Abstract
Salvia yunnanensis is a medicinal plant commonly used in the southwest of China. In this study, we sequenced the complete chloroplast (cp) genome sequence of S. yunnanensis to investigate its phylogenetic relationship in the family Lamiaceae. The total length of the cp genome was 151,338 bp, with 38.0% overall GC content and exhibited typical quadripartite structure, a pair of IRs (inverted repeats) of 25,578 bp each were separated by a small single-copy (SSC) region of 17,564 bp and a large single-copy (LSC) region of 82,618 bp. The cp genome contained 114 genes, including 80 protein coding genes, 30 tRNA genes, and 4 rRNA genes. The phylogenetic analysis indicated S. yunnanensis was closely related to S. miltiorrhiza, which afforded a scientific evidence that S. yunnanensis could be used as original species of Radix et Rhizoma Saliviae Miltiorrhizae (Danshen).
Keywords: Salvia yunnanensis, chloroplast, Illumina sequencing, phylogeny
Salvia L. is a great genus of the Lamiaceae family, which includes 900 species in the world (Topçu 2006). Most of them are widespread in temperate regions and tropical mountains including Central and South America, Central Asia/Mediterranean and Eastern Asia (Li et al. 2013). There are 84 species in China (Li and Ian 1994) and the majority of them are distributed in southwest China, notably in the Hengduan Mountain region. In some areas of southwest China, Salvia yunnanensis is a local medicine commonly used as the surrogate of the traditional Chinese medicine Radix et Rhizoma Saliviae Miltiorrhizae (Danshen, Salvia miltiorrhiza) for the treatment of various cardiovascular diseases (Jiangsu New Medical College 1977). However, up to now, for such a medicinal plant, studies have focussed on describing its chemical compositions (Xu et al. 2006; Xiang et al. 2013; Xia et al. 2015) and DNA barcoding analysis (Wang and Wang 2005; Wang et al. 2007), with little involvement in its genomes. Here, we report the chloroplast (cp) genome sequence of S. yunnanensis and find its internal relationships within the family Lamiaceae.
Fresh and clean leaf materials of S. yunnanensis were collected from Dali county, Yunnan, China (N25.62°, E100.48°), and the plant materials and a voucher specimen (No. LYTE01) were deposited at Tourism and Culture College of Yunnan University (Lijiang). Total genomic DNA was extracted using the improved CTAB method (Doyle 1987; Yang et al. 2014), and sequenced with Illumina Hiseq 2500 (Novogene, Tianjing, China) platform with pair-end (2 × 300 bp) library. About 5.19 Gb of raw reads with 17,290,260 paired-end reads were obtained from high-throughput sequencing. The raw data was filtered using Trimmomatic v.0.32 with default settings (Bolger et al. 2014). Then paired-end reads of clean data were assembled into circular contigs using GetOrganelle.py (Jin et al. 2018) with Salvia miltiorrhiza (No. NC_020431) as reference. Finally, the plastome was annotated by the Plastid Genome Annotator (PGA) (Qu et al. 2019) with manual adjustment using Geneious v. 7.1.3 (Kearse et al. 2012).
The circular genome map was generated with OGDRAW v.1.3.1 (Greiner et al. 2019). Then the annotated cp genome was submitted to the GenBank under the accession number MN341012. The total length of the cp genome was 151,338 bp, with 38.0% overall GC content. With typical quadripartite structure, a pair of IRs (inverted repeats) of 25,578 bp was separated by a small single-copy (SSC) region of 17,564 bp and a large single-copy (LSC) region of 82,618 bp. The cp genome contained 114 genes, including 80 protein-coding genes, 30 tRNA genes, and 4 rRNA genes. Of these, 20 genes were duplicated in the inverted repeat regions, 11 genes, and 6 tRNA genes contain one intron, while two genes (ycf3 and clpP) have two introns.
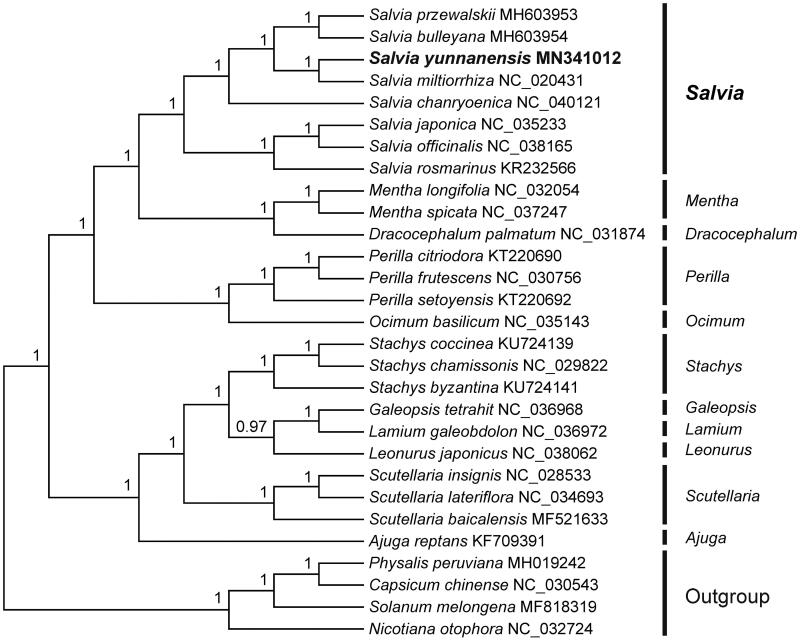
To investigate its taxonomic status, a total of 24 cp genome sequences of Lamiaceae species were downloaded from the NCBI database used for phylogenetic analysis. After using MAFFT V.7.149 for aligning (Katoh and Standley 2013), jModelTest v.2.1.7 (Darriba et al. 2012) was used to determine the model and the GTR + G model was the best-fitting for the cp genomes. Then Bayesian inference (BI) was performed by MrBayes v.3.2.6 (Ronquist et al. 2012) with four Solanaceae family species (NC_032724, NC_030543, MH019242, and MF818319) as outgroups. The results showed that S. yunnanensis was closely related to S. miltiorrhiza (Figure 1). Meanwhile, the present study afforded scientific evidence for resource development of Radix et Rhizoma Saliviae Miltiorrhizae and would be beneficial to taxonomy and phylogeny of Lamiaceae.
Figure 1.
Bayesian inference (BI) tree of 25 species within the family Lamiaceae based on the complete plastome sequences using four Solanaceae family species (NC_032724, NC_030543, MH019242, and MF818319) as outgroups.
Acknowledgments
The authors are highly grateful to the published genome data in the public database. The authors also thank Qi Chen in Dali University for helping in data analysis of complete chloroplast genome.
Disclosure statement
The authors declare no conflicts of interest and are responsible for the content of the article.
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle J. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15. [Google Scholar]
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–W64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiangsu New Medical College 1977. [Dictionary of Chinese Materia Medical]. Shanghai (China): Science and Technology Press of Shanghai. Chinese. [Google Scholar]
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. bioRxiv.:1–11. DOI: 10.1101/256479. [DOI] [Google Scholar]
- Katoh K, Standley D. 2013. MAFFT multiple sequence alignment software version improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HW, Ian CH. 1994. Lamiaceae In: Wu ZY, Raven PH, editors. Flora of China. 17th ed. Beijing (China): Science Press. [Google Scholar]
- Li MH, Li QQ, Liu YZ, Gui ZH, Zhang N, Huang LQ, et al. . 2013. Pharmacophylogenetic study on plants of genus Salvia L. from China. Chin Herbal Med. 2013:164–181. [Google Scholar]
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Meth. 15:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP, et al. . 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topçu G. 2006. Bioactive triterpenoids from Salvia species. J Nat Prod. 69(3):482–487. [DOI] [PubMed] [Google Scholar]
- Wang H, Wang Q. 2005. Analysis of rDNA ITS sequences of Radix et Rhizoma Salviae Miltiorrhizae and plants of Salvia L. Chin Trad Herb Drugs. 36:1381–1385. [Google Scholar]
- Wang Y, Li D-h, Zhang Y-T. 2007. Analysis of ITS sequence of some medicinal plants and their related species in Salvia. Yao Xue Xue Bao. 42(12):1309–1313. [PubMed] [Google Scholar]
- Xia F, Wu CY, Yang XW, Li X, Xu G. 2015. Diterpenoids from the Roots of Salvia yunnanensis. Nat Prod Bioprospect. 5(6):307–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang C, Zhu LP, Zhuang WT, Zhang LD, He J, Li P. 2013. Chemical constituents of stems and leaves of Salvia yunnanensis and their anti-angiogeneic activities. China J Chin Materia. 38:835–838. [PubMed] [Google Scholar]
- Xu G, Peng L-Y, Lu L, Weng Z-Y, Zhao Y, Li X-L, Zhao Q-S, Sun H-D. 2006. Two new abietane diterpenoids from Salvia yunnanensis. Planta Med. 72(1):84–86. [DOI] [PubMed] [Google Scholar]
- Yang JB, Li DZ, Li HT. 2014. Highly effective sequencing whole chloroplast genomes of angiosperms by nine novel universal primer pairs. Mol Ecol Resour. 14:1024–1031. [DOI] [PubMed] [Google Scholar]