], resulting in accumulation of dysfunctional and fragmented mitochondria as well as extensive defects in cellular energy metabolism[41].
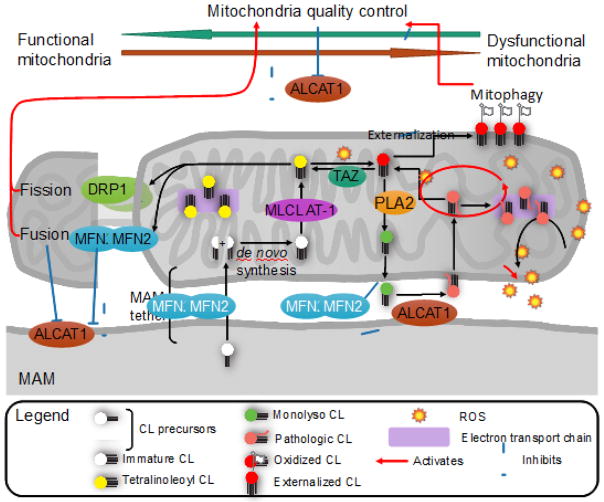
FIGURE 2. CL remodeling and mitochondrial quality control.
CL remodeling is critical for mitochondrial function and mitophagy through regulation of oxidative stress and mitochondrial dynamics. Nascent CL is synthesized de novo from phosphatidylglycerol and CDP-diacylglycerol without acyl chain specificity and is remodeled by tafazzin (TAZ) in the intermembrane space through successive transacylation to tetralinoleoyl CL (TLCL). TLCL is required for optimal electron transport chain function and minimal production of reactive oxygen species (ROS). When CL is damaged by oxidative stress, it is remodeled by hydrolysis of the altered fatty acid by phospholipase A2 (PLA2) to monolysoCL (MLCL) followed by re-acylation by lysoCL acyltransferase 1 (ALCAT1) at the mitochondria-associated membrane (MAM) or by monolysocardiolipin acyltransferase (MLCL AT) at inner mitochondrial membrane. In contrast to MLCL AT which demonstrates exclusive preference to linoleoyl-CoA, ALCAT1 catalyzes the pathologic remodeling of CL with long-chain highly unsaturated fatty acyl-CoAs, increasing the generation of oxidative stress and decreasing mitochondrial fusion/fission proteins DRP1 and MFN2. This oxidative stress not only reacts with other CL to damage more CL, but also depletes MFN2 and further increases ALCAT1 gene expression. Reduction in MFN2 disrupts the MAM tether, reducing lipid shuttling and lipid synthesis at the MAM, and inhibits mitochondrial fusion as a quality control mechanism. ALCAT1 also inhibits autophagic clearance of damaged mitochondria through mitophagy that requires externalization of oxidized CL by phospholipid scramblase 3 (PLS3). LC3 binding to CL is species specific, and has the greatest affinity for TLCL. ALCAT1-mediated CL remodeling results in a double hit to mitochondrial quality control by disrupting mitochondrial fusion and cargo recognition by LC3, and reduces autophagy by disrupting MAM integrity.
5. 1. CL and mitochondrial dynamics
Mitochondria form dynamic networks that are crucial for quality control and mitophagy. These dynamics are mediated by a set of GTPases that control mitochondrial trafficking, fusion, and fission. Dynamin related protein 1 (DRP1) is necessary for fission[73], while optic atrophy 1 (OPA1), mitochondrial genome maintenance 1 (MGM1), MFN1 and MFN2 are necessary for fusion[74–76] (mechanisms of these GTPases reviewed [77]). Experimentally, mitochondrial networks can be modulated by overexpressing or knocking down these GTPases[78]. Underlying the mechanisms of mitochondrial dynamics is the requirement of an intact membrane potential[79], which is decreased upon CL depletion[80]. Thus, CL content can profoundly affect mitochondrial fusion and fission. Additionally, CL was found to be directly required for activation of the GTPases DRP1 and MGM1[81, 82]. Work done by Bustillo-Zabalbeitia has shown that CL is required for enhancing DRP1 activity[81]. DRP1 specifically associates with CL through a lysine-rich variable domain[81]. The interaction with CL promotes DRP1 self-assembly into multimeric complexes and enhances GTPase activity[81]. Similarly, MGM1, the yeast homolog of OPA1 that mediates IMM fusion requires CL for optimal function. MGM1 binds tightly to CL-containing membranes, and the binding stimulates the dimerization of MGM1[82].
Emerging evidence also implicates a key role of CL acyl composition in mitochondrial dynamics through modulation of MFN2 expression. Accordingly, overexpression of ALCAT1, which remodels CL with long-chain highly unsaturated fatty acids, significantly reduced MFN2 protein and mRNA expression [5]. By contrast, ALCAT1 deficient mouse embryonic fibroblasts (MEFs) had increased MFN2 protein and mRNA expression[5]. Phenotypically, ALCAT1 overexpression resulted in significant mitochondrial fragmentation, which could be reversed by transient overexpression of MFN1 and MFN2[5]. This loss was mediated by oxidative stress, as H2O2 treatment in cell culture further depleted MFN1 and MFN2 protein levels, while pre-incubation with diphenyleneiodonium, an NADPH oxidase inhibitor, blocked H2O2-induced decreases in MFN1 and MFN2[5]. Restoring mitochondrial dynamics by MFN2 overexpression also rescued mitochondrial respiratory defects caused by depletion of CL[5]. Furthermore, ALCAT1 overexpression caused significant dilation of the MAM, which is consistent with decreased MFN2 expression.
5.2. CL and mitophagosome cargo recognition
Mitophagy requires the fission of mitochondria and subsequent packaging into autophagosomes. Mitochondria are typically excluded during starvation-induced autophagy by increased fusion activity[83]. Upon depolarization or damage, mitochondria undergo asymmetric fission, marking the damaged organelle for degradation[48]. There are a handful of autophagic adaptors identified to be involved in recognizing damaged mitochondria, including PTEN-induced putative kinase 1 (PINK1)-Parkin [84], BCL2/Adenovirus E1B 19kDa interacting protein 3 (BNIP3)-BNIP3-like (NIX)[85], and Ambra1[86], PINK1-Parkin-dependent mitophagy has been the best studied to date (mitophagy pathways reviewed here[87, 88]). CL itself can serve as a mitophagy signal when it becomes externalized and bind directly to Atg8/LC3. The requirement for CL in PINK1-Parkin, BNIP3-NIX, or Ambra1-mediated mitophagy has not been specifically excluded. However, CL depletion can inhibit mitophagy, leading to an accumulation of damaged mitochondria.
CL externalization onto the OMM can serve as a unique signal for mitophagy (Figure 2). Specifically, this externalization occurs in the presence of mitochondrial damage and stress. Upon treatment with depolarizing compounds such as carbonyl cyanide m-chlorophenyl hydrazine (CCCP) and rotenone, a complex I inhibitor, an increase of CL in OMM can be detected[6]. This translocation from the IMM to the outer leaflet of the OMM is dependent on the mitochondrial protein nucleotide diphosphate kinase (NDPK-D) and phospholipid scramblase 3 (PLS3)[6, 89]. The current hypothesis suggest that NDPK-D acts as a rotating hexamer transferring CL from the IMM to OMM while PLS3 equilibrates CL between inner and outer membrane leaflets, but the mechanisms of both are poorly understood[89]. Molecular modeling of the LC3 crystal structure and subsequent analysis of mutant LC3 indicated that it could directly bind to CL via its N-terminal α-helices[6]. This CL-binding domain of LC3 was not critical for non-selective autophagy, but was required for optimal mitophagy[6]. As expected, disruption of CL synthesis or CL externalization by silencing CRD1, NDPK-D, or PLS3 significantly reduced mitophagy without noticeably affecting autophagy [6, 89].
CL remodeling has a significant impact on the ability for LC3 to recognize the phospholipid. Specifically, LC3 binds most avidly to TLCL[6]. Thus, defects in physiological CL remodeling to TLCL will reduce LC3 affinity for externalized CL. Accordingly, mitophagy defects have been demonstrated in TAZ knockdown and ALCAT1 overexpression models[5, 72]. Both of these models reduce total CL amounts, and also reduce the proportion of TLCL present in mitochondria[42, 90]. TAZ depletion significantly reduced mitochondria co-localization with GFP-LC3 and lysosomes, but did not reduce autophagosome biogenesis [72]. Furthermore, restoration of CL maturation by overexpressing TAZ was sufficient to rescue the mitophagy defect. Thus, consistent with the work done by Chu et al., TLCL depletion impaired autophagosome recognition of mitochondria [6].
5.3. CL acyl composition and autophagosome and lysosome fusion
A recent study also suggests a potential role for CL acyl composition in the fusion event between lysosomes and autophagosomes. Inhibition of stearyl-CoA desaturase (SCD1) in islet β-cells leads to defective autophagosome-lysosome fusion in autophagy[91]. SCD1 catalyzes the synthesis of monounsaturated fatty acids from saturated precursors, and is believed to mediate lipid bilayer fluidity. As expected, pharmacologic inhibition of SCD1 generally resulted in larger proportion of saturated fatty acyl chains in phospholipids observed[91]. By contrast, SCD1 inhibition significantly increased CL unsaturation[91]. The mechanism of SCD1 inhibition increasing CL unsaturation is likely due to broad changes in the fatty acids available as substrates. Loss of SCD1 activity shifted the relative composition of fatty acid pools available for downstream acyltransferases evidenced by reduced monounsaturated fatty acids in phospholipids while increasing saturated fatty acids and arachidonic acid content[92]. Additionally, Ogasawara et al. showed that inhibition of SCD1 impaired autophagosome biogenesis [93]. However, SCD1 inhibition also affected the content and composition of PE, PI, PC, and PS[91, 92], and their roles in autophagosome and lysosome fusion cannot be neglected. Together, the findings lend further support for the previous observations that increased unsaturation of CL from pathological remodeling causes defective mitophagy in aging and aging-related diseases [41, 42, 73, 94].
5.4. A surprising cross-talk between CL and lysosomal function
While the mitochondria-ER interface has been in the spotlight, recent studies also provide evidence supporting a link between mitochondrial and lysosomal function. Early observations of CL biosynthesis and remodeling enzymes in yeast hinted that CL had functions outside of mitochondrial metabolism. The yeast mutants taz1Δ, crd1Δ, and pgs1Δ (phosphatdiylglycerolphosphate synthase 1) exhibited significant respiratory defects and sensitivity to elevated temperatures, which increased in severity as one progressed up the CL biosynthetic pathway. The taz1Δ mutant, which lack mature TLCL, grew normally on glucose but showed temperature sensitivity to growth on ethanol [95]. The crd1Δ mutants, which could not synthesize CL, had greater defects in growth and mitochondrial respiration, and a subset of the reported crd1Δ mutants could not form colonies on glucose [96, 97]. The pgs1Δ mutants, which lack both PG and CL, exhibited cell wall defects, growth arrest, and depletion of mtDNA [98–100]. These yeast experiments suggest an extra-mitochondria role for CL in cell survival.
Surprisingly, one of the major defects observed in the crd1Δ was a swollen vacuole with decreased acidification [101], indicating that CL-mediated vacuolar function. Additionally, vacuolar type H+-ATPase (V-ATPase) activity and acidification was significantly reduced at elevated temperatures. The vacuole is most closely related to the late endosome/lysosome in higher eukaryotes, and is important for osmotic homeostasis in yeast. In addition to the proteolytic environment maintained by a low pH, vacuolar acidity provides a proton gradient with which the cell can sequester ions and amino acids into the vacuole. Chen et al. observed that the vacuolar defect in the crd1Δ mutant could be phenotypically rescued in hyperosmotic culture conditions [101]. Moreover, the crd1Δ mutant had elevated Na+ stress, and deletion of Na+/H+ exchanger 1 (NHX1), an endosomal cation exchanger, rescued the vacuolar swelling and colony formation defects [101]. This suggested that the abnormal vacuolar acidification impaired ion transport and homeostasis in the cell. The V-ATPase defect was also found to be dependent on retrograde regulation (RTG2) signaling, a retrograde regulator that is upregulated in response to mitochondrial dysfunction. In fact, crd1Δrtg2Δ double mutants were able to grow and form colonies on glucose as well [101]. Thus, these data show that CL is required for optimal vacuolar function.
Consistent with the findings, vacuolar acidification was found to have a profound effect on mitochondrial function. Hughes and Gottschling found that vacuolar acidification was associated with replicative capacity in yeast [102]. Yeast exhibit cellular senescence due to a limited replicative capacity. The report show that the vacuolar acidity in mother cells decreased within the first four divisions and remained low [102]. This was followed by a decrease in mitochondrial potential present as early as the seventh division, and mitochondrial fragmentation by the eighth division [102]. Mitochondria potential continued to drop and mitochondria fragmented and formed aggregates throughout the life of the yeast. However, using a suppression screen looking for delayed mitochondrial fragmentation, they identified that overexpression of vacuolar membrane ATPase (VMA1) and vacuolar pH (VPH2) could suppress age-induced mitochondrial dysfunction [102]. VMA1 is subunit A of the V-ATPase complex and VPH2 is an ER-localized enzyme that functions in V-ATPase complex assembly, both of which functionally increase V-ATPase activity. Thus, by restoring lysosomal acidification, age-induced mitochondrial dysfunction could be attenuated.
The mitochondrial dysfunction observed in aged yeast was dependent on vacuolar amino acid homeostasis. Loss of V-ATPase activity would reduce the function of vacuolar importers, including AVT1 and NHX1. AVT1, a vacuolar amino acid transporter, was found to mediate the vacuole-mitochondria cross-talk [102]. Specifically, overexpression of AVT1 partially decoupled vacuolar acidity and mitochondrial morphology defects. Deletion of AVT1 accelerated mitochondrial dysfunction independently of vacuolar pH changes [102]. Furthermore, AVT1 deletion blocked VMA1 and VPH2 rescue of mitochondrial dysfunction [102]. This demonstrated that amino acid homeostasis links vacuolar pH with mitochondrial dysfunction. It is notable that the pathways mediating calorie restriction, which is an effective intervention that extends lifespan in yeast to mammals, also increase vacuolar acidity and maintenance of mitochondrial function [102, 103]. This calorie-restriction induced longevity in yeast was dependent on V-ATPase activity [102]. The important role of V-ATPase in autophagy is further underscored by a recent study that demonstrates a key role for V-ATPase in regulating amino acid signaling to mTORC1, a master regulator of autophagy [104]. TORC1 is activated in response to nutrient, including branch-chain amino acids. Accordingly, V-ATPase forms a multiprotein complex including the Rag GTPases and Regulator on the lysosomal surface as part of mTOR signaling to amino acids [105, 106]. Thus, vacuolar amino acid homeostasis has a significant effect on yeast lifespan secondary to mitochondrial function and V-ATPase activity. Additionally, the defects caused by CL deficiency in yeast crd1Δ resemble those of mutants of FAB1, which encodes a lipid kinase that converts phosphatidylinositol 3-phosphate (PtdIns3P) to phosphatidylinositol 3,5-bisphosphate [PtdIns(3,5)P2]. PtdIns(3,5)P2 not only activates V-GTPase, but is also required for TORC1 activity on the vacuole in yeast [107, 108]. Moreover, inhibition of TORC1 activity with rapamycin significantly increases lifespan from nematodes, flies to rodents [109]. Furthermore, suppression of TOR activity by starvation or two mTOR catalytic inhibitors leads to activation of lysosomal function [110]. However, how CL regulates V-ATPase activity and lysosome function in mammalian cells remains elusive for future studies.
In search for direct evidence that support the interaction between mitochondria and lysosome, Christian Ungermann’s group identified a distinct mitochondria-vacuole tethered interface they named vCLAMP (vacuole and mitochondria patch) that could serve as a link between the two organelles [111]. Interestingly, respiratory growth conditions that stimulated ERMES formation suppressed vCLAMP formation. The vacuole-mitochondria tether was found to require vacuolar yeast protein two (YPT7) and vacuolar morphogenesis (VPS39). VPS39 localizes in punctate spots along the vacuole-mitochondria interface, and overexpression of VPS39 resulted in aggregation of mitochondria and vacuoles. Thus, vCLAMP is proposed to be the vacuolar equivalent of ERMES. Accumulating evidence also suggest overlapping functions between and reciprocal regulation of ERMES and vCLAMP. ERMES mutants, which cannot grow on glycerol, could be partially rescued by VPS39 overexpression, suggesting a potential overlap in function such as small-molecule and lipid transport into mitochondria [111, 112]. Additionally, loss of either contact site resulted in an expansion of the other. Thus, loss of vCLAMP through the yeast vsp39Δ mutant resulted in increased ERMES, which is regulated by mitochondrial distribution and morphology 34 (MDM34) foci[112]. MDM34 is a component of ERMES that tethers mitochondria with ER in yeast. Conversely, in mdm34Δ and other ERMES mutants, vCLAMP was found to completely surround mitochondria[112]. In support of this, deletion of MDM34 is synthetically lethal with VPS39 deletion[112]. The data suggest that ERMES and vCLAMP are in a dynamic equilibrium to maintain mitochondrial contact.
The significance of having mitochondrial contact sites like ERMES and vCLAMP for mitochondrial phospholipid shuttling in yeast was demonstrated by analyzing defects in PE biosynthesis. The biosynthetic pathway is outlined in Figure 3. Using a yeast psd2Δ (phosphatidylserine decarboxylase 2) mutant grown on ethanolamine-free substrate, Elbaz-Alon et al. were able to limit PE synthesis through the Golgi (via PSD2) and the Kennedy pathway to mitochondrial PSD1 only. When vCLAMP and ERMES were ablated, they found a significant increase in PI and PS, and a decrease in PE and CL[112]. PS accumulated in the ER, and the substrates were shunted to increase PI synthesis[112]. Since PS was no longer transported into the mitochondria, PE synthesis via psd1 significantly reduced. Similarly, loss of the contact sites impaired CL precursor import into the mitochondria. These findings were consistent with a loss of phospholipid shuttling to mitochondria. While the research on this mitochondrial contact site is in its infancy, the importance of vCLAMP and vacuolar-mitochondrial interaction on mitochondrial phospholipid metabolism and function has already been identified.
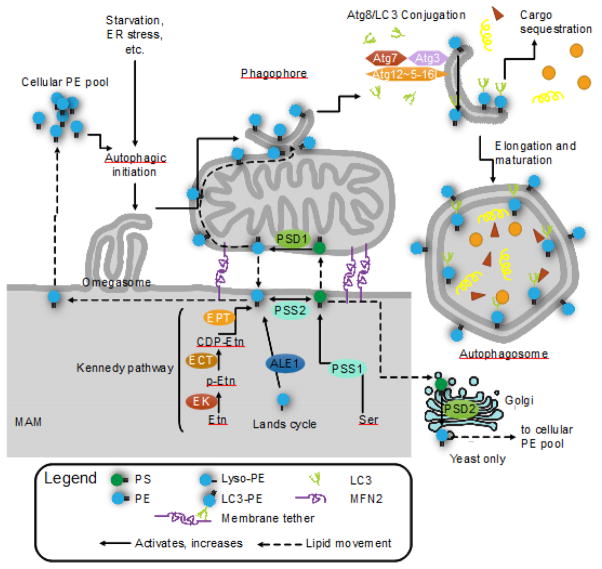
FIGURE 3. Mitochondrial phosphatidylethanolamine (PE) is required for autophagy.
PE is critical for the initiation of autophagy and elongation of autophagosomes. PE is synthesized through 4 pathways (5 for yeast): the CDP-ethanolamine/Kennedy pathway; baseexchange by phosphatidylserine synthase 2 (PSS2); re-acylation of lyso-PE via the Lands cycle (ALE1/LPLAT2); decarboxylation of phosphatidylserine (PS) by phosphatidylserine decarboxylase in the mitochondria (PSD1) or in the Golgi (PSD2, yeast only). PS is synthesized by PSS1, which catalyzes a base exchange with phosphatidylcholine (not pictured). The mitochondrial PSD1 and Kennedy pathway are the major sources of cellular PE. The cellular PE pool is divided between PE as a major component of most lipid bilayers, glycosylphosphatidylinositol (GPI) anchor synthesis and Atg8/LC3 lipidation. When cellular PE is limited, GPI anchor synthesis and Atg8/LC3 lipidation compete for substrates. Atg8/LC3 lipidation occurs on the phagophore, where Atg7 (E1), Atg3 (E2), and the Atg5 complex (Atg12~Atg5-Atg16L1) (E3-like) covalently link Atg8/LC3 onto PE, forming LC3-II. LC3-II is required selective autophagy and links autophagy receptors with an LC3-interacting region to the inner surface of the autophagosome. Knockdown of PSD1 significantly impairs autophagy and LC3 lipidation, indicating the importance of mitochondria-derived PE in autophagy over other sources. Exogenous ethanolamine (Etn) and PE can restore autophagy in PSD1 knockouts, and can stimulate autophagy and increase longevity in wild-type cells.
Given the highly conserved functions of vacuole and lysosome, it is tempting to speculate that CL may also mediate mitochondria-endo-lysosomal interactions. In mammalian systems, the contacts between mitochondria and the endo-lysosomal system are not as well defined. Functionally, defects in mitochondrial respiration have been shown to induce lysosomal dysfunction by altering lysosome pH and impairing lysosomal enzymatic activity[113]. Schiaffino’s group have recently identified direct interaction between mitochondria and lysosome-related organelles, melanosomes, mediated by MFN2[114]. MFN2 was shown to be required for melanosome-mitochondria contact, similar to that between mitochondria and the ER (MAM, section 3), and knockdown of MFN2 significantly impaired melanosome biogenesis and maturation[114]. With the recognition of mitochondrial interaction with lysosome-related organelles, it may only be a matter of time before a bona fide lysosome-mitochondria tether is identified. Since mitochondrial respiration has furthermore been linked to lysosomal function, pathological CL remodeling or depletion may have deleterious effects on the endo-lysosomal system that contribute to metabolic and aging-related diseases. Ultimately, these defects in vacuolar and lysosomal acidification and activity impinge back onto the autophagic pathway.
6. The dynamic roles of PE in autophagy
As previously mentioned, PE is an essential regulator of autophagy. PE is a major membrane component of most membranes, second only to PC in mammalian cells. PE is synthesized through four major pathways: in the MAM from CDP-ethanolamine pathway, through base exchange via phosphatidylserine synthase 2 (PSS2) at the ER, in mitochondria by phosphatidylserine decarboxylase 1 (PSD1/PISD) using PS imported at the MAM, and by re-acylation of lyso-PE via the Lands cycle[115, 116]. In yeast, an additional decarboxylase Psd2 is present in the Golgi and provides an alternate source of PE[117]. As an intermediate in phospholipid synthesis, disruption in PE levels directly affect downstream PC synthesis, as well as that of PS and PI. Aside from its function as a membrane component, PE is required for glycosylphosphatidylinositol (GPI) anchor synthesis. As shown in Figure 3, the importance of PE in autophagy has also been well defined not only as the lipid moiety conjugated to Atg8/LC3 that permit the protein’s insertion into autophagosomes, but also as PE abundance corresponds to autophagic activity and longevity[118]. Mitochondria-derived PE (and PS) may be a component of autophagosome membranes, as NBD-tagged PS may be converted into PE by PSD1 and localizes to LC3-positive structures[29]. The role of PE in autophagy has been illustrated in Figure 3.
PE is the limiting factor for autophagic activity and reductions in cellular PE limits substrate availability for autophagy and GPI anchor synthesis[119]. GPI anchors are post-translational modifications required for protein interactions with membrane rafts, and is critical for signaling and trafficking. Due to fact that GPI anchor synthesis and Atg8/LC3 lipidation share a common PE pool, autophagy is increased when GPI anchor synthesis is inhibited in Aspergillus fumigatus[120]. Furthermore, ethanolamine increases autophagy independently of mTOR and AMPK signaling, suggesting that PE may directly stimulate autophagy or activate other regulatory pathways[118]. By contrast, when PE levels are decreased experimentally, autophagy drops as well[118]. In yeast, psd1Δpsd2Δ mutants which had significantly reduced PE and normal levels of Atg8, Atg8 failed to be recruited to preautophagosomal structures[121].
The requirement of PE in autophagy as a membrane component may be due to its physical properties as a neutral, non-bilayer forming phospholipid[122]. PE tends to form inverted hexagonal lipid phase with a negative curvature that forms the inner leaflets of vesicles and promotes membrane fusion[123], which is critical in autophagosome biogenesis as well as fusion with endolysosomal vesicles. Proteins with amphipathic α-helices, alkyl chains, and lipidated residues can sense and insert into membrane defects formed by membrane curvature[124]. Furthermore, lipidated Atg8 can stabilize high curvature membranes[125], and may contribute to autophagosome activity and protein selectivity on the membrane surface. Selective exclusion of proteins may explain the relative lack of transmembrane proteins on autophagosome membranes[126], and may control recruitment of protein complexes such as the Atg5 complex. Thus, the role of PE in autophagy is not only a lipidation substrate for Atg8/LC3, but also a key structural component of the autophagosome membrane.
The major routes of PE synthesis result in two distinct pools of PE, one at the ER and one within mitochondria[116, 127]. Because of the importance of MAM on PE biosynthesis, it can be envisaged that disruption of MAM integrity would also inhibit autophagy via reduced mitochondrial PE levels[128]. The major defect would occur with PS transport across the MAM to the mitochondria for decarboxylation by PSD1[11]. However, it is possible that the Lands cycle is also reduced upon disrupting the MAM. This is performed by broad-specificity lysophospholipid acyltransferases: known as ALE1 in yeast[129]; and lysophospholipid acyltransferase 2 (LPLAT2) in mammals[115].
The enzymatically modified PE may also play a role in modulating the endo-lysosomal pathway. 12/15-lipoxygenase (LOX), known for its role in eicosanoid synthesis, has been shown to have activity towards PE[130]. Oxidized PE with 12-hydroxyeicosatetraenoic acid conjugated more effectively to Atg8 compared to di-oleoyl PE, suggesting that LOX and the oxidation of PE may be a novel link between inflammatory processes and autophagy in yeast[131]. With regards to LC3, oxidized PE and non-oxidized PE were conjugated with equal efficiency, demonstrating that oxidized PE is a viable substrate for lipidation for LC3[131]. While the effect of this on autophagy in mammals has yet to be determined, macrophages derived from LOX-deficient mice exhibit increased numbers of vacuoles and lysosomes on electron microscopy that are reminiscent of those seen in lysosomal storage diseases[131]. These data provide evidence not only showing that LOX may mediate an intersection between inflammatory signaling and autophagy, but also that modifying acyl chains through oxidation may provide an additional layer of control over autophagic activity.
Mitochondrial PE, specifically, has been found to be necessary for autophagosome formation. Though the ER also synthesizes PE, depletion of mitochondria-specific PE inhibits autophagosome formation. In yeast psd1Δ mutants, PE levels are significantly reduced with a corresponding decrease in autophagy[118]. On the other hand, overexpression of psd1, or supplementation with exogenous PE and ethanolamine significantly increased autophagy in yeast, Drosophila, and human cell lines[118]. This requirement for mitochondrial PE provides a compelling explanation for the observation that in the maturation process of an autophagosome, its membranes are continuous with that of a mitochondrion[29]. This is not to discount the influence of Golgi-derived PE. In yeast, it was found that psd2 is required for ER-stress induced autophagy, and that supplementation with exogenous ethanolamine was insufficient to rescue psd2Δ mutants[132]. However, the literature on Golgi-derived PE and its application in the mammalian system is limited, as a mammalian homolog of PSD2 in the Golgi has not been identified.
It is possible that a disruption in PE levels may affect autophagy indirectly through mitochondrial dysfunction. A 20–30% depletion of PE via Psd1 knockdown was sufficient to significantly impair mitochondrial function despite having unaltered CL levels[133]. Furthermore, low levels of PE lead to reduced mitochondrial function and an accumulation of α-synuclein, suggesting a defect in autophagy[134, 135]. Supplementation of ethanolamine in Psd1 knockdown models was able to reduce ER stress associated with low levels of cellular PE, but was notably unable to restore mitochondrial function[134]. Genetic manipulations in yeast have suggested that PE and CL have some functional redundancy—both exhibit negative curvature and are major components of IMM—as psd1Δ and crd1Δ, yeast CL synthase, are independently viable but synthetically lethal when combined[118, 136]. In mammalian systems, PISD knockout in mice resulted in embryonic lethality and caused significant mitochondrial dysfunction in MEFs derived from these mice[137]. This demonstrated that mitochondrial PE is essential for survival, and that the remaining three PE synthetic pathways could not compensate for the PISD defect. Together, the evidence clearly show a requirement for PE in autophagy as a membrane component, in Atg8/LC3 lipidation, and as a stimulus for autophagy.
7. PG and autophagy
PG is synthesized in the mitochondria and is the immediate precursor for de novo CL synthesis, yet relatively little is known about its role in autophagy and mitophagy. PG is synthesized from CDP-diacylglycerol (CDP-DAG), which is formed by the ER-resident phosphatidate cytidylyltransferase 1[138]. CDP-DAG is then transported into the mitochondria, where phosphatidylglycerolphosphate (PGP) is formed by addition of glycerolphosphate by PGS1[139]. At the final step, phosphatidylglycerophosphatase and protein-tyrosine phosphatase 1 (PTPMT1) dephosphorylates PGP to form PG[139, 140]. PG is well studied in plants and cyanobacteria where it is necessary for electron transport chain of the thylakoid in chloroplast, while it is dispensable in eukaryotes and higher organisms. Strong parallels can be drawn between PG in photosynthesis in Arabidopsis thaliana and CL in oxidative phosphorylation, providing insight into the structure and functions of PG.
7.1. PG and photosynthetic electron transport chain function
PG comprises of approximately 16% of total lipid in leaf extracts[141], and is concentrated in the thylakoid comprising of approximately 40% of the phospholipid content[142]. The thylakoid could be easily understood as the inner membrane of the chloroplast, where the electron transport chain resides. Just as depleting CL resulted in dysfunctional mitochondria, knocking out PGP synthase (pgsA) in A. thaliana resulted in significant dysfunction of chloroplasts and loss of photosynthetic activity[143, 144]. pgsA mutants grown on PG-supplemented medium and then transferred to medium without PG exponentially lost approximately 40% of their photosynthetic activity over the course of 90 minutes[144]. Photosynthetic activity could be rapidly restored through administration of PG into the medium, while untreated mutants continued to lose photosynthetic activity over time[144]. Mechanistically, Kruse et al. found that PG is required for the photosystem II (PSII) dimerization, and that dimerization was dependent on the acyl composition of PG[145]. As PG is necessary for PSII dimerization and function, so too is CL necessary for maintaining the respiratory supercomplex [145–147].
7.2. PG in eukaryotes
PG is not essential for growth in eukaryotes, and its function as well as subcellular localization is largely unknown outside of the mitochondria and as a precursor for CL synthesis[148]. An in vitro study of lipid bilayer behavior showed that PG could partially substitute for CL[149], notable since CL is not essential for growth in yeast per se [98, 150] and that crd1-null mutants have elevated PG levels[80]. In clinical medicine, PG is an important component of surfactant in the lungs, secreted by type II pneumocytes, and lack of PG is an indicator of lung immaturity. However, its function with regards to the rest of physiology is poorly understood.
Recently, comparative gene identification-58 (CGI-58), an enzyme with lysophosphatidylglycerol acyltransferase activity, was found to significantly affect mitophagy and autophagy[151]. CGI-58 is located on lipid droplets, and acts as a co-activator of adipose triglyercide lipase (ATGL)[152, 153]. This interaction can be modulated by perilipin 1 binding to CGI-58 to attenuate ATGL-mediated lipolysis under basal conditions[154]. However, unlike ATGL, where knockout animals become obese[155], CGI-58-deficient mice have severe lipid accumulation and skin barrier defects, resulting in neonatal death within hours after birth[156]. These, in addition to the acyl-transferase activity, suggest that CGI-58 has ATGL-independent actions.
Work done by our lab found that CGI-58 expression could modulate cellular PG levels. CGI-58 had acyltransferase activity for lysophosphatidic acid and lysophosphatidylglycerol, and had greater specificity for oleoyl-CoA (C18:1) than for palmitoyl-CoA (C16:0)[151]. Subcellular fractionation demonstrated CGI-58 protein expression in mitochondrial and microsomal fractions. Furthermore, overexpression or knockdown of CGI-58 in C2C12 mouse myoblasts could correspondingly increase or decrease cellular PG levels, providing a model for dynamically manipulating PG in a mammalian system. Increasing PG by overexpressing CGI-58 resulted in increased autophagic initiation by upregulating AMPK activation while inhibiting mTOR complex I (mTORC1) activity. Lastly, CGI-58 upregulation increased mitophagy due to an increase in mitochondrial fission from upregulation of DRP1 combined with an increase in PINK1 levels on mitochondria. It remains elusive whether the importance of CGI-58 in autophagy and mitophagy is a result of PG levels or other uncharacterized enzymatic functions. Further mechanistic studies are necessary to determine how PG and CGI-58 modulate AMPK and mTORC1 signaling. In further support for a role of PG in mitophagy, targeted deletion of ALCAT1 significantly increases PG level and stimulates mitophagy [41, 42, 157]
8. Mitochondrial phospholipids at the crossroad of mitophagy and diseases
In the past decade, research on the consequences of defective autophagy on human health and disease has exploded. Autophagy is required for cellular homeostasis and removal of dysfunctional organelles, and when any of the steps of autophagy- initiation, autophagosome biogenesis, elongation, maturation, lysosome fusion, and dissolution- are disrupted pathology often follows (Figure 4). The effects of altered autophagy are tissue-specific, and may depend on disease progression such as in cancer and neurodegenerative disease. Decreased autophagy, for instance, may result in accumulation of protein aggregates or dysfunctional mitochondria. Extensive research into congenital and acquired neurodegenerative diseases have shown that defective clearance of intracellular inclusions results in neurotoxicity[158, 159]. Defective autophagy and mitophagy, have gained notoriety as a culprit behind neurodegenerative diseases such as Parkinson’s disease (discussed in section 8.3)[160], amyotrophic lateral sclerosis[161], Huntington’s disease[162], and Alzheimer’s disease[163]. The result is neuronal cell death with clinical presentations of learning disability and cognitive decline.
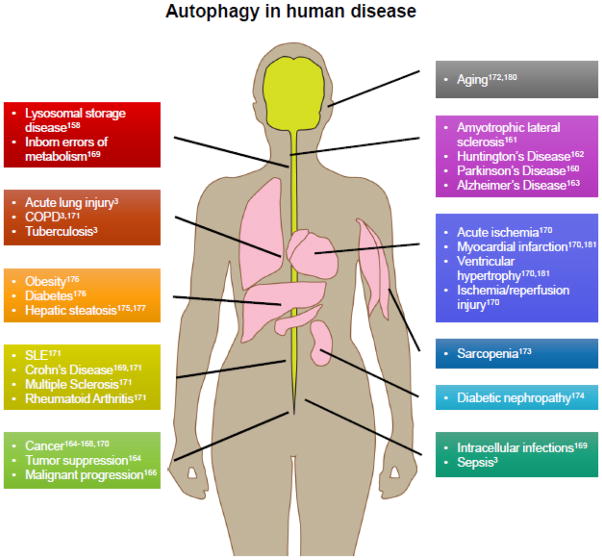
FIGURE 4. Autophagy in disease.
Aberrations in autophagy is implicated in the pathogenesis of many diseases. Please refer to the cited references for more detailed treatments of autophagy and specific diseases. Autophagy is required for clearance of protein aggregates, dysfunctional organelles, and intracellular pathogens. Impaired autophagy is implicated in the severe neurologic phenotypes of congenital disorders as well as impaired clearance of invasive bacteria. Impaired autophagy is associated with aging and aging-associated diseases such as cancer, metabolic disorders, sarcopenia, cardiovascular disease, pulmonary disease, and neurodegenerative conditions. Upregulated autophagy is a cell survival mechanism that is exploited in cancer increasing viability and resistance to chemotherapy, and is implicated in autoimmunity by promoting cell survival in lymphocytes in the face of apoptotic signaling.
The role of autophagy in cancer is complex. In healthy cells autophagy appears to act as a tumor suppressing mechanism by degrading damaged proteins and reducing the burden of oxidative stress[164]. This prevents accumulation of damage that may facilitate cancer initiation or progression[164]. Yet, once a cancer develops it can exploit autophagy and lead to tumor cell survival, growth, resistance to chemotherapy[165, 166]. However, autophagy is such a potent cell survival mechanism that tumors can become “addicted”, rendering it a potential therapeutic target against cancers. For instance, pharmacologically inhibiting autophagy in vitro radiosensitizes tumor cells[167, 168], suggesting that autophagic inhibition would be an effective adjunct therapy in combination with standard treatments. When autophagy is inhibited in vivo, the effects are mixed, demonstrating that we still have a lot to learn with regards to autophagy in cancer[167, 168]. Research now is focused on understanding the mechanisms by which autophagy causes resistance or sensitivity to clinical therapies.
Autophagy has been implicated in autoimmune and inflammatory disease[3, 169]. Polymorphisms in autophagy-related genes have been identified as a risk factor in autoimmune diseases such as systemic lupus erythematosus, Crohn’s disease, multiple sclerosis, and rheumatoid arthritis[170, 171]. Though the mechanisms of these polymorphisms are not precisely known, evidence suggest that generally autophagy allows survival of regulatory T cells that modulate the immune response towards autoimmunity[170, 171]. To better understand the role of autophagy in autoimmunity, the polymorphisms need be explored in model systems.
Furthermore, autophagy is generally decreased as we age[172], and is implicated in the pathogenesis of many aging-related disorders. These include sarcopenia[173], metabolic diseases[41, 42, 174], obesity[175, 176], hepatosteatosis[175, 177], cardiomyopathy[40, 178], and cardiovascular diseases[157, 179]. At the center of these aging-related diseases, dysfunctional energy metabolism and mitochondria have been shown to drive disease progression, providing further evidence linking mitochondria to autophagy and aging (reviewed elsewhere[180, 181]). Thus, the evidence in aggregate supports robust autophagy in healthy aging highlights the importance of mitochondrial lipids, specifically their requirement for autophagosome elongation and mitochondrial function and quality control.
8.1. CL remodeling by ALCAT1 as a key regulator of defective autophagy in aging-related metabolic diseases
Depletion of CL and pathological remodeling of its acyl composition have been implicated in the pathogenesis of aging and aging-related diseases[182–184]. These changes have broad impacts on mitochondrial function by affecting the stability and activity of membrane proteins, MAM structure, mitochondrial fusion and fission, and mitophagy (Figure 2)[54, 185–190]. TLCL is the most prevalent CL species in mitochondria-rich tissues like cardiac and skeletal muscles that depend heavily on mitochondria for ATP production. This is highlighted in Barth syndrome, an X-linked mutation in the TAZ gene that results in dilated cardiomyopathy, skeletal muscle weakness, and early mortality due to heart failure (further discussed in section 8.2)[191]. In mammalian cells, particularly the heart, TLCL is required for mitochondrial membrane structure and optimum oxidative phosphorylation activity. Consequently, loss of TLCL is frequently seen in human heart failure[192] as well as experimental models of heart disease including spontaneously hypertensive heart failure[192, 193] and ischemia/reperfusion injury[194].
Although the molecular mechanisms underlying defective mitophagy in aging related diseases remains poorly elucidated, recent progress in the field has identified a key role of ALCAT1 in regulating defective mitophagy in the pathogenesis. ALCAT1 protein and mRNA expression are potently induced by ROS in response to oxidative stress associated with aging-related diseases, including obesity, diabetes, hypertrophic cardiomyopathy, and hepatosteatosis [41, 42, 157]. In response to oxidative stress, ALCAT1 catalyzes the remodeling the remodeling CL with acyl-CoAs dominated by long-chain highly unsaturated fatty acids, such as arachidonic acids and DHA. Although high unsaturation content in CL significantly increases the mitochondrial membrane fluidity that can presumably help mitochondria to better cope with oxidative stress in the short term, long-chain highly unsaturated fatty acids also render CL highly sensitive to damage by ROS from oxidative stress due to CL’s exclusive location in mitochondria where ROS are generated. Oxidized CL is especially toxic, as it induces the release of cytochrome c, an intermediate in the apoptosis pathway, from the IMM[195, 196]. Consequently, overexpression of ALCAT1 in skeletal and cardiac cell lines leads to CL depletion and multiple defects in mitochondrial dysfunction, including lipid peroxidation, mtDNA mutation and depletion, and defective oxidative phosphorylation [42, 197].
As discussed in section 5, CL is required for multiple processes of mitophagy. Externalization of oxidized CL serves as the key recognition signal by autophagosome for damaged mitochondria to be cleared through mitophagy (Fig. 2). In addition to mitochondrial dysfunction, CL remodeling by ALCAT1 also caused severe dilation of MAM and defective mitophagy, leading to accumulation of dysfunctional mitochondria. Consequently, targeted deletion of ALCAT1 not only significantly increased TLCL and PG levels in the heart, but also restored mitophagy, leading to increased mitochondrial function, mtDNA copy number, and the expression of MLCL AT[42]. ALCAT1 deficiency also decreased mitochondrial ROS production and lipid peroxidation[42], and attenuated mitochondrial fragmentation associated with aging-related diseases by increasing MFN2 expression [5]. Accordingly, ALCAT1−/− mice are fully protected from the onset of various aging-related diseases, including high-fat diet induced obesity, type 2 diabetes, hepatosteatosis, and hypertrophic cardiomyopathy[41, 42, 157].
8.2. TAZ and Barth syndrome
Barth syndrome is a lethal disease which was initially identified by a depletion in TLCL[191]. In Barth syndrome and experimental models of TAZ deficiency, mature TLCL is significantly reduced while MLCL and other CL species increased[90, 198, 199]. As one would expect, the consequences of this are a decrease in mitophagy and a concomitant increase in mitochondrial dysfunction[72, 200]. On a cellular level, the major defect visualized is accumulation of swollen mitochondria[201]. This suggested that either there is excessive mitochondrial biogenesis in the cells or that mitophagy was significantly impaired. As discussed in section 5.2, a defect in mitophagy was observed in TAZ knockdown models. This combined with the increased mitochondrial dysfunction from TLCL depletion is the one-two punch that can explain the observation of cells full of swollen mitochondria seen from patients.
Oxidative stress in TAZ depletion models has been suggested to partially mediate disease pathology. Yeast taz1Δ mutants exhibit increased protein carbonylation, a marker of oxidative damage, during respiratory growth conditions[202]. This has been replicated in induced pluripotent stem cells derived from Barth syndrome patient-derived fibroblasts, where isolated mitochondria produce more ROS compared to mitochondria isolated from control fibroblasts[200]. In cultured rodent cardiomyocytes, treatment with mitoTEMPO, a mitochondria-targeted antioxidant, attenuated some of the mitochondrial damage accumulated from TAZ knockdown[203]. Furthermore, mitoTEMPO prevented hypertrophy and contractile dysfunction, suggesting that blocking mitochondrial ROS production may ameliorate cardiac dysfunction in Barth syndrome[203]. However, it remains to be seen whether mitochondria-targeted antioxidants can attenuate cardiac dysfunction in live animals models of Barth syndrome.
Clinically, antioxidants have had no significant effects despite a wealth of supporting animal data. Meta-analyses of clinical trials showed that antioxidant supplementation had a negligible effect on chronic diseases and mortality in the general population[204–206]. Unfortunately, the general consensus is that the experimental designs, antioxidants used, and the dose and potency of these compounds have been poorly controlled between studies, rendering it difficult to draw concrete conclusions[204–206]. It may be that the right antioxidant must be matched according to tissue and subcellular localization of the disease process, and that general antioxidant supplementation is ineffective in preventing chronic diseases.
8.3. Mitophagy and Parkinson’s Disease
PD is a common neurodegenerative disease caused by the degradation of dopaminergic neurons in the substantia nigra pars compacta (SNpc), resulting in progressive motor symptoms of tremor, bradykinesia, rigidity, and postural instability[207]. It is an aging-associated disease characterized by α-synuclein (α-syn) aggregates and Lewy body deposition within the SNpc. PD is diagnosed clinically by the presence of the primary motor symptoms and may be confirmed post-mortem. The causes of the disease has not been identified as a majority of cases are sporadic, approximately 5% to 10% of cases have been linked to genetic mutations such as α-syn, PINK1 and parkin discussed in section 5.2[208, 209]. Mutations in α-syn that block its degradation or increase its expression and accumulation result in autosomal dominant PD[210]. Aggregates of α-syn are neurotoxic with evidence pointing towards disruption of ER, mitochondria, and lysosomal functions[211–213]. Oxidative stress has also been implicated in the pathogenesis of PD, as demonstrated by the neurotoxic effects of 1-methyl-4-phenylpyridinium (MPP+), rotenone, paraquat, and other pesticides [214–216]. Furthermore, MPP+, rotenone, and paraquat cause significant mitochondrial dysfunction and production of mitochondrial H2O2 and apoptosis in cell culture[217–219]. However, the difficulty in studying PD is that a majority of our understanding comes from rare genetic causes and samples collected post-mortem after the disease has wreaked havoc through the brain.
The cellular mechanisms leading up to neuronal death are under debate, as is the specific vulnerability of SNpc neurons to apoptosis in PD[208]. Mitochondrial dysfunction and the decline of mitophagy with aging has been implicated in neurodegeneration seen in PD. This is supported by the observations that mitochondrial oxidants have been linked to the risk of developing PD as well as the functions of PINK1 and parkin discussed in section 5.2. Oxidative damage to mitochondria begets the production of additional ROS, compounding the initial insult. It has been hypothesized that dopaminergic SNpc neurons are particularly sensitive because of its extensive axonal arborization with unmyelinated fibers[220–222]. The energetic costs of maintaining such a network would be large, especially given its unmyelinated nature[223, 224]. Decreases in the energy balance due to oxidative stress or mitochondrial dysfunction may result in neuronal failure and cell death. In healthy individuals, mitochondrial quality control mechanisms could restore dysfunctional mitochondrial networks and cellular bioenergetics, but this capacity is lost with aging. While CL remodeling has not been directly implicated in PD disease process, we suspect that the defects in mitophagy and mitochondrial dynamics associated with pathological CL remodeling by ALCAT1 may mediate neuronal dysfunction and eventual cell death. It should be noted that SNpc dopaminergic neurons are not the only neurons lost with aging and damage, but are causative in the motor findings. In support for the speculation, ALCAT1 deficiency significantly increased expression of PINK1 in multiple tissues of aging-related diseases [41, 157].
9. Concluding remarks
Autophagy itself requires the interaction of autophagosome with other organelles, and at the center of this are the MAM and vCLAMP. These mitochondria contact sites serve as major signaling hubs for energy sensing and lipid metabolism. Evidence is accumulating that support the MAM or adjacent membranes as the location for omegasome formation and autophagosome biogenesis[36]. This is plausible as the MAM serves as a major phospholipid synthesis, remodeling, and shuttling site that can directly provide phospholipids to the nascent omegasome[225, 226]. Importantly, it appears that CL remodeling, mitochondria function, and MAM integrity are tightly linked via MFN2[42, 227] and that disruption of one necessarily affects the other. Interrupting the mitochondria-ER coupling blocks PS, PC, and PI shuttling to mitochondria, altering PE, PG, and CL metabolism, resulting in defective mitophagy.
Recent studies have shed light on the importance of mitochondria-derived phospholipids in autophagy and mitophagy. While the key role of PE in autophagy is underscored by its role in GPI anchor synthesis and Atg8/LC3 lipidation, PE levels are also strongly correlated with autophagic activity because LC3-II is the limiting factor in autophagosome elongation. Disruptions in CL have major defects in mitophagy and autophagy as a consequence of mitochondrial dysfunction[42, 54, 192, 194, 200, 202], impaired mitochondrial dynamics[5], and of reduced mitophagy[5, 72]. Early work in modulating PG levels through CGI-58 expression has found that CGI-58 overexpression can increase autophagy and mitophagy. However, whether these effects are through PG, CL, or CGI-58 directly have yet to be elucidated.
The next leap in the understanding of autophagic membrane dynamics requires direct visualization of phospholipids in living cells to capture the transient autophagosomes. This has been pioneered by Lippincott-Schwartz’s group discussed above but should to be expanded in examine other phospholipids and confirm the proposed organelle sources for autophagic membranes. While nucleic acids[228, 229] and proteins have long been amenable to visualization by fluorophores, lipids remain stubbornly difficult to visualize in cell culture. This is partly due to toxicity from exogenous lipids and to rapid degradation upon up-taking by living cells, adding great uncertainty to the localization and physiologic relevance of exogenous lipids. However, developing a robust cell-based imaging technique for individual phospholipids will open the door for new questions involving protein-lipid interactions involved with autophagy, mitophagy, and beyond. What is the mechanism for non-vesicular lipid shuttling between organelles? How are vesicles selected for inclusion into nascent autophagosomes? How do the distributions and subcellular localizations of phospholipids affect mitochondrial function? Do mitochondrial phospholipids distribute to other organelles, and for what other cellular processes are they necessary? These questions will become more feasible as the techniques for imaging lipids and studying their interactions with other molecules in vitro and in vivo become more mature.
Review Highlights.
Macroautophagy requires mitochondrial phospholipids for optimal function
Pathological cardiolipin remodeling leads to impaired autophagy
Defects in autophagy are implicated in the pathogenesis of aging-related diseases
Acknowledgments
The authors acknowledge funding support from the National Institutes of Health (2R01DK076685-06A1, Y.S.), American Diabetes Association (1-14-BS-185, Y.S.), by a grant from the Barth Syndrome Foundation (to Y.S.), and support from a T32 training grant through the National Institute of General Medical Sciences (T32GM108563, supporting P.H.). We thank Drs. Leonard S. Jefferson and Hong-Gang Wang for critical reading of the manuscript.
ABBREVIATIONS
- ALCAT1
lysocardiolipin acyltransferase 1
- ALE1
lysophospholipid acyltransferase (yeast)
- AMPK
AMP-activated protein kinase
- Atg
autophagy-related gene
- ATGL
adipose triglyceride lipase
- AVT1
amino acid vacuolar transport
- BNIP3
BCL2/adenovirus E1B 19kDa interacting protein 3
- CCCP
carbonyl cyanide m-chlorophenyl hydrazine
- CDP-DAG
CDP-diacylglycerol
- CDS
phosphatidate cytidylyltransferase 1
- CGI-58
comparative gene identification-589
- CL
cardiolipin
- CRD1
cardiolipin synthase
- ER
endoplasmic reticulum
- ERMES
ER-mitochondria encounter structure (yeast)
- ETC
electron transport chain
- Etn
ethanolamine
- GFP-LC3
green fluorescent protein-LC3 fusion protein
- GPI
glycosylphosphatidylinositol
- IMM
inner mitochondrial membrane
- LC3
microtubule-associated protein 1A/1B-light chain 3 (mammalian)
- LOX
12/15-lipoxygenase
- LPLAT2
lysophospholipid acyltransferase 2 (mammalian)
- MAM
mitochondria-associated membrane (mammalian)
- MDM34
mitochondrial distribution and morphology 34
- MGM1
mitochondrial genome maintenance 1
- MEF
mouse embryonic fibroblast
- MFN1
mitofusin 1
- MFN2
mitofusin 2
- MLCL
monolysocardiolipin
- mtDNA
mitochondrial DNA
- mTOR
mechanistic target of rapamycin
- mTORC1
mTOR complex I
- NBD
nitrobenzoxadiazole
- NHX1
Na+/H+ exchanger 1
- NIX
BCL2/adenovirus E1B 19 kDa protein-interacting protein 3-like
- NDPK-D
nucleotide diphosphate kinase, D
- OMM
outer mitochondrial membrane
- OPA1
optic atrophy 1
- PE
phosphatidylethanolamine
- PG
phosphatidylglycerol
- PGP
phosphatidylglycerolphosphate
- PGS1
phosphatidylglycerolphosphate synthase 1
- PI(3)P
phosphatidylinositol-3-phosphate
- PI
phosphatidylinositol
- PI3KC3
phosphatidylinositol 3-kinase class III
- PINK1
PTEN-induced putative kinase 1
- PISD
phosphatidylserine decarboxylase, mitochondrial (mammalian)
- PLA2
phospholipase A2
- PLS3
phospholipid scramblase 3
- PS
phosphatidylserine
- PSD1
phosphatidylserine decarboxylase 1 (yeast)
- PSD2
phosphatidylserine decarboxylase 2
- PSII
photosystem II
- PSS1
phosphatidylserine synthase 1
- PSS2
phosphatidylserine synthase 2
- PTMP1
phosphatidylglycerophosphatase and protein-tyrosine phosphatase 1
- ROS
reactive oxygen species
- RTG2
retrograde regulation
- SCD1
stearyl-CoA desaturase
- TAZ
tafazzin (mammalian)
- TAZ1
tafazzin (yeast)
- TLCL
tetralinoleoyl cardiolipin
- vCLAMP
vacuole and mitochondria patch
- VMA1
vacuolar membrane ATPase
- VPH2
vacuolar pH
- VPS34
phosphatidylinositol 3-kinase
- VPS39
vacuolar morphogenesis
- WIPI2
WD repeat domain phosphoinositide-interacting protein 2
- YPT7
yeast protein two
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cebollero E, Reggiori F, Kraft C. Reticulophagy and ribophagy: regulated degradation of protein production factories. International journal of cell biology. 2012;2012:182834. doi: 10.1155/2012/182834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Till A, Lakhani R, Burnett SF, Subramani S. Pexophagy: the selective degradation of peroxisomes. International journal of cell biology. 2012;2012:512721. doi: 10.1155/2012/512721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mizumura K, Choi AM, Ryter SW. Emerging role of selective autophagy in human diseases. Frontiers in pharmacology. 2014;5:244. doi: 10.3389/fphar.2014.00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu K, Czaja MJ. Regulation of lipid stores and metabolism by lipophagy. Cell death and differentiation. 2013;20:3–11. doi: 10.1038/cdd.2012.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li J, Liu X, Wang H, Zhang W, Chan DC, Shi Y. Lysocardiolipin acyltransferase 1 (ALCAT1) controls mitochondrial DNA fidelity and biogenesis through modulation of MFN2 expression. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:6975–6980. doi: 10.1073/pnas.1120043109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chu CT, Ji J, Dagda RK, Jiang JF, Tyurina YY, Kapralov AA, Tyurin VA, Yanamala N, Shrivastava IH, Mohammadyani D, Qiang Wang KZ, Zhu J, Klein-Seetharaman J, Balasubramanian K, Amoscato AA, Borisenko G, Huang Z, Gusdon AM, Cheikhi A, Steer EK, Wang R, Baty C, Watkins S, Bahar I, Bayir H, Kagan VE. Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nature cell biology. 2013;15:1197–1205. doi: 10.1038/ncb2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia-Prat L, Martinez-Vicente M, Perdiguero E, Ortet L, Rodriguez-Ubreva J, Rebollo E, Ruiz-Bonilla V, Gutarra S, Ballestar E, Serrano AL, Sandri M, Munoz-Canoves P. Autophagy maintains stemness by preventing senescence. Nature. 2016;529:37–42. doi: 10.1038/nature16187. [DOI] [PubMed] [Google Scholar]
- 8.Kanninen TT, de Andrade Ramos BR, Witkin SS. The role of autophagy in reproduction from gametogenesis to parturition. European journal of obstetrics, gynecology, and reproductive biology. 2013;171:3–8. doi: 10.1016/j.ejogrb.2013.07.020. [DOI] [PubMed] [Google Scholar]
- 9.Marino G, Niso-Santano M, Baehrecke EH, Kroemer G. Self-consumption: the interplay of autophagy and apoptosis. Nature reviews. Molecular cell biology. 2014;15:81–94. doi: 10.1038/nrm3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amaya MJ, Oliveira AG, Schroeder LK, Allgeyer ES, Bewersdorf J, Nathanson MH. Apical localization of inositol 1,4,5-trisphosphate receptors is independent of extended synaptotagmins in hepatocytes. PloS one. 2014;9:e114043. doi: 10.1371/journal.pone.0114043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Voelker DR. Bridging gaps in phospholipid transport. Trends in biochemical sciences. 2005;30:396–404. doi: 10.1016/j.tibs.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 12.Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annual review of cell and developmental biology. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 13.Suzuki K, Akioka M, Kondo-Kakuta C, Yamamoto H, Ohsumi Y. Fine mapping of autophagy-related proteins during autophagosome formation in Saccharomyces cerevisiae. Journal of cell science. 2013;126:2534–2544. doi: 10.1242/jcs.122960. [DOI] [PubMed] [Google Scholar]
- 14.Dall’Armi C, Devereaux KA, Di Paolo G. The role of lipids in the control of autophagy. Current biology : CB. 2013;23:R33–45. doi: 10.1016/j.cub.2012.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marat AL, Haucke V. Phosphatidylinositol 3-phosphates-at the interface between cell signalling and membrane traffic. The EMBO journal. 2016;35:561–579. doi: 10.15252/embj.201593564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wirth M, Joachim J, Tooze SA. Autophagosome formation--the role of ULK1 and Beclin1-PI3KC3 complexes in setting the stage. Seminars in cancer biology. 2013;23:301–309. doi: 10.1016/j.semcancer.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 17.Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nature cell biology. 2009;11:1433–1437. doi: 10.1038/ncb1991. [DOI] [PubMed] [Google Scholar]
- 18.Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. The Journal of cell biology. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Polson HE, de Lartigue J, Rigden DJ, Reedijk M, Urbe S, Clague MJ, Tooze SA. Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy. 2010;6:506–522. doi: 10.4161/auto.6.4.11863. [DOI] [PubMed] [Google Scholar]
- 20.Dooley HC, Razi M, Polson HE, Girardin SE, Wilson MI, Tooze SA. WIPI2 links LC3 conjugation with PI3P, autophagosome formation, and pathogen clearance by recruiting Atg12-5-16L1. Molecular cell. 2014;55:238–252. doi: 10.1016/j.molcel.2014.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, Noda T, Ohsumi Y. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- 22.Xie Z, Nair U, Klionsky DJ. Atg8 controls phagophore expansion during autophagosome formation. Molecular biology of the cell. 2008;19:3290–3298. doi: 10.1091/mbc.E07-12-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, Klionsky DJ, Ohsumi M, Ohsumi Y. A protein conjugation system essential for autophagy. Nature. 1998;395:395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- 24.Mizushima N, Sugita H, Yoshimori T, Ohsumi Y. A new protein conjugation system in human. The counterpart of the yeast Apg12p conjugation system essential for autophagy. The Journal of biological chemistry. 1998;273:33889–33892. doi: 10.1074/jbc.273.51.33889. [DOI] [PubMed] [Google Scholar]
- 25.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. The EMBO journal. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sou YS, Waguri S, Iwata J, Ueno T, Fujimura T, Hara T, Sawada N, Yamada A, Mizushima N, Uchiyama Y, Kominami E, Tanaka K, Komatsu M. The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Molecular biology of the cell. 2008;19:4762–4775. doi: 10.1091/mbc.E08-03-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishihara N, Hamasaki M, Yokota S, Suzuki K, Kamada Y, Kihara A, Yoshimori T, Noda T, Ohsumi Y. Autophagosome requires specific early Sec proteins for its formation and NSF/SNARE for vacuolar fusion. Molecular biology of the cell. 2001;12:3690–3702. doi: 10.1091/mbc.12.11.3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ge L, Zhang M, Schekman R. Phosphatidylinositol 3-kinase and COPII generate LC3 lipidation vesicles from the ER-Golgi intermediate compartment. eLife. 2014;3:e04135. doi: 10.7554/eLife.04135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK, Lippincott-Schwartz J. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141:656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ravikumar B, Moreau K, Jahreiss L, Puri C, Rubinsztein DC. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nature cell biology. 2010;12:747–757. doi: 10.1038/ncb2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamamoto H, Kakuta S, Watanabe TM, Kitamura A, Sekito T, Kondo-Kakuta C, Ichikawa R, Kinjo M, Ohsumi Y. Atg9 vesicles are an important membrane source during early steps of autophagosome formation. The Journal of cell biology. 2012;198:219–233. doi: 10.1083/jcb.201202061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mari M, Griffith J, Rieter E, Krishnappa L, Klionsky DJ, Reggiori F. An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. The Journal of cell biology. 2010;190:1005–1022. doi: 10.1083/jcb.200912089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stolz A, Ernst A, Dikic I. Cargo recognition and trafficking in selective autophagy. Nature cell biology. 2014;16:495–501. doi: 10.1038/ncb2979. [DOI] [PubMed] [Google Scholar]
- 34.Kornmann B, Currie E, Collins SR, Schuldiner M, Nunnari J, Weissman JS, Walter P. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 2009;325:477–481. doi: 10.1126/science.1175088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen H, Detmer SA, Ewald AJ, Griffin EE, Fraser SE, Chan DC. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. The Journal of cell biology. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hamasaki M, Furuta N, Matsuda A, Nezu A, Yamamoto A, Fujita N, Oomori H, Noda T, Haraguchi T, Hiraoka Y, Amano A, Yoshimori T. Autophagosomes form at ER-mitochondria contact sites. Nature. 2013;495:389–393. doi: 10.1038/nature11910. [DOI] [PubMed] [Google Scholar]
- 37.de Brito OM, Scorrano L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 2008;456:605–610. doi: 10.1038/nature07534. [DOI] [PubMed] [Google Scholar]
- 38.Sugiura A, Nagashima S, Tokuyama T, Amo T, Matsuki Y, Ishido S, Kudo Y, McBride HM, Fukuda T, Matsushita N, Inatome R, Yanagi S. MITOL regulates endoplasmic reticulum-mitochondria contacts via Mitofusin2. Molecular cell. 2013;51:20–34. doi: 10.1016/j.molcel.2013.04.023. [DOI] [PubMed] [Google Scholar]
- 39.Ding Y, Gao H, Zhao L, Wang X, Zheng M. Mitofusin 2-deficiency suppresses cell proliferation through disturbance of autophagy. PloS one. 2015;10:e0121328. doi: 10.1371/journal.pone.0121328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao T, Huang X, Han L, Wang X, Cheng H, Zhao Y, Chen Q, Chen J, Cheng H, Xiao R, Zheng M. Central role of mitofusin 2 in autophagosome-lysosome fusion in cardiomyocytes. The Journal of biological chemistry. 2012;287:23615–23625. doi: 10.1074/jbc.M112.379164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang L, Liu X, Nie J, Zhang J, Kimball SR, Zhang H, Zhang WJ, Jefferson LS, Cheng Z, Ji Q, Shi Y. ALCAT1 controls mitochondrial etiology of fatty liver diseases, linking defective mitophagy to steatosis. Hepatology. 2015;61:486–496. doi: 10.1002/hep.27420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li J, Romestaing C, Han X, Li Y, Hao X, Wu Y, Sun C, Liu X, Jefferson LS, Xiong J, Lanoue KF, Chang Z, Lynch CJ, Wang H, Shi Y. Cardiolipin remodeling by ALCAT1 links oxidative stress and mitochondrial dysfunction to obesity. Cell metabolism. 2010;12:154–165. doi: 10.1016/j.cmet.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang L, Li P, Fu S, Calay ES, Hotamisligil GS. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell metabolism. 2010;11:467–478. doi: 10.1016/j.cmet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tubbs E, Theurey P, Vial G, Bendridi N, Bravard A, Chauvin MA, Ji-Cao J, Zoulim F, Bartosch B, Ovize M, Vidal H, Rieusset J. Mitochondria-associated endoplasmic reticulum membrane (MAM) integrity is required for insulin signaling and is implicated in hepatic insulin resistance. Diabetes. 2014;63:3279–3294. doi: 10.2337/db13-1751. [DOI] [PubMed] [Google Scholar]
- 45.Lang T, Reiche S, Straub M, Bredschneider M, Thumm M. Autophagy and the cvt pathway both depend on AUT9. Journal of bacteriology. 2000;182:2125–2133. doi: 10.1128/jb.182.8.2125-2133.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Noda T, Kim J, Huang WP, Baba M, Tokunaga C, Ohsumi Y, Klionsky DJ. Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. The Journal of cell biology. 2000;148:465–480. doi: 10.1083/jcb.148.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Longatti A, Lamb CA, Razi M, Yoshimura S, Barr FA, Tooze SA. TBC1D14 regulates autophagosome formation via Rab11- and ULK1-positive recycling endosomes. The Journal of cell biology. 2012;197:659–675. doi: 10.1083/jcb.201111079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, Stiles L, Haigh SE, Katz S, Las G, Alroy J, Wu M, Py BF, Yuan J, Deeney JT, Corkey BE, Shirihai OS. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. The EMBO journal. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Campello S, Strappazzon F, Cecconi F. Mitochondrial dismissal in mammals, from protein degradation to mitophagy. Biochimica et biophysica acta. 2014;1837:451–460. doi: 10.1016/j.bbabio.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 50.Alexeyev M, Shokolenko I, Wilson G, LeDoux S. The maintenance of mitochondrial DNA integrity--critical analysis and update. Cold Spring Harbor perspectives in biology. 2013;5:a012641. doi: 10.1101/cshperspect.a012641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gilkerson RW, Schon EA, Hernandez E, Davidson MM. Mitochondrial nucleoids maintain genetic autonomy but allow for functional complementation. The Journal of cell biology. 2008;181:1117–1128. doi: 10.1083/jcb.200712101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Twig G, Graf SA, Wikstrom JD, Mohamed H, Haigh SE, Elorza A, Deutsch M, Zurgil N, Reynolds N, Shirihai OS. Tagging and tracking individual networks within a complex mitochondrial web with photoactivatable GFP. American journal of physiology. Cell physiology. 2006;291:C176–184. doi: 10.1152/ajpcell.00348.2005. [DOI] [PubMed] [Google Scholar]
- 53.Gomes LC, Di Benedetto G, Scorrano L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nature cell biology. 2011;13:589–598. doi: 10.1038/ncb2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fry M, Green DE. Cardiolipin requirement for electron transfer in complex I and III of the mitochondrial respiratory chain. The Journal of biological chemistry. 1981;256:1874–1880. [PubMed] [Google Scholar]
- 55.Gebert N, Joshi AS, Kutik S, Becker T, McKenzie M, Guan XL, Mooga VP, Stroud DA, Kulkarni G, Wenk MR, Rehling P, Meisinger C, Ryan MT, Wiedemann N, Greenberg ML, Pfanner N. Mitochondrial cardiolipin involved in outer-membrane protein biogenesis: implications for Barth syndrome. Current biology : CB. 2009;19:2133–2139. doi: 10.1016/j.cub.2009.10.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wriessnegger T, Gubitz G, Leitner E, Ingolic E, Cregg J, de la Cruz BJ, Daum G. Lipid composition of peroxisomes from the yeast Pichia pastoris grown on different carbon sources. Biochimica et biophysica acta. 2007;1771:455–461. doi: 10.1016/j.bbalip.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 57.Manganelli V, Capozzi A, Recalchi S, Signore M, Mattei V, Garofalo T, Misasi R, Degli Esposti M, Sorice M. Altered Traffic of Cardiolipin during Apoptosis: Exposure on the Cell Surface as a Trigger for “Antiphospholipid Antibodies”. Journal of immunology research. 2015;2015:847985. doi: 10.1155/2015/847985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sorice M, Circella A, Misasi R, Pittoni V, Garofalo T, Cirelli A, Pavan A, Pontieri GM, Valesini G. Cardiolipin on the surface of apoptotic cells as a possible trigger for antiphospholipids antibodies. Clinical and experimental immunology. 2000;122:277–284. doi: 10.1046/j.1365-2249.2000.01353.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sorice M, Circella A, Cristea IM, Garofalo T, Di Renzo L, Alessandri C, Valesini G, Esposti MD. Cardiolipin and its metabolites move from mitochondria to other cellular membranes during death receptor-mediated apoptosis. Cell death and differentiation. 2004;11:1133–1145. doi: 10.1038/sj.cdd.4401457. [DOI] [PubMed] [Google Scholar]
- 60.Khalifat N, Fournier JB, Angelova MI, Puff N. Lipid packing variations induced by pH in cardiolipin-containing bilayers: the driving force for the cristae-like shape instability. Biochimica et biophysica acta. 2011;1808:2724–2733. doi: 10.1016/j.bbamem.2011.07.013. [DOI] [PubMed] [Google Scholar]
- 61.Chen D, Zhang XY, Shi Y. Identification and functional characterization of hCLS1, a human cardiolipin synthase localized in mitochondria. The Biochemical journal. 2006;398:169–176. doi: 10.1042/BJ20060303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Houtkooper RH, Akbari H, van Lenthe H, Kulik W, Wanders RJ, Frentzen M, Vaz FM. Identification and characterization of human cardiolipin synthase. FEBS letters. 2006;580:3059–3064. doi: 10.1016/j.febslet.2006.04.054. [DOI] [PubMed] [Google Scholar]
- 63.Schlame M, Ren M, Xu Y, Greenberg ML, Haller I. Molecular symmetry in mitochondrial cardiolipins. Chemistry and physics of lipids. 2005;138:38–49. doi: 10.1016/j.chemphyslip.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 64.Schlame M, Acehan D, Berno B, Xu Y, Valvo S, Ren M, Stokes DL, Epand RM. The physical state of lipid substrates provides transacylation specificity for tafazzin. Nature chemical biology. 2012;8:862–869. doi: 10.1038/nchembio.1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ades LC, Gedeon AK, Wilson MJ, Latham M, Partington MW, Mulley JC, Nelson J, Lui K, Sillence DO. Barth syndrome: clinical features and confirmation of gene localisation to distal Xq28. American journal of medical genetics. 1993;45:327–334. doi: 10.1002/ajmg.1320450309. [DOI] [PubMed] [Google Scholar]
- 66.Schlame M. Cardiolipin remodeling and the function of tafazzin. Biochimica et biophysica acta. 2013;1831:582–588. doi: 10.1016/j.bbalip.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 67.Hsu YH, Dumlao DS, Cao J, Dennis EA. Assessing phospholipase A2 activity toward cardiolipin by mass spectrometry. PloS one. 2013;8:e59267. doi: 10.1371/journal.pone.0059267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kinsey GR, McHowat J, Beckett CS, Schnellmann RG. Identification of calcium-independent phospholipase A2gamma in mitochondria and its role in mitochondrial oxidative stress. American journal of physiology. Renal physiology. 2007;292:F853–860. doi: 10.1152/ajprenal.00318.2006. [DOI] [PubMed] [Google Scholar]
- 69.Beranek A, Rechberger G, Knauer H, Wolinski H, Kohlwein SD, Leber R. Identification of a cardiolipin-specific phospholipase encoded by the gene CLD1 (YGR110W) in yeast. The Journal of biological chemistry. 2009;284:11572–11578. doi: 10.1074/jbc.M805511200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Taylor WA, Hatch GM. Purification and characterization of monolysocardiolipin acyltransferase from pig liver mitochondria. The Journal of biological chemistry. 2003;278:12716–12721. doi: 10.1074/jbc.M210329200. [DOI] [PubMed] [Google Scholar]
- 71.Taylor WA, Hatch GM. Identification of the human mitochondrial linoleoyl-coenzyme A monolysocardiolipin acyltransferase (MLCL AT-1) The Journal of biological chemistry. 2009;284:30360–30371. doi: 10.1074/jbc.M109.048322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hsu P, Liu X, Zhang J, Wang HG, Ye JM, Shi Y. Cardiolipin remodeling by TAZ/tafazzin is selectively required for the initiation of mitophagy. Autophagy. 2015;11:643–652. doi: 10.1080/15548627.2015.1023984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee YJ, Jeong SY, Karbowski M, Smith CL, Youle RJ. Roles of the mammalian mitochondrial fission and fusion mediators Fis1, Drp1, and Opa1 in apoptosis. Molecular biology of the cell. 2004;15:5001–5011. doi: 10.1091/mbc.E04-04-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Santel A, Fuller MT. Control of mitochondrial morphology by a human mitofusin. Journal of cell science. 2001;114:867–874. doi: 10.1242/jcs.114.5.867. [DOI] [PubMed] [Google Scholar]
- 75.Santel A, Frank S, Gaume B, Herrler M, Youle RJ, Fuller MT. Mitofusin-1 protein is a generally expressed mediator of mitochondrial fusion in mammalian cells. Journal of cell science. 2003;116:2763–2774. doi: 10.1242/jcs.00479. [DOI] [PubMed] [Google Scholar]
- 76.Bach D, Pich S, Soriano FX, Vega N, Baumgartner B, Oriola J, Daugaard JR, Lloberas J, Camps M, Zierath JR, Rabasa-Lhoret R, Wallberg-Henriksson H, Laville M, Palacin M, Vidal H, Rivera F, Brand M, Zorzano A. Mitofusin-2 determines mitochondrial network architecture and mitochondrial metabolism. A novel regulatory mechanism altered in obesity. The Journal of biological chemistry. 2003;278:17190–17197. doi: 10.1074/jbc.M212754200. [DOI] [PubMed] [Google Scholar]
- 77.Chan DC. Fusion and fission: interlinked processes critical for mitochondrial health. Annual review of genetics. 2012;46:265–287. doi: 10.1146/annurev-genet-110410-132529. [DOI] [PubMed] [Google Scholar]
- 78.Fransson S, Ruusala A, Aspenstrom P. The atypical Rho GTPases Miro-1 and Miro-2 have essential roles in mitochondrial trafficking. Biochemical and biophysical research communications. 2006;344:500–510. doi: 10.1016/j.bbrc.2006.03.163. [DOI] [PubMed] [Google Scholar]
- 79.Mattenberger Y, James DI, Martinou JC. Fusion of mitochondria in mammalian cells is dependent on the mitochondrial inner membrane potential and independent of microtubules or actin. FEBS letters. 2003;538:53–59. doi: 10.1016/s0014-5793(03)00124-8. [DOI] [PubMed] [Google Scholar]
- 80.Jiang F, Ryan MT, Schlame M, Zhao M, Gu Z, Klingenberg M, Pfanner N, Greenberg ML. Absence of cardiolipin in the crd1 null mutant results in decreased mitochondrial membrane potential and reduced mitochondrial function. The Journal of biological chemistry. 2000;275:22387–22394. doi: 10.1074/jbc.M909868199. [DOI] [PubMed] [Google Scholar]
- 81.Bustillo-Zabalbeitia I, Montessuit S, Raemy E, Basanez G, Terrones O, Martinou JC. Specific interaction with cardiolipin triggers functional activation of Dynamin-Related Protein 1. PloS one. 2014;9:e102738. doi: 10.1371/journal.pone.0102738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.DeVay RM, Dominguez-Ramirez L, Lackner LL, Hoppins S, Stahlberg H, Nunnari J. Coassembly of Mgm1 isoforms requires cardiolipin and mediates mitochondrial inner membrane fusion. The Journal of cell biology. 2009;186:793–803. doi: 10.1083/jcb.200906098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rambold AS, Kostelecky B, Elia N, Lippincott-Schwartz J. Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:10190–10195. doi: 10.1073/pnas.1107402108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, Springer W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nature cell biology. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 85.Shi RY, Zhu SH, Li V, Gibson SB, Xu XS, Kong JM. BNIP3 interacting with LC3 triggers excessive mitophagy in delayed neuronal death in stroke. CNS neuroscience & therapeutics. 2014;20:1045–1055. doi: 10.1111/cns.12325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Strappazzon F, Nazio F, Corrado M, Cianfanelli V, Romagnoli A, Fimia GM, Campello S, Nardacci R, Piacentini M, Campanella M, Cecconi F. AMBRA1 is able to induce mitophagy via LC3 binding, regardless of PARKIN and p62/SQSTM1. Cell death and differentiation. 2015;22:419–432. doi: 10.1038/cdd.2014.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Durcan TM, Fon EA. The three ‘P’s of mitophagy: PARKIN, PINK1, and post-translational modifications. Genes & development. 2015;29:989–999. doi: 10.1101/gad.262758.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wei H, Liu L, Chen Q. Selective removal of mitochondria via mitophagy: distinct pathways for different mitochondrial stresses. Biochimica et biophysica acta. 2015;1853:2784–2790. doi: 10.1016/j.bbamcr.2015.03.013. [DOI] [PubMed] [Google Scholar]
- 89.Kagan VE, Jiang J, Huang Z, Tyurina YY, Desbourdes C, Cottet-Rousselle C, Dar HH, Verma M, Tyurin VA, Kapralov AA, Cheikhi A, Mao G, Stolz D, St Croix CM, Watkins S, Shen Z, Li Y, Greenberg ML, Tokarska-Schlattner M, Boissan M, Lacombe ML, Epand RM, Chu CT, Mallampalli RK, Bayir H, Schlattner U. NDPK-D (NM23-H4)-mediated externalization of cardiolipin enables elimination of depolarized mitochondria by mitophagy. Cell death and differentiation. 2016 doi: 10.1038/cdd.2015.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Soustek MS, Falk DJ, Mah CS, Toth MJ, Schlame M, Lewin AS, Byrne BJ. Characterization of a transgenic short hairpin RNA-induced murine model of Tafazzin deficiency. Human gene therapy. 2011;22:865–871. doi: 10.1089/hum.2010.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Janikiewicz J, Hanzelka K, Dziewulska A, Kozinski K, Dobrzyn P, Bernas T, Dobrzyn A. Inhibition of SCD1 impairs palmitate-derived autophagy at the step of autophagosome-lysosome fusion in pancreatic beta-cells. Journal of lipid research. 2015;56:1901–1911. doi: 10.1194/jlr.M059980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dobrzyn A, Dobrzyn P, Miyazaki M, Sampath H, Chu K, Ntambi JM. Stearoyl-CoA desaturase 1 deficiency increases CTP:choline cytidylyltransferase translocation into the membrane and enhances phosphatidylcholine synthesis in liver. The Journal of biological chemistry. 2005;280:23356–23362. doi: 10.1074/jbc.M502436200. [DOI] [PubMed] [Google Scholar]
- 93.Ogasawara Y, Itakura E, Kono N, Mizushima N, Arai H, Nara A, Mizukami T, Yamamoto A. Stearoyl-CoA desaturase 1 activity is required for autophagosome formation. The Journal of biological chemistry. 2014;289:23938–23950. doi: 10.1074/jbc.M114.591065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Han X, Yang J, Yang K, Zhao Z, Abendschein DR, Gross RW. Alterations in myocardial cardiolipin content and composition occur at the very earliest stages of diabetes: a shotgun lipidomics study. Biochemistry. 2007;46:6417–6428. doi: 10.1021/bi7004015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gu Z, Valianpour F, Chen S, Vaz FM, Hakkaart GA, Wanders RJ, Greenberg ML. Aberrant cardiolipin metabolism in the yeast taz1 mutant: a model for Barth syndrome. Molecular microbiology. 2004;51:149–158. doi: 10.1046/j.1365-2958.2003.03802.x. [DOI] [PubMed] [Google Scholar]
- 96.Jiang F, Gu Z, Granger JM, Greenberg ML. Cardiolipin synthase expression is essential for growth at elevated temperature and is regulated by factors affecting mitochondrial development. Molecular microbiology. 1999;31:373–379. doi: 10.1046/j.1365-2958.1999.01181.x. [DOI] [PubMed] [Google Scholar]
- 97.Zhong Q, Gohil VM, Ma L, Greenberg ML. Absence of cardiolipin results in temperature sensitivity, respiratory defects, and mitochondrial DNA instability independent of pet56. The Journal of biological chemistry. 2004;279:32294–32300. doi: 10.1074/jbc.M403275200. [DOI] [PubMed] [Google Scholar]
- 98.Chang SC, Heacock PN, Clancey CJ, Dowhan W. The PEL1 gene (renamed PGS1) encodes the phosphatidylglycero-phosphate synthase of Saccharomyces cerevisiae. The Journal of biological chemistry. 1998;273:9829–9836. doi: 10.1074/jbc.273.16.9829. [DOI] [PubMed] [Google Scholar]
- 99.Dzugasova V, Obernauerova M, Horvathova K, Vachova M, Zakova M, Subik J. Phosphatidylglycerolphosphate synthase encoded by the PEL1/PGS1 gene in Saccharomyces cerevisiae is localized in mitochondria and its expression is regulated by phospholipid precursors. Current genetics. 1998;34:297–302. doi: 10.1007/s002940050399. [DOI] [PubMed] [Google Scholar]
- 100.Zhong Q, Li G, Gvozdenovic-Jeremic J, Greenberg ML. Up-regulation of the cell integrity pathway in saccharomyces cerevisiae suppresses temperature sensitivity of the pgs1Delta mutant. The Journal of biological chemistry. 2007;282:15946–15953. doi: 10.1074/jbc.M701055200. [DOI] [PubMed] [Google Scholar]
- 101.Chen S, Tarsio M, Kane PM, Greenberg ML. Cardiolipin mediates cross-talk between mitochondria and the vacuole. Molecular biology of the cell. 2008;19:5047–5058. doi: 10.1091/mbc.E08-05-0486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hughes AL, Gottschling DE. An early age increase in vacuolar pH limits mitochondrial function and lifespan in yeast. Nature. 2012;492:261–265. doi: 10.1038/nature11654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bishop NA, Guarente L. Genetic links between diet and lifespan: shared mechanisms from yeast to humans. Nature reviews. Genetics. 2007;8:835–844. doi: 10.1038/nrg2188. [DOI] [PubMed] [Google Scholar]
- 104.Efeyan A, Comb WC, Sabatini DM. Nutrient-sensing mechanisms and pathways. Nature. 2015;517:302–310. doi: 10.1038/nature14190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Efeyan A, Zoncu R, Sabatini DM. Amino acids and mTORC1: from lysosomes to disease. Trends in molecular medicine. 2012;18:524–533. doi: 10.1016/j.molmed.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jin N, Mao K, Jin Y, Tevzadze G, Kauffman EJ, Park S, Bridges D, Loewith R, Saltiel AR, Klionsky DJ, Weisman LS. Roles for PI(3,5)P2 in nutrient sensing through TORC1. Molecular biology of the cell. 2014;25:1171–1185. doi: 10.1091/mbc.E14-01-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Li SC, Diakov TT, Xu T, Tarsio M, Zhu W, Couoh-Cardel S, Weisman LS, Kane PM. The signaling lipid PI(3,5)P(2) stabilizes V(1)-V(o) sector interactions and activates the V-ATPase. Molecular biology of the cell. 2014;25:1251–1262. doi: 10.1091/mbc.E13-10-0563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, Pahor M, Javors MA, Fernandez E, Miller RA. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhou J, Tan SH, Nicolas V, Bauvy C, Yang ND, Zhang J, Xue Y, Codogno P, Shen HM. Activation of lysosomal function in the course of autophagy via mTORC1 suppression and autophagosome-lysosome fusion. Cell research. 2013;23:508–523. doi: 10.1038/cr.2013.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Honscher C, Mari M, Auffarth K, Bohnert M, Griffith J, Geerts W, van der Laan M, Cabrera M, Reggiori F, Ungermann C. Cellular metabolism regulates contact sites between vacuoles and mitochondria. Developmental cell. 2014;30:86–94. doi: 10.1016/j.devcel.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 112.Elbaz-Alon Y, Rosenfeld-Gur E, Shinder V, Futerman AH, Geiger T, Schuldiner M. A dynamic interface between vacuoles and mitochondria in yeast. Developmental cell. 2014;30:95–102. doi: 10.1016/j.devcel.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 113.Baixauli F, Acin-Perez R, Villarroya-Beltri C, Mazzeo C, Nunez-Andrade N, Gabande-Rodriguez E, Ledesma MD, Blazquez A, Martin MA, Falcon-Perez JM, Redondo JM, Enriquez JA, Mittelbrunn M. Mitochondrial Respiration Controls Lysosomal Function during Inflammatory T Cell Responses. Cell metabolism. 2015;22:485–498. doi: 10.1016/j.cmet.2015.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Daniele T, Hurbain I, Vago R, Casari G, Raposo G, Tacchetti C, Schiaffino MV. Mitochondria and melanosomes establish physical contacts modulated by Mfn2 and involved in organelle biogenesis. Current biology : CB. 2014;24:393–403. doi: 10.1016/j.cub.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 115.Shindou H, Shimizu T. Acyl-CoA:lysophospholipid acyltransferases. The Journal of biological chemistry. 2009;284:1–5. doi: 10.1074/jbc.R800046200. [DOI] [PubMed] [Google Scholar]
- 116.Vance JE, Tasseva G. Formation and function of phosphatidylserine and phosphatidylethanolamine in mammalian cells. Biochimica et biophysica acta. 2013;1831:543–554. doi: 10.1016/j.bbalip.2012.08.016. [DOI] [PubMed] [Google Scholar]
- 117.Calzada E, Onguka O, Claypool SM. Phosphatidylethanolamine Metabolism in Health and Disease. International review of cell and molecular biology. 2016;321:29–88. doi: 10.1016/bs.ircmb.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Rockenfeller P, Koska M, Pietrocola F, Minois N, Knittelfelder O, Sica V, Franz J, Carmona-Gutierrez D, Kroemer G, Madeo F. Phosphatidylethanolamine positively regulates autophagy and longevity. Cell death and differentiation. 2015;22:499–508. doi: 10.1038/cdd.2014.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wilson-Zbinden C, dos Santos AX, Stoffel-Studer I, van der Vaart A, Hofmann K, Reggiori F, Riezman H, Kraft C, Peter M. Autophagy competes for a common phosphatidylethanolamine pool with major cellular PE-consuming pathways in Saccharomyces cerevisiae. Genetics. 2015;199:475–485. doi: 10.1534/genetics.114.169797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yan J, Du T, Zhao W, Hartmann T, Lu H, Lu Y, Ouyang H, Jiang X, Sun L, Jin C. Transcriptome and biochemical analysis reveals that suppression of GPI-anchor synthesis leads to autophagy and possible necroptosis in Aspergillus fumigatus. PloS one. 2013;8:e59013. doi: 10.1371/journal.pone.0059013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Nebauer R, Rosenberger S, Daum G. Phosphatidylethanolamine, a limiting factor of autophagy in yeast strains bearing a defect in the carboxypeptidase Y pathway of vacuolar targeting. The Journal of biological chemistry. 2007;282:16736–16743. doi: 10.1074/jbc.M611345200. [DOI] [PubMed] [Google Scholar]
- 122.Jouhet J. Importance of the hexagonal lipid phase in biological membrane organization. Frontiers in plant science. 2013;4:494. doi: 10.3389/fpls.2013.00494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chernomordik LV, Kozlov MM. Mechanics of membrane fusion. Nature structural & molecular biology. 2008;15:675–683. doi: 10.1038/nsmb.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hatzakis NS, Bhatia VK, Larsen J, Madsen KL, Bolinger PY, Kunding AH, Castillo J, Gether U, Hedegard P, Stamou D. How curved membranes recruit amphipathic helices and protein anchoring motifs. Nature chemical biology. 2009;5:835–841. doi: 10.1038/nchembio.213. [DOI] [PubMed] [Google Scholar]
- 125.Knorr RL, Nakatogawa H, Ohsumi Y, Lipowsky R, Baumgart T, Dimova R. Membrane morphology is actively transformed by covalent binding of the protein Atg8 to PE-lipids. PloS one. 2014;9:e115357. doi: 10.1371/journal.pone.0115357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Tooze SA. The role of membrane proteins in mammalian autophagy. Seminars in cell & developmental biology. 2010;21:677–682. doi: 10.1016/j.semcdb.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 127.Bleijerveld OB, Brouwers JF, Vaandrager AB, Helms JB, Houweling M. The CDP-ethanolamine pathway and phosphatidylserine decarboxylation generate different phosphatidylethanolamine molecular species. The Journal of biological chemistry. 2007;282:28362–28372. doi: 10.1074/jbc.M703786200. [DOI] [PubMed] [Google Scholar]
- 128.Kornmann B, Walter P. ERMES-mediated ER-mitochondria contacts: molecular hubs for the regulation of mitochondrial biology. Journal of cell science. 2010;123:1389–1393. doi: 10.1242/jcs.058636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Riekhof WR, Wu J, Jones JL, Voelker DR. Identification and characterization of the major lysophosphatidylethanolamine acyltransferase in Saccharomyces cerevisiae. The Journal of biological chemistry. 2007;282:28344–28352. doi: 10.1074/jbc.M705256200. [DOI] [PubMed] [Google Scholar]
- 130.Maskrey BH, Bermudez-Fajardo A, Morgan AH, Stewart-Jones E, Dioszeghy V, Taylor GW, Baker PR, Coles B, Coffey MJ, Kuhn H, O’Donnell VB. Activated platelets and monocytes generate four hydroxyphosphatidylethanolamines via lipoxygenase. The Journal of biological chemistry. 2007;282:20151–20163. doi: 10.1074/jbc.M611776200. [DOI] [PubMed] [Google Scholar]
- 131.Morgan AH, Hammond VJ, Sakoh-Nakatogawa M, Ohsumi Y, Thomas CP, Blanchet F, Piguet V, Kiselyov K, O’Donnell VB. A novel role for 12/15-lipoxygenase in regulating autophagy. Redox biology. 2015;4:40–47. doi: 10.1016/j.redox.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Muthukumar K, Nachiappan V. Phosphatidylethanolamine from phosphatidylserine decarboxylase2 is essential for autophagy under cadmium stress in Saccharomyces cerevisiae. Cell biochemistry and biophysics. 2013;67:1353–1363. doi: 10.1007/s12013-013-9667-8. [DOI] [PubMed] [Google Scholar]
- 133.Tasseva G, Bai HD, Davidescu M, Haromy A, Michelakis E, Vance JE. Phosphatidylethanolamine deficiency in Mammalian mitochondria impairs oxidative phosphorylation and alters mitochondrial morphology. The Journal of biological chemistry. 2013;288:4158–4173. doi: 10.1074/jbc.M112.434183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wang S, Zhang S, Liou LC, Ren Q, Zhang Z, Caldwell GA, Caldwell KA, Witt SN. Phosphatidylethanolamine deficiency disrupts alpha-synuclein homeostasis in yeast and worm models of Parkinson disease. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E3976–3985. doi: 10.1073/pnas.1411694111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lopes da Fonseca T, Villar-Pique A, Outeiro TF. The Interplay between Alpha-Synuclein Clearance and Spreading. Biomolecules. 2015;5:435–471. doi: 10.3390/biom5020435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gohil VM, Thompson MN, Greenberg ML. Synthetic lethal interaction of the mitochondrial phosphatidylethanolamine and cardiolipin biosynthetic pathways in Saccharomyces cerevisiae. The Journal of biological chemistry. 2005;280:35410–35416. doi: 10.1074/jbc.M505478200. [DOI] [PubMed] [Google Scholar]
- 137.Steenbergen R, Nanowski TS, Beigneux A, Kulinski A, Young SG, Vance JE. Disruption of the phosphatidylserine decarboxylase gene in mice causes embryonic lethality and mitochondrial defects. The Journal of biological chemistry. 2005;280:40032–40040. doi: 10.1074/jbc.M506510200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lykidis A, Jackson PD, Rock CO, Jackowski S. The role of CDP-diacylglycerol synthetase and phosphatidylinositol synthase activity levels in the regulation of cellular phosphatidylinositol content. The Journal of biological chemistry. 1997;272:33402–33409. doi: 10.1074/jbc.272.52.33402. [DOI] [PubMed] [Google Scholar]
- 139.Kiyasu JY, Pieringer RA, Paulus H, Kennedy EP. The biosynthesis of phosphatidylglycerol. The Journal of biological chemistry. 1963;238:2293–2298. [PubMed] [Google Scholar]
- 140.Zhang J, Guan Z, Murphy AN, Wiley SE, Perkins GA, Worby CA, Engel JL, Heacock P, Nguyen OK, Wang JH, Raetz CR, Dowhan W, Dixon JE. Mitochondrial phosphatase PTPMT1 is essential for cardiolipin biosynthesis. Cell metabolism. 2011;13:690–700. doi: 10.1016/j.cmet.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Essigmann B, Guler S, Narang RA, Linke D, Benning C. Phosphate availability affects the thylakoid lipid composition and the expression of SQD1, a gene required for sulfolipid biosynthesis in Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:1950–1955. doi: 10.1073/pnas.95.4.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Block MA, Dorne AJ, Joyard J, Douce R. Preparation and characterization of membrane fractions enriched in outer and inner envelope membranes from spinach chloroplasts. II. Biochemical characterization. The Journal of biological chemistry. 1983;258:13281–13286. [PubMed] [Google Scholar]
- 143.Hagio M, Sakurai I, Sato S, Kato T, Tabata S, Wada H. Phosphatidylglycerol is essential for the development of thylakoid membranes in Arabidopsis thaliana. Plant & cell physiology. 2002;43:1456–1464. doi: 10.1093/pcp/pcf185. [DOI] [PubMed] [Google Scholar]
- 144.Hagio M, Gombos Z, Varkonyi Z, Masamoto K, Sato N, Tsuzuki M, Wada H. Direct evidence for requirement of phosphatidylglycerol in photosystem II of photosynthesis. Plant physiology. 2000;124:795–804. doi: 10.1104/pp.124.2.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kruse O, Hankamer B, Konczak C, Gerle C, Morris E, Radunz A, Schmid GH, Barber J. Phosphatidylglycerol is involved in the dimerization of photosystem II. The Journal of biological chemistry. 2000;275:6509–6514. doi: 10.1074/jbc.275.9.6509. [DOI] [PubMed] [Google Scholar]
- 146.Zhang M, Mileykovskaya E, Dowhan W. Cardiolipin is essential for organization of complexes III and IV into a supercomplex in intact yeast mitochondria. The Journal of biological chemistry. 2005;280:29403–29408. doi: 10.1074/jbc.M504955200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zhang M, Mileykovskaya E, Dowhan W. Gluing the respiratory chain together. Cardiolipin is required for supercomplex formation in the inner mitochondrial membrane. The Journal of biological chemistry. 2002;277:43553–43556. doi: 10.1074/jbc.C200551200. [DOI] [PubMed] [Google Scholar]
- 148.van Meer G, de Kroon AI. Lipid map of the mammalian cell. Journal of cell science. 2011;124:5–8. doi: 10.1242/jcs.071233. [DOI] [PubMed] [Google Scholar]
- 149.Khalifat N, Rahimi M, Bitbol AF, Seigneuret M, Fournier JB, Puff N, Arroyo M, Angelova MI. Interplay of packing and flip-flop in local bilayer deformation. How phosphatidylglycerol could rescue mitochondrial function in a cardiolipin-deficient yeast mutant. Biophysical journal. 2014;107:879–890. doi: 10.1016/j.bpj.2014.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Jiang F, Rizavi HS, Greenberg ML. Cardiolipin is not essential for the growth of Saccharomyces cerevisiae on fermentable or non-fermentable carbon sources. Molecular microbiology. 1997;26:481–491. doi: 10.1046/j.1365-2958.1997.5841950.x. [DOI] [PubMed] [Google Scholar]
- 151.Zhang J, Xu D, Nie J, Han R, Zhai Y, Shi Y. Comparative gene identification-58 (CGI-58) promotes autophagy as a putative lysophosphatidylglycerol acyltransferase. The Journal of biological chemistry. 2014;289:33044–33053. doi: 10.1074/jbc.M114.573857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lu X, Yang X, Liu J. Differential control of ATGL-mediated lipid droplet degradation by CGI-58 and G0S2. Cell cycle. 2010;9:2719–2725. doi: 10.4161/cc.9.14.12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lass A, Zimmermann R, Haemmerle G, Riederer M, Schoiswohl G, Schweiger M, Kienesberger P, Strauss JG, Gorkiewicz G, Zechner R. Adipose triglyceride lipase-mediated lipolysis of cellular fat stores is activated by CGI-58 and defective in Chanarin-Dorfman Syndrome. Cell metabolism. 2006;3:309–319. doi: 10.1016/j.cmet.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 154.Subramanian V, Rothenberg A, Gomez C, Cohen AW, Garcia A, Bhattacharyya S, Shapiro L, Dolios G, Wang R, Lisanti MP, Brasaemle DL. Perilipin A mediates the reversible binding of CGI-58 to lipid droplets in 3T3-L1 adipocytes. The Journal of biological chemistry. 2004;279:42062–42071. doi: 10.1074/jbc.M407462200. [DOI] [PubMed] [Google Scholar]
- 155.Haemmerle G, Lass A, Zimmermann R, Gorkiewicz G, Meyer C, Rozman J, Heldmaier G, Maier R, Theussl C, Eder S, Kratky D, Wagner EF, Klingenspor M, Hoefler G, Zechner R. Defective lipolysis and altered energy metabolism in mice lacking adipose triglyceride lipase. Science. 2006;312:734–737. doi: 10.1126/science.1123965. [DOI] [PubMed] [Google Scholar]
- 156.Radner FP, Streith IE, Schoiswohl G, Schweiger M, Kumari M, Eichmann TO, Rechberger G, Koefeler HC, Eder S, Schauer S, Theussl HC, Preiss-Landl K, Lass A, Zimmermann R, Hoefler G, Zechner R, Haemmerle G. Growth retardation, impaired triacylglycerol catabolism, hepatic steatosis, and lethal skin barrier defect in mice lacking comparative gene identification-58 (CGI-58) The Journal of biological chemistry. 2010;285:7300–7311. doi: 10.1074/jbc.M109.081877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Liu X, Ye B, Miller S, Yuan H, Zhang H, Tian L, Nie J, Imae R, Arai H, Li Y, Cheng Z, Shi Y. Ablation of ALCAT1 mitigates hypertrophic cardiomyopathy through effects on oxidative stress and mitophagy. Molecular and cellular biology. 2012;32:4493–4504. doi: 10.1128/MCB.01092-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Bosch ME, Kielian T. Neuroinflammatory paradigms in lysosomal storage diseases. Frontiers in neuroscience. 2015;9:417. doi: 10.3389/fnins.2015.00417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ebrahimi-Fakhari D, Saffari A, Wahlster L, Lu J, Byrne S, Hoffmann GF, Jungbluth H, Sahin M. Congenital disorders of autophagy: an emerging novel class of inborn errors of neuro-metabolism. Brain : a journal of neurology. 2016;139:317–337. doi: 10.1093/brain/awv371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Klein C, Westenberger A. Genetics of Parkinson’s disease. Cold Spring Harbor perspectives in medicine. 2012;2:a008888. doi: 10.1101/cshperspect.a008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Lee JK, Shin JH, Lee JE, Choi EJ. Role of autophagy in the pathogenesis of amyotrophic lateral sclerosis. Biochimica et biophysica acta. 2015;1852:2517–2524. doi: 10.1016/j.bbadis.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 162.Martinez-Vicente M, Talloczy Z, Wong E, Tang G, Koga H, Kaushik S, de Vries R, Arias E, Harris S, Sulzer D, Cuervo AM. Cargo recognition failure is responsible for inefficient autophagy in Huntington’s disease. Nature neuroscience. 2010;13:567–576. doi: 10.1038/nn.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Boland B, Kumar A, Lee S, Platt FM, Wegiel J, Yu WH, Nixon RA. Autophagy induction and autophagosome clearance in neurons: relationship to autophagic pathology in Alzheimer’s disease. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2008;28:6926–6937. doi: 10.1523/JNEUROSCI.0800-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Chen HY, White E. Role of autophagy in cancer prevention. Cancer prevention research. 2011;4:973–983. doi: 10.1158/1940-6207.CAPR-10-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Yang ZJ, Chee CE, Huang S, Sinicrope FA. The role of autophagy in cancer: therapeutic implications. Molecular cancer therapeutics. 2011;10:1533–1541. doi: 10.1158/1535-7163.MCT-11-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, Mukherjee C, Shi Y, Gelinas C, Fan Y, Nelson DA, Jin S, White E. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Ko A, Kanehisa A, Martins I, Senovilla L, Chargari C, Dugue D, Marino G, Kepp O, Michaud M, Perfettini JL, Kroemer G, Deutsch E. Autophagy inhibition radiosensitizes in vitro, yet reduces radioresponses in vivo due to deficient immunogenic signalling. Cell death and differentiation. 2014;21:92–99. doi: 10.1038/cdd.2013.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Palumbo S, Comincini S. Autophagy and ionizing radiation in tumors: the “survive or not survive” dilemma. Journal of cellular physiology. 2013;228:1–8. doi: 10.1002/jcp.24118. [DOI] [PubMed] [Google Scholar]
- 169.Lapaquette P, Guzzo J, Bretillon L, Bringer MA. Cellular and Molecular Connections between Autophagy and Inflammation. Mediators of inflammation. 2015;2015:398483. doi: 10.1155/2015/398483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. The New England journal of medicine. 2013;368:1845–1846. doi: 10.1056/NEJMc1303158. [DOI] [PubMed] [Google Scholar]
- 171.Yang Z, Goronzy JJ, Weyand CM. Autophagy in autoimmune disease. Journal of molecular medicine. 2015;93:707–717. doi: 10.1007/s00109-015-1297-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Rubinsztein DC, Marino G, Kroemer G. Autophagy and aging. Cell. 2011;146:682–695. doi: 10.1016/j.cell.2011.07.030. [DOI] [PubMed] [Google Scholar]
- 173.Fan J, Kou X, Jia S, Yang X, Yang Y, Chen N. Autophagy as a Potential Target for Sarcopenia. Journal of cellular physiology. 2016;231:1450–1459. doi: 10.1002/jcp.25260. [DOI] [PubMed] [Google Scholar]
- 174.Ding Y, Choi ME. Autophagy in diabetic nephropathy. The Journal of endocrinology. 2015;224:R15–30. doi: 10.1530/JOE-14-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Lavallard VJ, Gual P. Autophagy and non-alcoholic fatty liver disease. BioMed research international. 2014;2014:120179. doi: 10.1155/2014/120179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Stienstra R, Haim Y, Riahi Y, Netea M, Rudich A, Leibowitz G. Autophagy in adipose tissue and the beta cell: implications for obesity and diabetes. Diabetologia. 2014;57:1505–1516. doi: 10.1007/s00125-014-3255-3. [DOI] [PubMed] [Google Scholar]
- 177.Madrigal-Matute J, Cuervo AM. Regulation of Liver Metabolism by Autophagy. Gastroenterology. 2016;150:328–339. doi: 10.1053/j.gastro.2015.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Ren SY, Xu X. Role of autophagy in metabolic syndrome-associated heart disease. Biochimica et biophysica acta. 2015;1852:225–231. doi: 10.1016/j.bbadis.2014.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Orogo AM, Gustafsson AB. Therapeutic targeting of autophagy: potential and concerns in treating cardiovascular disease. Circulation research. 2015;116:489–503. doi: 10.1161/CIRCRESAHA.116.303791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Martinez-Lopez N, Athonvarangkul D, Singh R. Autophagy and aging. Advances in experimental medicine and biology. 2015;847:73–87. doi: 10.1007/978-1-4939-2404-2_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Dutta D, Calvani R, Bernabei R, Leeuwenburgh C, Marzetti E. Contribution of impaired mitochondrial autophagy to cardiac aging: mechanisms and therapeutic opportunities. Circulation research. 2012;110:1125–1138. doi: 10.1161/CIRCRESAHA.111.246108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.He Q, Han X. Cardiolipin remodeling in diabetic heart. Chemistry and physics of lipids. 2014;179:75–81. doi: 10.1016/j.chemphyslip.2013.10.007. [DOI] [PubMed] [Google Scholar]
- 183.Lee HJ, Mayette J, Rapoport SI, Bazinet RP. Selective remodeling of cardiolipin fatty acids in the aged rat heart. Lipids in health and disease. 2006;5:2. doi: 10.1186/1476-511X-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Petrosillo G, Matera M, Casanova G, Ruggiero FM, Paradies G. Mitochondrial dysfunction in rat brain with aging Involvement of complex I, reactive oxygen species and cardiolipin. Neurochemistry international. 2008;53:126–131. doi: 10.1016/j.neuint.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 185.Paradies G, Ruggiero FM, Petrosillo G, Quagliariello E. Age-dependent decline in the cytochrome c oxidase activity in rat heart mitochondria: role of cardiolipin. FEBS letters. 1997;406:136–138. doi: 10.1016/s0014-5793(97)00264-0. [DOI] [PubMed] [Google Scholar]
- 186.Nicolay K, de Kruijff B. Effects of adriamycin on respiratory chain activities in mitochondria from rat liver, rat heart and bovine heart. Evidence for a preferential inhibition of complex III and IV. Biochimica et biophysica acta. 1987;892:320–330. doi: 10.1016/0005-2728(87)90236-2. [DOI] [PubMed] [Google Scholar]
- 187.Sedlak E, Robinson NC. Phospholipase A(2) digestion of cardiolipin bound to bovine cytochrome c oxidase alters both activity and quaternary structure. Biochemistry. 1999;38:14966–14972. doi: 10.1021/bi9914053. [DOI] [PubMed] [Google Scholar]
- 188.Gomez B, Jr, Robinson NC. Phospholipase digestion of bound cardiolipin reversibly inactivates bovine cytochrome bc1. Biochemistry. 1999;38:9031–9038. doi: 10.1021/bi990603r. [DOI] [PubMed] [Google Scholar]
- 189.Ostrander DB, Zhang M, Mileykovskaya E, Rho M, Dowhan W. Lack of mitochondrial anionic phospholipids causes an inhibition of translation of protein components of the electron transport chain. A yeast genetic model system for the study of anionic phospholipid function in mitochondria. The Journal of biological chemistry. 2001;276:25262–25272. doi: 10.1074/jbc.M103689200. [DOI] [PubMed] [Google Scholar]
- 190.Zeczycki TN, Whelan J, Hayden WT, Brown DA, Shaikh SR. Increasing levels of cardiolipin differentially influence packing of phospholipids found in the mitochondrial inner membrane. Biochemical and biophysical research communications. 2014;450:366–371. doi: 10.1016/j.bbrc.2014.05.133. [DOI] [PubMed] [Google Scholar]
- 191.Barth PG, Scholte HR, Berden JA, Van der Klei-Van Moorsel JM, Luyt-Houwen IE, Van ’t Veer-Korthof ET, Van der Harten JJ, Sobotka-Plojhar MA. An X-linked mitochondrial disease affecting cardiac muscle, skeletal muscle and neutrophil leucocytes. Journal of the neurological sciences. 1983;62:327–355. doi: 10.1016/0022-510x(83)90209-5. [DOI] [PubMed] [Google Scholar]
- 192.Sparagna GC, Chicco AJ, Murphy RC, Bristow MR, Johnson CA, Rees ML, Maxey ML, McCune SA, Moore RL. Loss of cardiac tetralinoleoyl cardiolipin in human and experimental heart failure. Journal of lipid research. 2007;48:1559–1570. doi: 10.1194/jlr.M600551-JLR200. [DOI] [PubMed] [Google Scholar]
- 193.Saini-Chohan HK, Holmes MG, Chicco AJ, Taylor WA, Moore RL, McCune SA, Hickson-Bick DL, Hatch GM, Sparagna GC. Cardiolipin biosynthesis and remodeling enzymes are altered during development of heart failure. Journal of lipid research. 2009;50:1600–1608. doi: 10.1194/jlr.M800561-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Paradies G, Petrosillo G, Pistolese M, Di Venosa N, Federici A, Ruggiero FM. Decrease in mitochondrial complex I activity in ischemic/reperfused rat heart: involvement of reactive oxygen species and cardiolipin. Circulation research. 2004;94:53–59. doi: 10.1161/01.RES.0000109416.56608.64. [DOI] [PubMed] [Google Scholar]
- 195.Ott M, Robertson JD, Gogvadze V, Zhivotovsky B, Orrenius S. Cytochrome c release from mitochondria proceeds by a two-step process. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:1259–1263. doi: 10.1073/pnas.241655498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Petrosillo G, Ruggiero FM, Pistolese M, Paradies G. Reactive oxygen species generated from the mitochondrial electron transport chain induce cytochrome c dissociation from beef-heart submitochondrial particles via cardiolipin peroxidation. Possible role in the apoptosis. FEBS letters. 2001;509:435–438. doi: 10.1016/s0014-5793(01)03206-9. [DOI] [PubMed] [Google Scholar]
- 197.Watkins SM, Carter LC, German JB. Docosahexaenoic acid accumulates in cardiolipin and enhances HT-29 cell oxidant production. Journal of lipid research. 1998;39:1583–1588. [PubMed] [Google Scholar]
- 198.Schlame M, Kelley RI, Feigenbaum A, Towbin JA, Heerdt PM, Schieble T, Wanders RJ, DiMauro S, Blanck TJ. Phospholipid abnormalities in children with Barth syndrome. Journal of the American College of Cardiology. 2003;42:1994–1999. doi: 10.1016/j.jacc.2003.06.015. [DOI] [PubMed] [Google Scholar]
- 199.Xu Y, Condell M, Plesken H, Edelman-Novemsky I, Ma J, Ren M, Schlame M. A Drosophila model of Barth syndrome. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:11584–11588. doi: 10.1073/pnas.0603242103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Dudek J, Cheng IF, Balleininger M, Vaz FM, Streckfuss-Bomeke K, Hubscher D, Vukotic M, Wanders RJ, Rehling P, Guan K. Cardiolipin deficiency affects respiratory chain function and organization in an induced pluripotent stem cell model of Barth syndrome. Stem cell research. 2013;11:806–819. doi: 10.1016/j.scr.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 201.Xu Y, Sutachan JJ, Plesken H, Kelley RI, Schlame M. Characterization of lymphoblast mitochondria from patients with Barth syndrome. Laboratory investigation; a journal of technical methods and pathology. 2005;85:823–830. doi: 10.1038/labinvest.3700274. [DOI] [PubMed] [Google Scholar]
- 202.Chen S, He Q, Greenberg ML. Loss of tafazzin in yeast leads to increased oxidative stress during respiratory growth. Molecular microbiology. 2008;68:1061–1072. doi: 10.1111/j.1365-2958.2008.06216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.He Q, Harris N, Ren J, Han X. Mitochondria-targeted antioxidant prevents cardiac dysfunction induced by tafazzin gene knockdown in cardiac myocytes. Oxidative medicine and cellular longevity. 2014;2014:654198. doi: 10.1155/2014/654198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Myung SK, Ju W, Cho B, Oh SW, Park SM, Koo BK, Park BJ G. Korean Meta-Analysis Study. Efficacy of vitamin and antioxidant supplements in prevention of cardiovascular disease: systematic review and meta-analysis of randomised controlled trials. Bmj. 2013;346:f10. doi: 10.1136/bmj.f10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Kris-Etherton PM, Lichtenstein AH, Howard BV, Steinberg D, Witztum JL P.A. Nutrition Committee of the American Heart Association Council on Nutrition. Metabolism, Antioxidant vitamin supplements and cardiovascular disease. Circulation. 2004;110:637–641. doi: 10.1161/01.CIR.0000137822.39831.F1. [DOI] [PubMed] [Google Scholar]
- 206.Jerome-Morais A, Diamond AM, Wright ME. Dietary supplements and human health: for better or for worse? Molecular nutrition & food research. 2011;55:122–135. doi: 10.1002/mnfr.201000415. [DOI] [PubMed] [Google Scholar]
- 207.Jankovic J. Parkinson’s disease: clinical features and diagnosis. Journal of neurology, neurosurgery, and psychiatry. 2008;79:368–376. doi: 10.1136/jnnp.2007.131045. [DOI] [PubMed] [Google Scholar]
- 208.Ryan BJ, Hoek S, Fon EA, Wade-Martins R. Mitochondrial dysfunction and mitophagy in Parkinson’s: from familial to sporadic disease. Trends in biochemical sciences. 2015;40:200–210. doi: 10.1016/j.tibs.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 209.Deas E, Wood NW, Plun-Favreau H. Mitophagy and Parkinson’s disease: the PINK1-parkin link. Biochimica et biophysica acta. 2011;1813:623–633. doi: 10.1016/j.bbamcr.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Goedert M NEURODEGENERATION. Alzheimer’s and Parkinson’s diseases: The prion concept in relation to assembled Abeta, tau, and alpha-synuclein. Science. 2015;349:1255555. doi: 10.1126/science.1255555. [DOI] [PubMed] [Google Scholar]
- 211.Smith WW, Jiang H, Pei Z, Tanaka Y, Morita H, Sawa A, Dawson VL, Dawson TM, Ross CA. Endoplasmic reticulum stress and mitochondrial cell death pathways mediate A53T mutant alpha-synuclein-induced toxicity. Human molecular genetics. 2005;14:3801–3811. doi: 10.1093/hmg/ddi396. [DOI] [PubMed] [Google Scholar]
- 212.Stefanis L, Larsen KE, Rideout HJ, Sulzer D, Greene LA. Expression of A53T mutant but not wild-type alpha-synuclein in PC12 cells induces alterations of the ubiquitin-dependent degradation system, loss of dopamine release, and autophagic cell death. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2001;21:9549–9560. doi: 10.1523/JNEUROSCI.21-24-09549.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Orth M, Tabrizi SJ, Schapira AH, Cooper JM. Alpha-synuclein expression in HEK293 cells enhances the mitochondrial sensitivity to rotenone. Neuroscience letters. 2003;351:29–32. doi: 10.1016/s0304-3940(03)00941-8. [DOI] [PubMed] [Google Scholar]
- 214.Kamel F. Epidemiology. Paths from pesticides to Parkinson’s. Science. 2013;341:722–723. doi: 10.1126/science.1243619. [DOI] [PubMed] [Google Scholar]
- 215.Jackson-Lewis V, Przedborski S. Protocol for the MPTP mouse model of Parkinson’s disease. Nature protocols. 2007;2:141–151. doi: 10.1038/nprot.2006.342. [DOI] [PubMed] [Google Scholar]
- 216.Elbaz A, Clavel J, Rathouz PJ, Moisan F, Galanaud JP, Delemotte B, Alperovitch A, Tzourio C. Professional exposure to pesticides and Parkinson disease. Annals of neurology. 2009;66:494–504. doi: 10.1002/ana.21717. [DOI] [PubMed] [Google Scholar]
- 217.Yang W, Tiffany-Castiglioni E. Paraquat-induced apoptosis in human neuroblastoma SH-SY5Y cells: involvement of p53 and mitochondria. Journal of toxicology and environmental health. Part A. 2008;71:289–299. doi: 10.1080/15287390701738467. [DOI] [PubMed] [Google Scholar]
- 218.Anantharam V, Kaul S, Song C, Kanthasamy A, Kanthasamy AG. Pharmacological inhibition of neuronal NADPH oxidase protects against 1-methyl-4-phenylpyridinium (MPP+)-induced oxidative stress and apoptosis in mesencephalic dopaminergic neuronal cells. Neurotoxicology. 2007;28:988–997. doi: 10.1016/j.neuro.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Li N, Ragheb K, Lawler G, Sturgis J, Rajwa B, Melendez JA, Robinson JP. Mitochondrial complex I inhibitor rotenone induces apoptosis through enhancing mitochondrial reactive oxygen species production. The Journal of biological chemistry. 2003;278:8516–8525. doi: 10.1074/jbc.M210432200. [DOI] [PubMed] [Google Scholar]
- 220.Matsuda W, Furuta T, Nakamura KC, Hioki H, Fujiyama F, Arai R, Kaneko T. Single nigrostriatal dopaminergic neurons form widely spread and highly dense axonal arborizations in the neostriatum. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2009;29:444–453. doi: 10.1523/JNEUROSCI.4029-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Prensa L, Parent A. The nigrostriatal pathway in the rat: A single-axon study of the relationship between dorsal and ventral tier nigral neurons and the striosome/matrix striatal compartments. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2001;21:7247–7260. doi: 10.1523/JNEUROSCI.21-18-07247.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Bolam JP, Pissadaki EK. Living on the edge with too many mouths to feed: why dopamine neurons die. Movement disorders : official journal of the Movement Disorder Society. 2012;27:1478–1483. doi: 10.1002/mds.25135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Attwell D, Laughlin SB. An energy budget for signaling in the grey matter of the brain. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 2001;21:1133–1145. doi: 10.1097/00004647-200110000-00001. [DOI] [PubMed] [Google Scholar]
- 224.Pissadaki EK, Bolam JP. The energy cost of action potential propagation in dopamine neurons: clues to susceptibility in Parkinson’s disease. Frontiers in computational neuroscience. 2013;7:13. doi: 10.3389/fncom.2013.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Yla-Anttila P, Vihinen H, Jokitalo E, Eskelinen EL. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy. 2009;5:1180–1185. doi: 10.4161/auto.5.8.10274. [DOI] [PubMed] [Google Scholar]
- 226.Vance JE. MAM (mitochondria-associated membranes) in mammalian cells: lipids and beyond. Biochimica et biophysica acta. 2014;1841:595–609. doi: 10.1016/j.bbalip.2013.11.014. [DOI] [PubMed] [Google Scholar]
- 227.de Brito OM, Scorrano L. Mitofusin-2 regulates mitochondrial and endoplasmic reticulum morphology and tethering: the role of Ras. Mitochondrion. 2009;9:222–226. doi: 10.1016/j.mito.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 228.Bann DV, Parent LJ. Application of live-cell RNA imaging techniques to the study of retroviral RNA trafficking. Viruses. 2012;4:963–979. doi: 10.3390/v4060963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Wiegant J, Brouwer AK, Tanke HJ, Dirks RW. Visualizing nucleic acids in living cells by fluorescence in vivo hybridization. Methods in molecular biology. 2010;659:239–246. doi: 10.1007/978-1-60761-789-1_17. [DOI] [PubMed] [Google Scholar]