Abstract
Bupleurum chinense DC. is a traditional medicinal herb species widely distributed in most provinces of China. In this study, we constructed and annotated a complete circular chloroplast (cp) genome of B. chinense. The cp genome of B. chinense is 155,545 bp in length, including two inverted repeat (IR) regions of 26,305 bp, separated by a large single-copy (LSC) region of 85,430 bp and a small single-copy (SSC) region of 17,505 bp. The GC content of whole cp genome is 37.68%. The genome contains 113 different genes, including 79 protein-coding genes, 30 tRNA genes, and 4 rRNA genes. A maximum-likelihood phylogenomic analysis showed that Bupleurum formed a monophyletic group, and was sister to other groups of Apiaceae.
Keywords: Bupleurum chinense, chloroplast genome, phylogeny
Bupleurum chinense DC. is a perennial plant in the Apiaceae family (Lin et al. 2008). The genus Bupleurum includes approximately 200 species, many of which have been pharmaceutically used for thousands of years, mainly in Asia and Europe (Pan 2006). In addition to the authentic species of B. chinense, there are more than 20 other species in Bupleurum also habitually utilized as Bupleuri Radix in some local areas. Knowing the high demand for Bupleuri Radix and the diversity of species that can be – both rightly and wrongly – used for this herb, the resources of B. chinense are very scarce (Liu et al. 2011). Here, we assembled the complete chloroplast (cp) genome sequence of B. chinense based on the next-generation sequencing method. The primary purpose of this study was to analyze the structure of the complete cp genome of B. chinense and to resolve the phylogenetic relationships of B. chinense and other species in Apiaceae.
Fresh samples of B. chinense was collected from Zhashui county (Shaanxi province, China; 109°20′27. 24″E, 33°48′13. 68″N). Voucher specimen was stored in the herbarium of Institute of Chinese Materia Medica (CMMI), China Academy of Chinese Medical Sciences. The specimen voucher number is 611026LY0126. Total genomic DNA was extracted from the dry materials using the mCTAB approach (Li et al. 2013).
Using the Illumina HiSeq Xten platform, a 400 bp (insertion size) double-ended library was constructed by splicing DNA. The readings from the paired-end were qualitatively assessed and assembled with SPAdes version 3.13.1 (Bankevich et al. 2012). Plann is used for initial annotation, Sequin for corrections (Huang and Cronk 2015). The complete cp genomic sequence annotated had been submitted to GenBank under the accession number of MN337347 for B. chinense.
The structure of cp genome of B. chinense was circular, and the size was 155,545 bp. It included two inverted repeat (IR) regions of 26,305 bp, separated by a large single-copy (LSC) region of 85,430 bp and a small single-copy (SSC) region of 17,505 bp. The whole cp genome GC content is 37.68%. There are 113 different genes in the cp genome of B. chinense, including 79 protein-coding genes, 30 tRNA genes, and 4 rRNA genes.
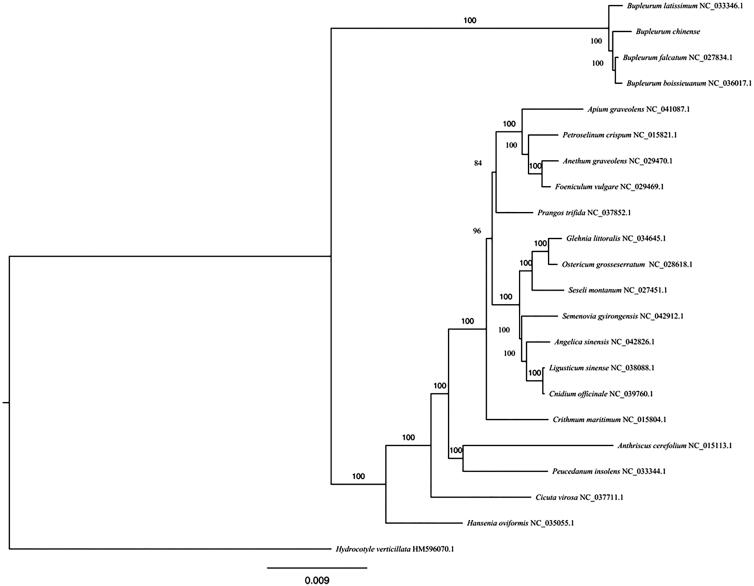
We chose 21 species within the family Apiaceae and Hydrocotyle verticillata (Araliaceae) as an outgroup to establish a phylogenetic tree by the maximum likelihood (ML) analysis using RAxML with 1000 bootstrap replicates (Stamatakis 2014). The phylogenetic tree showed that B. chinense and other species of Bupleurum formed a monophyletic group, and was sister to other groups of Apiaceae (Figure 1). The complete cp genome of B. chinense provided a lot of genetic information for species conservation and identification as well as the phylogenetic studies of Apiaceae.
Figure 1.
The best ML phylogeny recovered from 22 complete chloroplast sequences by RAxML. Bootstrap support values are given at each node.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DI, Cronk Q. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3:1500026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wang S, Jing Y, Wang L, Zhou S. 2013. A modified CTAB protocol for plant DNA extraction. Chinese Bull Bot. 48:72–78. [Google Scholar]
- Lin WY, Chen LR, Lin TY. 2008. Rapid authentication of Bupleurum species using an array of immobilized sequence-specific oligonucleotide probes. Planta Med. 74:464–469. [DOI] [PubMed] [Google Scholar]
- Liu XJ, Hu J, Li ZY, Qin XM, Zhang LZ, Guo XQ. 2011. Species classification and quality assessment of Chaihu (Radix Bupleuri) based on high-performance liquid chromatographic fingerprint and combined chemometrics methods. Arch Pharm Res. 34:961–969. [DOI] [PubMed] [Google Scholar]
- Pan SL. 2006. Bupleurum species: scientific evaluation and clinical applications. Boca Raton (FL): Taylor and Francis Group; p. 1–6. [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]