Abstract
Cinnamomum aromaticum has long been recognized and cultivated in tropical and subtropical Asia for their aromatic bark to produce cinnamon. We reported for the first time the complete plastid genome of C. aromaticum and reconstructed its phylogenetic position. The complete plastid genome is 152,754 bp in length with a quadripartite organization: a large single copy (LSC) region of 93,706 bp and a small single copy (SSC) region of 18,916 bp. Each of the two inverted repeat regions (IRa and IRb) is 20,066 bp. We recovered 128 functional genes, including 84 protein-coding genes, 36 tRNA genes and 8 rRNA genes. The phylogenetic analysis suggested that C. aromaticum and two samples of C. camphora forms a strongly supported clade, which is sister to another cinnamon species of C. verum native to Sri Lanka with strong ultrafast bootstrap support.
Keywords: China, chloroplast genome, phylogeny, laurel family
Several species of the genus Cinnamomum in the Lauraceae family, e.g., C. aromaticum Nees, C. verum J. Presl, C. citriodorum Thwaites, have long been recognized for their economical importance as source of spice. Of these species, C. aromaticum (= C. cassia (L.) Bercht. & Presl. and C. cassia D. Don), also known as Chinese cinnamon or Chinese cassia originated in southern China, is the most widely cultivated species in tropical and subtropical Asia for their aromatic bark to produce cinnamon (Wang and Tang 2006; Wu et al. 2008; Huang et al. 2016). For a better understanding of the plastid genome characterization and its phylogenetic relationships with other Cinnamomum species, we generated the plastid genome of C. aromaticum using genome skimming method.
The fresh leaf tissues of C. aromaticum were collected from South China Botanical Garden, Guangzhou, China (113.36°E, 23.19°N). Voucher specimens (XPW504) were deposited in the IBCS. We isolated the whole genomic DNA using a modified CTAB method (Doyle and Doyle 1987). We fragmented the isolated total genomic DNA into 300-500 bp in length to construct libraries following the manufacturer’s manual (Illumina). Paired-end (PE) sequencing was conducted on the Illumina HiSeq X-Ten instrument at Beijing Genomics Institute (BGI). We used GetOrganelle pipeline (Jin et al. 2018) to assemble the plastome. The GetOrganelle automatically recruits plastid-like reads by using Bowtie2 (Langmead and Salzberg 2012), and assembled the filtered reads using SPAdes (Bankevich et al. 2012). We generated the complete circular chloroplast genome by Bandage (Wick et al. 2015). We employed Plastid Genome Annotator (PGA) (Qu et al. 2019) and Geneious v11.0 (Kearse et al. 2012) to annotate the plastome and to verify the accuracy of the assembly. The annotated plastome has been deposited in GenBank (MN173819).
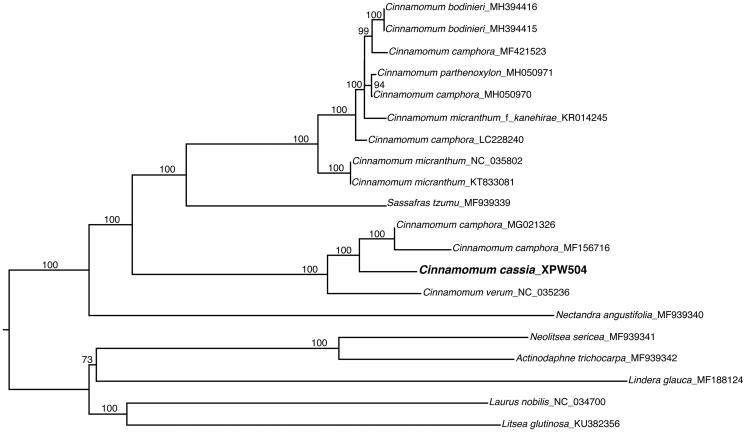
To reconstruct the phylogenetic tree of C. aromaticum, we included 19 plastid genomes in previous publications and unpublished data in GenBank (Figure 1) (Song 2016; Wu et al. 2016; Chen et al. 2017, 2019; Song et al. 2017; Zeng et al. 2018). We aligned the data matrix using MAFFT v.1.3.7 (Katoh and Standley 2013) with default parameters. The maximum likelihood tree was build in IQ-tree (Trifinopoulos et al. 2016) using models recommended by ModelFinder (Kalyaanamoorthy et al. 2017). The branch supports were estimated using 1000 integrations of ultrafast bootstrap (Hoang et al. 2018).
Figure 1. Phylogenetic position of Cinnamomum aromaticum inferred from likelihood method based on 19 complete plastome sequences from Lauraceae. Numbers on the nods are nanparametric bootstrap values from 1000 replicates.
The complete plastid genome of C. aromaticum was 152,754 bp in length and showed a typical quadripartite organization: a large single copy (LSC) region of 93,706 bp and a small single copy (SSC) region of 18,916 bp, respectively. These two regions were separated by two inverted repeat regions (IRa and IRb), each of 20,066 bp in length. A total of 128 functional genes were recovered, consisting of 84 protein-coding genes, 36 tRNA genes and 8 rRNA genes. The phylogenetic analysis suggested that C. aromaticum and two samples of C. camphora (L.) J. Presl forms a strongly supported clade, which is sister to another cinnamon species of C. verum native to Sri Lanka with strong bootstrap support. This study demonstrated the potential power of genomic data in resolving the phylogenetic relationships among cinnamon species and in answering questions regarding their genetic diversity.
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Disclosure statement
The authors report no conflicts of interest.
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Zheng Y, Liu S, Zhong Y, Wu Y, Li J, Xu L-A, Xu M. 2017. The complete chloroplast genome of Cinnamomum camphora and its comparison with related Lauraceae species. PEERJ. 5:e3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Liu C, Han L, Song Y, Tang L. 2019. The plastid genome of an oil plants Cinnamomum chago (Lauraceae). Mitochondrial DNA Part B. 4(1):1733–1734. [Google Scholar]
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15. [Google Scholar]
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 35(2):518–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JF, Li L, van der Werff H, Li HW, Rohwer JG, Crayn DM, Meng HH, van der Merwe M, Conran JG, Li J. 2016. Origins and evolution of cinnamon and camphor: a phylogenetic and historical biogeographical analysis of the Cinnamomum group (Lauraceae). Mol Phylogenet Evol. 96:33–44. [DOI] [PubMed] [Google Scholar]
- Jin J, Yu W, Yang J, Song Y, Yi T, Li D. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. BioRxiv.256479. 10.1101/256479 [DOI] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9(4):357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y. 2016. Phylogenomic analysis of Phoebe and Machilus in the family Lauraceae [dissertation]. Beijing: University of Chinese Academy of Sciences. [Google Scholar]
- Song Y, Yu W, Tan Y, Liu B, Yao X, Jin J, Padmanaba M, Yang J, Corlett RT. 2017. Evolutionary comparisons of the chloroplast genome in Lauraceae and insights into loss events in the Magnoliids. Genome Biol Evol. 9(9):2354–2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifinopoulos J, Nguyen LT, von Haeseler A, Minh BQ. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44(W1):W232–W235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Tang Y. 2006. Resources of cinnamon and cassia plants and their nomenclatures in China. Subtrop Plant Sci. 35(3):45–47. [Google Scholar]
- Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 31(20):3350–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CC, Ho CK, Chang SH. 2016. The complete chloroplast genome of Cinnamomum kanehirae Hayata (Lauraceae). Mitochondrial DNA Part A. 27(4):2681–2682. [DOI] [PubMed] [Google Scholar]
- Wu ZY, Raven PH, Hong DY, editors. 2008. Flora of China. Vol. 7, Cinnamomum Schaeffer. Beijing and Missouri (MO): Science Press and Missouri Botanical Garden Press. [Google Scholar]
- Zeng CX, Hollingsworth PM, Yang J, He ZS, Zhang ZR, Li DZ, Yang JB. 2018. Genome skimming herbarium specimens for DNA barcoding and phylogenomics. Plant Methods. 14(1):43. [DOI] [PMC free article] [PubMed] [Google Scholar]