Abstract
We assembled and annotated the first complete mitochondrial genome of a species from the subfamily Microweiseinae, Coccidophilus cariba Gordon, a predator of scale insect pest. The circular mitogenome consists of 15,343 bp in length, including 13 PCGs, 22 tRNA, and 2 rRNA genes, and exhibits the typical ladybird mitogenome structure. A phylogenetic analysis with the published mitogenomes of other 12 ladybirds is presented, which confirms the position of Microweiseinae as a sister group of Coccinellinae. The C. cariba mitogenome could be a useful source to future studies in the relationships inside the subfamily and the genus.
Keywords: Illumina, ladybird, Microweiseinae, mitogenome, phylogeny
Coccinellidae comprises around 360 genera and 6000 species (Vandenberg 2002) and has been recently divided into the subfamilies Microweiseinae and Coccinellinae (Seago et al. 2011). To date, 28 mitochondrial genomes, complete or partial have been published in Genbank, all belonging to the Coccinellinae. The Microweiseinae Coccidophilus cariba Gordon is a scale predator and is the only member of Coccidophilus living in the West Indies (Gordon 1978).
The specimen sequenced was collected in Guaeloupe in 1997 (Ravine Chaude, GP 1417) and a voucher was deposited in the MNHN (MNHN-EC-7788). DNA was extracted from the whole specimen, following the non-destructive protocol described in Gilbert et al. (2007), and then quantified using a Qubit fluorometer (Life Technologies, Paisley, UK). Genomic DNA was indexed and libraries prepared using the NEBNext Ultra II library prep kit (New England BioLabs; NEB), with a modified version of Meyer and Kircher (2010) protocol. DNA was sequenced at the Genome and Transcriptome Platform of Toulouse (Genotoul, France) using Illumina HiSeq 3000 technology (150 bp paired‐end). Quality and length distribution of the sequences were inspected prior and post-clean using FASTQC (Andrews 2010). Low-quality reads and adaptor contamination were then trimmed using the BBDuk plugin as implemented in Geneious prime 2019.1.3 (Biomatters Ltd., Auckland, New Zealand). To separate the mitochondrial genome sequences from the rest of the sequence data, an iterative read mapping strategy was employed using Geneious workflow Align/Assemble ‘Map to Reference’ and the mitochondrial sequence of Henosepilachna pusillanima as reference (Geneious mapper, custom sensitivity with a maximum mismatch of 30%, fine-tuning 25 times). We repeat this operation with a mismatch of 10% and we conclude with a final de novo step (maximum mismatch of 30%). 7,514 overlapped reads (mean 109.1 bp) were used to generate a consensus sequence and to create a circular molecule. We obtain a mitogenome of 15,343 bp long, with a read coverage of 53×. The identity and position of 13 PCGs, 22 tRNA, and 2 rRNA genes were determined using the MITOS web server (Bernt et al. 2013; http://mitos.bioinf.uni-leipzig.de), in combination with the annotation with the mitogenome of Henosepilachna pusillanima in Geneious. The gene order is similar to other Coccinellidae mitochondrial genomes. The mitochondrial genome was submitted to GenBank (accession number # MN447521) with the submission tool implemented in Geneious.
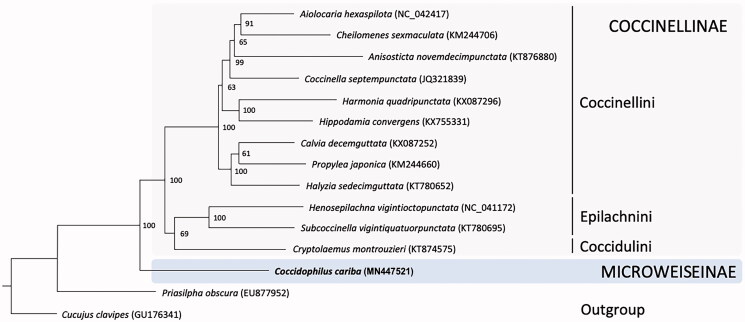
The phylogenetic position of C. cariba was inferred from 12 mitochondrial genome sequences of Coccinellidae available in Genbank, with Priasilpha obscura (Priasilphidae) and Cucujus clavipes (Cucujidae) as outgroups. The 13 PCGs and the 2 rRNA was extracted from each genome, aligned separately with the MAFFT algorithm (Katoh and Standley 2013) implemented in MEGA version X (Kumar et al. 2018), then concatenated. Phylogenetic analysis was performed with using Iq-Tree 1.5 (Nguyen et al. 2015) as implemented in the W-Iq-Tree web server (http://iqtree.cibiv.univie.ac.at; Trifinopoulos et al. 2016). All PCGs and rRNA alignment were partitioned, the models of substitution were automatically selected, and an ultrafast bootstrap with 1,000 iterations was performed. The resulting topology (Figure 1) confirms the position of Microweiseinae as sister group of Coccinellinae (Seago et al. 2011; Robertson et al. 2015).
Figure 1.
Phylogenetic relationships of Coccinellidae, based on 13 mitochondrial genomes, reconstructed with Iq-Tree 1.5 as implemented on the W-Iq-Tree web server. Ultrafast bootstrap support from 1000 replicates is indicated at the right of each node. For each species is showed the GenBank accession number of the mitochondrial genome.
Acknowledgments
The authors thank to the ‘Service de Systématique Moléculaire, SSM’ (UMS 2700 CNRS-MNHN) where the molecular work was realised. We are grateful to Pascaline CHIFFLET-BELLE for technical support in the laboratory phase, the staff of SSM for the support in the molecular data, and Tony ROBILLARD for his advice on improving the manuscript. The financial support in the doctoral studies to KS was provided by the government agency Colciencias-Colombia (program number 756‐2016).
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data [accessed 2019 July 2019]. http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319. [DOI] [PubMed] [Google Scholar]
- Gilbert MTP, Moore W, Melchior L, Worobey M. 2007. DNA extraction from dry museum beetles without conferring external morphological damage. PLoS One. 2:e272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon RD. 1978. West Indian Coccinellidae II (Coleoptera): some scale predators with keys to genera and species. Coleopts Bull. 32:205–218. [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT Multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M, Kircher M. 2010. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb Protoc. 2010:pdb.prot5448. [DOI] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson JA, Ślipiński A, Moulton M, Shockley FW, Giorgi A, Lord NP, Mckenna DD, Tomaszewska W, Forrester J, Miller KB, et al. 2015. Phylogeny and classification of Cucujoidea and the recognition of a new superfamily Coccinelloidea (Coleoptera: Cucujiformia). Syst Entomol. 40:745–778. [Google Scholar]
- Seago AE, Giorgi JA, Li J, Ślipiński A. 2011. Phylogeny, classification and evolution of ladybird beetles (Coleoptera: Coccinellidae) based on simultaneous analysis of molecular and morphological data. Mol Phylogenet Evol. 60:137–151. [DOI] [PubMed] [Google Scholar]
- Trifinopoulos J, Nguyen LT, von Haeseler A, Minh BQ. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44:W232–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenberg NJ. 2002. Coccinellidae Latreille 1807 Vol. 2 In: Arnett RH Jr., Thomas MC, Skelley PE, Frank JH, editors. American Beetles. Polyphaga: Scarabaeoidea through Curculionoidea. Boca Raton (FL): CRC Press; p. 371–389. [Google Scholar]