Abstract
The complete mitochondrial genome of Glycera chirori Izuka (Annelida: Polychaeta) was presented, which is a circular molecule of 15,930 bp nucleotides. It encodes 37 genes, including 13 PCGs, 22 tRNAs, and two rRNAs. The length of non-coding regions is 1428 bp, and the longest one (1346 bp) is speculated as the control region, which is located between trnA and trnL2 and is longer than most species in Glycera. The complete mitogenome of G. chirori Izuka consists of 31.2% A, 23.6% C, 12.9% G, and 32.2% T, which has T vs. A skew (−0.02) and C vs. G skew (−0.29), respectively. Phylogenetic analysis indicates the classification status of G. chirori Izuka and the relationship with other species in Glycera, which is closer with Glycera unicornis and Glycera fallax (bootstrap = 100). By comparisons, the gene arrangement of G. chirori Izuka and other seven species in Glycera are identical and they also cluster together in phylogenetic tree with higher support rate, which indicates the conservativeness between gene arrangement and phylogenetic analysis in Glycera. In conclusion, the complete mitochondrial genome of G. chirori Izuka can provide supportive data for further molecular and evolutionary analysis of Glycera.
Keywords: Glycera chirori Izuka, mitochondrial genome, gene arrangement, phylogeny
Glycera is one of three genera in Glyceridae and is also the most species-rich genus at present (Richter et al. 2015). There are some reports on this genus, which involved geographical distribution, biological activity, and molecular basis (Schüller 2011; von Reumont et al. 2014; Richter et al. 2015). Glycera chirori Izuka (Annelida: Polychaeta: Glyceridae) is one of the most common species in Glyceridae (Sun and Yang 1994), distributing in China's coastal water and Japan's sea area and living in the intertidal and subtidal zones (Wang and Song 2017).
The specimen of G. chirori Izuka was collected from intertidal of Multi-Island Sea, Lanshan District, Rizhao, Shandong Province, China (N: 30.71, E: 122.78). The total DNA was stored at Marine Museum of Jiangsu Ocean University (Accession number: Gchi-002). We used ultrasonic to interrupt long DNA fragments to 2–3 kb to build a library and get the complete mitochondrial genome sequence by sequencing and assembly with SeqMan 7.1.0 software (Swindell and Plasterer 1997). Gene annotation was obtained with MITOS (Bernt et al. 2013) and tRNAscan-SE (Chan and Lowe 2019).
The total length of mitogenome of G. chirori Izuka is 15,930 bp, which is a circular molecule, encoding 13 PCGs, two rRNA, and 22 tRNA genes on one strand (GenBank accession number: MK858188) (Boore 1999; Shen et al. 2009). AT and GC skews of the whole genome are −0.02 and −0.29, respectively (Perna and Kocher 1995). The length of all non-coding regions is 1428 bp. The Tandem Repeats Finder (Benson 1999) was used to speculate the control region (1346 bp), locating between trnA and trnL2, which is longer than most species in Glycera (Richter et al. 2015).
Gene arrangement was a useful tool to elucidate the evolution and phylogenetic relationship between homologous species (Boore 1999; Weigert et al. 2016). Compared with the other 12 species of Glycera in the GenBank database, the gene arrangement of G. chirori Izuka is identical in 13 PCGs and two rRNA genes and this arrangement pattern (15 genes) is consistent with most species in Annelida (Jennings and Halanych 2005; Zhong et al. 2008; Weigert et al. 2016). Besides, only a few tRNA genes show translocation for their higher variability (Weigert et al. 2016).
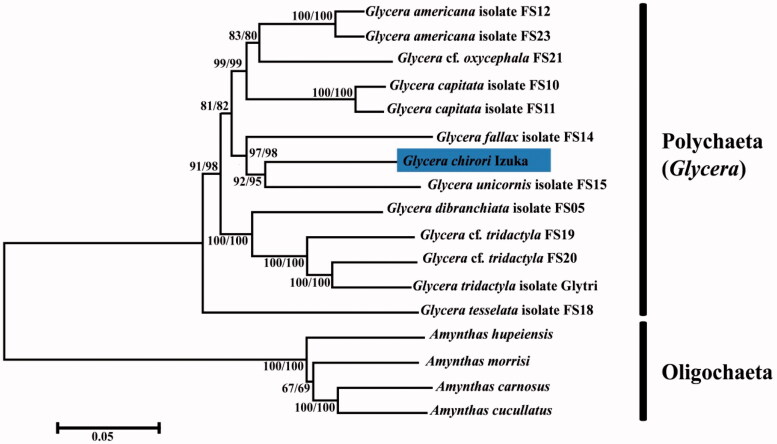
We used MEGA 7.0.25 (Kumar et al. 2016) to construct phylogenetic trees based on neighbour-joining (NJ) and maximum-likelihood (ML) methods, using 13 PCGs’s amino acid data from 13 species (Glycera) and four Oligochaeta species (outgroup) (Figure 1). As result showed, G. chirori Izuka clusters with Glycera unicoins isolated FS15 (BP = 92/95), and the two species cluster with Glycera fallax isolated FS14 (BP = 97/98). And then this group is clustered with the other two groups from other five species with high support rate (BP = 99). Significantly, G. chirori Izuka and other six species grouped on phylogenetic tree share the same gene arrangement, which indicated that gene arrangement and phylogenetic tree in Glycera complement each other. Thus, G. chirori Izuka could be a valid support to reflect the conservativeness between gene arrangement and phylogenetic analysis in Glycera.
Figure 1.
Phylogenetic trees constructed from neighbour-joining (NJ) and maximum-likelihood (ML) methods of 13 PCGs (amino acid data). The numerical values at the node represent the bootstrap value from ML and NJ methods, respectively.
The accession numbers of the genomes used for comparison were KT989321 (G. americana isolate FS12), KT989330 (G. americana isolate FS23), KT989329 (G cf. oxycephola FS21), KT989319 (G. capitata isolate FS10), KT989320 (G. capitata isolate FS11), KT989323 (G. fallax isolate FS14), KT989324 (G. unicornis isolate FS15), KT989318 (G. dibranchiata isolate FS15), KT989327 (G cf. tridactyla FS19), KT989328 (G cf. tridactyla FS20), KT989331 (G. tridactyla isolate Glytri), KT989326 (G. tesselata isolate FS18), NC_029864 (Amynthas hupeiensis), NC_029865 (Amynthas morrisi), NC_029863 (Amynthas carnosus), and NC_029866 (Amynthas cucullatus).
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27:573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319. [DOI] [PubMed] [Google Scholar]
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27:1767–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan PP, Lowe TM. 2019. tRNAscan-SE: searching for tRNA genes in genomic sequences. Methods Mol Biol. 1962:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings RM, Halanych KM. 2005. Mitochondrial genomes of Clymenella torquata (Maldanidae) and Riftia pachyptila (Siboglinidae): evidence for conserved gene order in Annelida. Mol Biol Evol. 22:210–222. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perna NT, Kocher TD. 1995. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J Mol Evol. 41:353–358. [DOI] [PubMed] [Google Scholar]
- Richter S, Schwarz F, Hering L, BÖggemann M, Bleidorn C. 2015. The utility of genome skimming for phylogenomic analyses as demonstrated for Glycerid relationships (Annelida, Glyceridae). Genome Biol Evol. 7:3443–3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüller M. 2011. Evidence for a role of bathymetry and emergence in speciation in the genus Glycera (Glyceridae, Polychaeta) from the deep Eastern Weddell Sea. Polar Biol. 34:549–564. [Google Scholar]
- Shen X, Ma XY, Ren JF, Zhao FQ. 2009. A close phylogenetic relationship between Sipuncula and Annelida evidenced from the complete mitochondrial genome sequence of Phascolosoma esculenta. BMC Genomics. 10:136–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun RP, Yang DJ. 1994. Common marine Polychaeta in China (in Chinese). B Biol. 29:7–9. [Google Scholar]
- Swindell SR, Plasterer TN. 1997. SEQMAN. Contig assembly. Methods Mol Biol. 70:75–89. [PubMed] [Google Scholar]
- von Reumont BM, Campbell LI, Richter S, Hering L, Sykes D, Hetmank J, Jenner RA, Bleidorn R. 2014. A Polychaete’s powerful punch: venom gland transcriptomics of Glycera reveals a complex cocktail of toxin homologs. Genome Biol Evol. 6:2406–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang MJ, Song XK. 2017. Atlas of biodiversity of marine protected areas in Shandong Sea area of Bohai Sea (in Chinese). Beijing: Navy Press; p. 44–45. [Google Scholar]
- Weigert A, Golombek A, Gerth M, Schwarz F, Struck TH, Bleidorn C. 2016. Evolution of mitochondrial gene order in Annelida. Mol Phylogenet Evol. 94:196–206. [DOI] [PubMed] [Google Scholar]
- Zhong M, Struck TH, Halanych KM. 2008. Phylogenetic information from three mitochondrial genomes of Terebelliformia (Annelida) worms and duplication of the methionine tRNA. Gene. 416:11–21. [DOI] [PubMed] [Google Scholar]