Abstract
Euphorbia helioscopia Linn. known as a traditional Chinese medicine of Euphorbiaceae, which contains terpenes, steroids, flavonoids, acetophenones, tannins, phenylpropanoids, cerebrosides and so on. Euphorbia helioscopia L. was used to treat malignant tumors and chronic obstructive pulmonary diseases such as cough, phlegm-turbidity, asthma, and chronic bronchitis. The complete chloroplast genome was assembled by Illumina paired-end reads data. The length of circular cp genome distribution in 160,041 bp, containing a large single-copy region (LSC) of 88,832 bp, a small single-copy region (SSC) of 17,145 bp and a pair of inverted repeat (IR) regions of 27,032 bp. In addition, 11 genes possess a single intron, while the other two genes (ycf3, clpP) have a couple of introns. The GC content of entire Euphorbia helioscopia L. cp genome, LSC, SSC and IR regions are 35.9, 33.1, 30.3, and 42.3%, respectively. From the NJ phylogenetic tree analysis showed that Euphorbia helioscopia L. and Euphorbia esula are closely related to each other within the family Euphorbiaceae.
Keywords: Euphorbia helioscopia, Euphorbiaceae, chloroplast genome, Illumina sequencing, phylogenetic analysis
Euphorbia helioscopia Linn. is a traditional Chinese medicine of Euphorbiaceae, which contains terpenes (iridoids, sesquiterpenoids, diterpenoids, triterpenoids), steroids, flavonoids, acetophenones, tannins, phenylpropanoids, cerebrosides and so on (Shi et al. 2008; Wang et al. 2018). Euphorbia helioscopia L. was used to treat malignant tumors and chronic obstructive pulmonary diseases such as cough, phlegm-turbidity, asthma and chronic bronchitis (Chen et al. 1979; Cheng et al. 2015). Modern pharmacological research shows that Euphorbia diterpenoids have anti-cancer effects (Wang et al. 2018).
The complete chloroplast (cp) genome is a conserved structure of four parts, the four parts as follows, a pair of inverted repeats (IRs), separated by a large single-copy region (LSC) and a small single-copy region (SSC) (Wolfe et al. 1992; Lee et al. 2007). Our study will be very important for futher studying the phylogenetic relationships between Euphorbia helioscopia L. and Euphorbiaceae.
The fresh leaves of Euphorbia helioscopia L. were collected in the Botanical Garden of Northwest University (34°16′N, 108°54′E; Shaanxi, NW China). A voucher specimen (FS190321) was deposited at the structural plant laboratory in International University. We used the modified CTAB method to extract the genomic DNA (Doyle and Doyle 1987) and then followed the manufacturer’s specification of Illumina HiSeq X Ten Sequencing System (Illumina, San Diego, CA) to construct a shotgun library.
The Euphorbia esula (GenBank: KY000001) was used as the initial reference to assemble cp genome with the program MITObim v 1.8 (https://github.com/chrishah/MITObim) (Hahn et al. 2013). Based on the web-based tool OGDRaw v1.2 (http://ogdraw.mpimp-golm.mpg.de/) to generate the complete cp genome (Lohse et al. 2013). The complete cp genome sequence has been submitted to GenBank (accession number MNI 199031).
The complete cp genome is a circular double-stranded DNA molecule with a typical quadripartite structure, including a pair of IRs, an LSC region, and an SSC region. Based on the sequencing results, we got 33,132,728 raw Paired-End Reads, the length distribution in 160,041 bp (GC content accounts for 35.9%). In addition, the length of LSC region, SSC region and IRs regions were distributed as 88,832 bp (GC, 33.1%), 17,145 bp (GC, 30.3%), and 27,032 bp (GC, 42.3%), respectively. It encodes 113 complete genes, containing 77 protein-coding genes, 32 transfer RNA genes, and 4 ribosomal RNA genes. 5 tRNA genes (tRNA-Ala, -Ile, -Leu, -Lys, and -Val) harbor a single intron. AtpF, ndhA, ndhB, rps12, rpl2, and rpoC1, these 6 PCG genes possess a single intron, clpP and ycf3 harbor two introns, while, 69 PCG genes no intron.
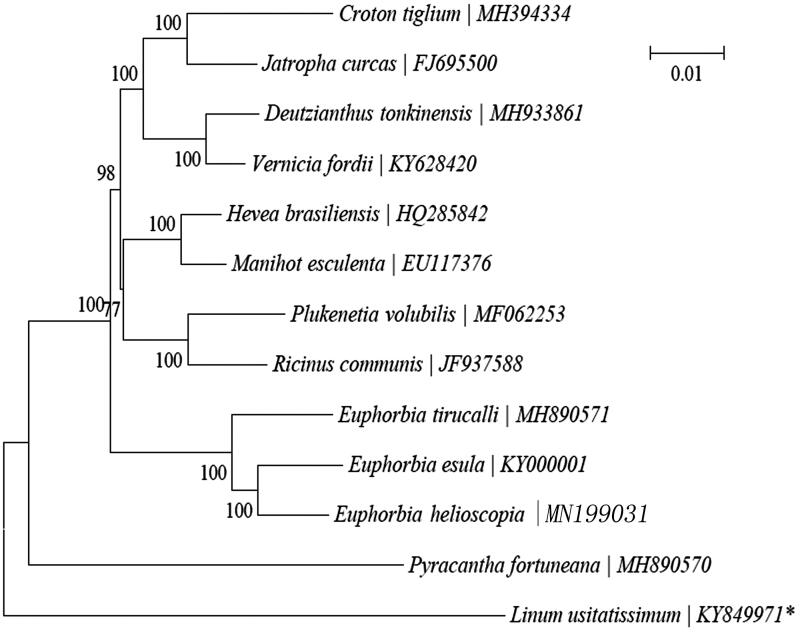
To study the phylogenetic position of Euphorbia helioscopia L. we constructed a neighbour-joining (NJ) phylogenetic tree (Figure 1) through MEGA7 with 1000 bootstrap replicates (http://www.megasoftware.net/) based on the concatenated coding sequences of 13 chloroplast PCGs for 12 plastid genomes from published species of Euphorbiaceae (Kumar et al. 2016). From Figure 1, we find that Euphorbia helioscopia L. (MNI199031) is closely related to Euphorbia esula (GenBank: KY000001).
Figure 1.
Phylogeny of 13 species within the order Euphorbia based on the neighbour-joining (NJ) analysis of chloroplast PCGs. The bootstrap values were based on 1000 resamplings and are placed next to the branches. *represents the outgroup of Euphorbia.
Disclosure statement
The authors report no conflicts of interest and are solely responsible for the content and writing of this paper.
References
- Chen Y, Tang ZJ, Jiang FX, Zhang XX, Lao AN. 1979. Studies on the active principles of Ze-Qi (Euphorbia helioscopia L.), a drug used for chronic bronchitis (I). Acta Pharm Sin. 14:91–95. [PubMed] [Google Scholar]
- Cheng J, Han W, Wang Z, Shao Y, Zhang Y, Xu X, Zhang Y, Wang Y, Li Z. 2015. Hepatocellular carcinoma growth is inhibited by Euphorbia helioscopia L. extract in nude mice xenografts. Biomed Res Int. 35:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15. [Google Scholar]
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads: a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HL, Jansen RK, Chumley TW, Kim KJ. 2007. Gene relocations within chloroplast genomes of Jasminum and Menodora (Oleaceae) are due to multiple, overlapping inversions. Mol Biol Evol. 24(5):1161–1180. [DOI] [PubMed] [Google Scholar]
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. Organellar Genome DRAW-a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucl Acids Res. 41(W1):W575–W581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi QW, Su XH, Kiyota H. 2008. Chemical and pharmacological research of the plants in genus Euphorbia. Chem Rev. 108(10):4295–4327. [DOI] [PubMed] [Google Scholar]
- Wang WP, Jiang K, Zhang P, Shen KK, Qu AJ, Yu AP, Tan CH. 2018. Highly oxygenated and structurally diverse diterpenoids from Euphorbia helioscopia. Phytochemistry. 145:93–102. [DOI] [PubMed] [Google Scholar]
- Wolfe KH, Morden CW, Ems SC, Palmer JD. 1992. Rapid evolution of the plastid translational apparatus in a nonphotosynthetic plant: loss or accelerated sequence evolution of tRNA and ribosomal protein genes. J Mol Evol. 35(4):304–317. [DOI] [PubMed] [Google Scholar]