Abstract
The complete chloroplast genome sequence of Yunnanopilia longistaminea, an endemic species in southwest China, is presented in this study. The total genome size of Y. longistaminea was 148,503 bp in length, with a typical quadripartite structure including a pair of inverted repeat (IRs, 28,075 bp) regions separated by a large single copy (LSC, 84,547bp) region and a small single copy (SSC, 7805 bp) region. The all GC content was 37.3%. The genome contains 117 genes, including 72 protein-coding genes, 37 tRNA genes, and 8 rRNA genes, 13 genes contain a single intron, and 3 genes have two introns. Further, a maximum-likelihood (ML) phylogenetic tree results that Y. longistaminea was closely related to the genera of Champereia manillana.
Keywords: Yunnanopilia longistaminea, endemic species, chloroplast genome, phylogenetic analysis
Yunnanopilia longistaminea is a tropical evergreen woody plant belongs to the genus Opiliaceae in the family Yunnanopilia which mainly distributed in southwest China (Pu 2014). It is especially valued for nutrition and medicine because the leaves are rich in Vitamin C, iditol and potassium (Xu et al. 2014). It is well known as an edible vegetable by the local ethnic minorities. However, due to its Small reserves, narrow distribution (Yang 2013) and the excessive excavation of people, it is already facing the danger of extinction in the wild (Shi 2011).
Chloroplast genomes are widely used for phylogeny (Xue et al. 2012), DNA barcoding (Dong et al. 2014), genome evolution and species conservation (Dong et al. 2013). Up to now, The chloroplast genome of many species has been reported, such as Champereia manillana (Yang et al. 2017), Zanthoxylurm bungeanum (Liu and Wei 2017) and Fabaceae (Kato et al. 2000). In this study, we reported the complete chloroplast genome sequence of Y. longistaminea based on the Illumina pair-end sequencing data.
Leaf samples of Y. longistaminea were collected from Puer, Yunnan, China (geospatial coordinates: 101°49′35″E, 23°58′12″N; altitude: 1046 m). The Total DNA samples (YL 1-3) was extracted with Magnetic beads plant genomic DNA preps Kit (TSINGKE Biological Technology, Beijing, China) and kept at the Key Laboratory for Forest Genetic and Tree Improvement and Propagation in Universities of Yunnan Province, Southwest Forestry University, Kunming, China. Total DNA was used the Illumina Hiseq X platform to sequenced. Approximately 5.73 GB of raw data were genomic paired-end library of 150 bp length. Then, the raw data were used to assemble the complete Cp genome made by the software of GetOrganelle (Jin et al. 2018) with Champereia manillana as the reference. Genome annotation was performed with the programme Geneious R8 (Biomatters Ltd, Auckland, New Zealand), and then manually confirmed by comparison with the homologues Champereia manillana. The CpDNA sequence of Y. longistaminea was submitted to GenBank (accession number: MN444856).
The complete Cp genome of Y. longistaminea is 148,503 bp in size, which comprising a large single copy (LSC) region with a size of 84,547 bp, a small single copy (SSC) region of with a size of 7805 bp, separated by a pair of inverted repeats of 28,075 bp. The GC content of all Y. longistaminea Cp genome is 37.3%, while the GC contant of LSC was 35.2%, and the corresponding values of SSC and IR regions are 27.8% and 41.9%, respectively. A total of 117 function genes are contained in the chloroplast genome of Y. longistaminea, including 72 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. 5 protein-coding genes, 8 tRNA genes, and all of rRNA genes were duplicated in IR regions. Among these function genes, 13 genes (rpoC1, rpl2, petB, rpl16, atpF, petD, rps16, trnK-UUU, trnI-GAU, trnA-UGC, trnG-UCC, trnV-UAC, trnL-UAA.) have a single intron and three genes (rps12, ycf3, and clpP) contain two introns.
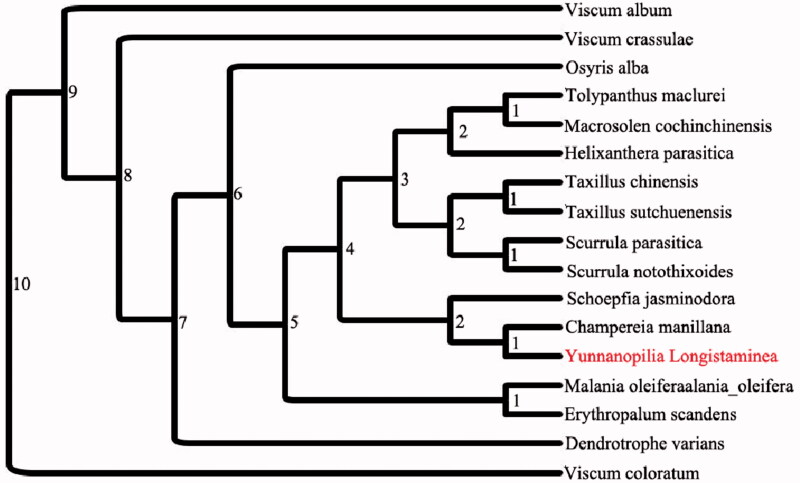
To determine the phylogenetic location of Y. longistaminea, maximum-likelihood (ML) phylogenetic tree was reconstructed within the family of Santalales with fully sequenced chloroplast genomes, 16 complete chloroplast genome of Santalales were obtained from GenBank, the plastomes of Viscum coloratum in the family of Santalales were used as out-group. The chloroplast genome sequences of Santalales were aligned using MAFFT version 7 software (Katoh and Standley 2013) to reconstruct the phylogeny of the family of Santalales. The ML method for phylogenetic was reconstructed by using RA × ML version 8 programme with 1000 bootstrap replicates which based on TVM + F + I + G4 model (Stamatakis 2014). The phylogenetic tree revealed that Y. longistaminea was closely related to Champereia manillana (Figure 1). Moreover, the chloroplast genome of Y. longistaminea will provide useful information for phylogenetic studies and genomic resources available of Opiliaceae species.
Figure. 1.
Phylogenetic relationships among 16 complete chloroplast genomes of Santalales. Bootstrap support values are given at the nodes. Chloroplast genome accession number used in this phylogeny analysis: Viscum album: KT003925.1; Viscum crassulae: KT070881.1; Osyris alba: KT070882.1; Dendrotrophe varians: MF592987.1; Tolypanthus maclurei: MH922027.1; Malania oleifera: NC_039426; Schoepfia jasminodora: NC_034228.1; Champereia manillana: NC-034931.1; Viscum coloratum: NC-035414.1; Taxillus chinensis: NC-036306.1; Taxillus sutchuenensis: NC_036307.1; Erythropalum scandens: NC_036759.1; Helixanthera parasitica: NC_039375.1; Macrosolen cochinchinensis: NC_039376.1; Scurrula parasitica: NC_040862.1; Scurrula notothixoides: NC_041305.1.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Dong W, Liu H, Xu C, Zuo Y, Chen Z, Zhou S. 2014. A chloroplast genomic strategy for designing taxon specific DNA mini-barcodes: a case study on ginsengs. BMC Genet. 15:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong W, Xu C, Cheng T, Zhou S. 2013. Complete chloroplast genome of Sedum sarmentosum and chloroplast genome evolution in Saxifragales. PLoS One. 8:e77965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for de novo assemble of a complete circular chloroplast genome using genome skimming data. BioRxiv [in Preprint]. doi: 10.1101/256479 [DOI] [Google Scholar]
- Kato T, Kaneko T, Sato S, Nakamura Y, Tabata S. 2000. Complete structure of the chloroplast genome of a legume, Lotus japonicus. DNA Res. 7:323–330. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YL, Wei AZ. 2017. The complete chloroplast genome sequence of an economically important plant, Zanthoxylum bungeanum (Rutaceae). Conserv Genet Resour. 9:25–27. [Google Scholar]
- Pu YM. 2014. To explore how to cultivate Melientha longistaminea. TAS Technol. 37:40–42. [Google Scholar]
- Shi N. 2011. Study on tissue culture of Melientha longistaminea. Yunnan Agric Sci Tech. 2:14–17. [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu LP, Tang HY, Jia P, Li Q, Liu Y, Zhang JZ. 2014. Research advances in Yunnanopilia longistaminea of Opiliaceae in Yunnan. Chin Wild Plant Resour. 33:44–46. [Google Scholar]
- Xue JH, Dong WP, Cheng T, Zhou SL. 2012. Nelumbonaceae: systematic position and species diversification revealed by the complete chloroplast genome. J Syst Evol. 50:477–487. [Google Scholar]
- Yang CB. 2013. Excellent cultivation technique of woody vegetable Melientha longistaminea. Yunnan Forestry. 34:65. [Google Scholar]
- Yang GS, Wang YH, Wang YH, Shen SK. 2017. The complete chloroplast genome of a vulnerable species Champereia manillana (Opiliaceae). Conserv Genet Resour. 9:415–418. [Google Scholar]