Abstract
Background
Tuberculous meningitis (TBM) is a severe form of extrapulmonary tuberculosis and its early diagnosis is very difficult leading to present with severe disability or die. The current study aimed to assess the accuracy of metagenomic next generation sequencing (mNGS) for TBM, and to identify a new test for the early diagnosis of TBM.
Methods
We searched for articles published in Embase, PubMed, Cochrane Library, China National Knowledge Infrastructure, and Wanfang Data up to June 30, 2020 for studies that assessed the efficacy of mNGS for the diagnosis of TBM. Then, the accuracy between mNGS and a composite reference standard (CRS) in these articles was compared using the meta-analysis approach.
Results
Four independent studies with 342 samples comparing mNGS and a CRS were included in this study. The sensitivity of mNGS for TBM diagnosis ranged from 27% to 84%. The combined sensitivity of mNGS was 61%, and the I2 value was 92%. Moreover, the specificity of mNGS for TBM diagnosis ranged from 96% to 100%. The combined specificity of mNGS was 98%, and the I2 value was 74%. The heterogeneity between studies in terms of sensitivity and specificity was significant. The area under the curve (AUC) of the summary receiver operating characteristic curve (SROC) of mNGS for TBM was 0.98.
Conclusions
The sensitivity of mNGS for TBM diagnosis was moderate. Furthermore, the specificity was extremely high, and the AUC of the SROC indicated a very good diagnostic efficacy. mNGS could be used as an early diagnostic method for TBM, however, the results should be treated with caution for the heterogeneity between studies was extremely significant.
Systematic review registration
INPLASY202070100.
Introduction
Tuberculosis (TB) is a serious threat to human health worldwide [1]. A high proportion of individuals are infected with tuberculosis annually, and some die from related diseases. In 2018, there were about 10 million new cases of TB worldwide and about 1.45 million deaths from TB [2]. Moreover, this condition is the leading cause of death from infectious diseases [2]. In addition to pulmonary tuberculosis (PTB), Mycobacterium tuberculosis (MTB) can also cause extrapulmonary tuberculosis (EPTB) [3]. Tuberculous meningitis (TBM) is a highly lethal type of EPTB. Although it accounts for a relatively small proportion (1%–5%) of new TB cases, 50% of individuals with TBM can die or present with severe disability [4]. Failure to facilitate early diagnosis and treatment is one of the primary causes for these serious complications [5]. Thus, early diagnosis can improve the prognosis of TBM. Conventional mycobacterium detection techniques do not meet the need for early detection [4]. Acid-fast bacilli (AFB) smear is the cheapest and most convenient method for the diagnosis of TB. However, its sensitivity in the cerebrospinal fluid (CSF) is extremely low (≤15%), especially in the absence of professional microscopists in the laboratory [6]. Mycobacterium culture in the CSF and AFB smear have extremely low sensitivity, and these methods are time-consuming. Therefore, culture results cannot be used as an indicator of early diagnosis [7]. Currently, in some cases, empirical antituberculous therapy is provided when TBM cannot be ruled out. Hence, the early and rapid diagnosis of TBM should be improved.
The advent of nucleic acid amplification tests (NAATs) has facilitated early and rapid TB diagnosis [8]. Xpert MTB/RIF (Cepheid, Sunnyvale, CA) is a representative NAAT that is extensively used for the diagnosis of TB [9]. Furthermore, according to the recommendation of the World Health Organization, it is used as the initial test for the diagnosis of TBM [10].
Recently, with the advancement of molecular diagnostic technology, metagenomic next generation sequencing (mNGS) technology was developed, and it can provide information on the genomic DNA sequences of microorganisms. The mNGS technology is a new detection method, which allows for an unbiased and detailed test of the total DNA or RNA content of all currently known pathogenic microorganisms [11]. Thus, it has been increasingly used [12]. mNGS could detect pathogenic microorganisms from a wide range of clinical specimens, including CSF [11, 13, 14]. Recent studies have shown that the sensitivity and specificity of mNGS for detecting pathogenic bacteria are significantly better than that of culture [15]. However, studies on the diagnostic efficacy of mNGS-based detection of MTB DNA for TBM were controversial [16, 17]. Moreover, relevant systematic evaluation and meta-analysis of the diagnostic efficacy of mNGS for TBM have not been conducted. Therefore, this study evaluated and compared the efficacy of mNGS for the diagnosis of TBM using CSF specimens versus a composite reference standard (CRS) to identify a new technique for the early diagnosis of TBM.
Methods
Design and registration
A systematic review and meta-analysis of the diagnostic accuracy of mNGS was conducted. The study protocol was registered on the International Platform of Registered Systematic Review and Meta-Analysis Protocols (INPLASY) (registration number: INPLASY202070100) [18]. This study was conducted in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses statements [19]. Ethical approval was not required for the study.
Information sources
We searched studies, which assessed the diagnostic accuracy of mNGS for TBM, in Embase, PubMed, Cochrane Library, China National Knowledge Infrastructure (CNKI), and Wanfang Database up to June 30, 2020. Moreover, the references cited in the reviews were evaluated for other possible studies.
Search strategy
Guocan Yu and Wuchen Zhao conducted the search strategies. There were no language restrictions in our search process. Search strategy of PubMed was listed as follows:
#1 "Tuberculosis, Meningeal"[Mesh] OR “Meningeal Tuberculoses” OR “Meningeal Tuberculosis” OR “Tuberculoses, Meningeal” OR “TB Meningitis” OR “TB Meningitides” OR “Tubercular Meningitis” OR “Meningitides, Tubercular” OR “Meningitis, Tubercular” OR “Tubercular Meningitides” OR “Meningitis, Tuberculous” OR “Meningitides, Tuberculous” OR “Tuberculous Meningitides” OR “Tuberculous Meningitis” OR “Tuberculosis Meningitis” OR “Meningitides, Tuberculosis” OR “Meningitis, Tuberculosis” OR “Tuberculosis Meningitides” OR “Tuberculous Hypertrophic Pachymeningitis” OR “Hypertrophic Pachymeningitides, Tuberculous” OR “Hypertrophic Pachymeningitis, Tuberculous” OR “Pachymeningitides, Tuberculous Hypertrophic” OR “Pachymeningitis, Tuberculous Hypertrophic” OR “Tuberculous Hypertrophic Pachymeningitides”
#2 “Extrapulmonary tuberculosis” OR “Extra pulmonary tuberculosis”
#3 "Meningitis"[Mesh] OR Meningitides OR Pachymeningitis OR Pachymeningitides
#4 "Cerebrospinal Fluid"[Mesh] OR “Cerebrospinal Fluids” OR “Fluid, Cerebrospinal” OR “Fluids, Cerebrospinal” OR “Cerebro Spinal Fluid” OR “Cerebro Spinal Fluids” OR “Fluid, Cerebro Spinal” OR “Fluids, Cerebro Spinal” OR “Spinal Fluid, Cerebro” OR “Spinal Fluids, Cerebro”
#5 #1 OR #2 OR #3 OR #4
#6 “Metagenomic Next-Generation Sequencing” OR mNGS
#7 #5 AND #6
Similar search formulae were used for the Cochrane Library, Embase, CNKI, and Wanfang databases.
Eligibility criteria
Type of study
We included different types of studies, such as case-control, retrospective, and prospective studies. Full text original studies that assessed the efficacy of mNGS for the diagnosis of TBM were included. The true-positive (TP), false-positive (FP), false-negative (FN), and true-negative (TN) values for the assay can be extracted or calculated directly from the studies. However, case reports, articles written in languages other than Chinese and English, studies with < 10 specimens, conference reports, and abstracts without full articles were excluded.
Patients
Studies with patients diagnosed with TBM via mNGS were included, and there were no restrictions on gender, age, and nations.
Index tests
mNGS was considered as the index test.
Main outcomes
Sensitivity and the specificity of mNGS were considered as the main outcome.
Reference standards
CRS was defined as the reference standard in this study. The reference standards in CRS comprised clinical symptoms, radiographic features, immunological index, biochemical test results, smears, culture, histopathology, and response to anti-tuberculosis drugs. Patients with some or all factors who had positive results were considered positive for TBM. Meanwhile, cases were considered as non-TBP if all the test results were negative.
Literature screening and selection
The primary search records were imported into the ENDNOTE X9.2 literature management software, according to the eligibility criteria. Two investigators (Guocan Yu and Wuchen Zhao) independently assessed the candidate articles by reviewing their titles and abstracts, followed by the full text, for inclusion. Discrepancies between the two investigators was resolved via a discussion with a third investigator (Hong Zheng).
Data extraction
We extracted data including those on first author name; publication year; country where the study was conducted; TP, FP, FN, and TN values for the assay; type of research; patient selection method; sample pre-treatment method; homogenization; and sample condition along with other parameters. The same researchers independently extracted relevant data from the included articles; the data obtained were then cross-checked. Discrepancies between the two information sets were settled via a discussion with a third investigator, similar to the method used during the literature selection phase.
Quality evaluation
Based on the reference standards, to independently assess study quality, the two investigators independently used a revised tool for the Quality Assessment of Diagnostic Accuracy Studies (QUADAS-2) [20]. The discrepancy between reviewers was resolved via a discussion with a third investigator (Hong Zheng). The Funnel chart was used to evaluate whether publication bias existed in the included studies.
Data synthesis and statistical analysis
We first obtained the TP, FP, FN, and TN values in each study. Next, using the bivariate random-effects models, the combined sensitivity and specificity with 95% confidence interval (CI) between mNGS and CRS were calculated and compared. The forest plots for the sensitivity and specificity of each study were generated. The areas under the curve (AUC) of the summary receiver operating characteristic (SROC) were subsequently calculated. I2 statistics was used to assess heterogeneity between the studies and the reference standard. A value of 0% indicated the absence of heterogeneity and a value > 50%, substantial heterogeneity [21]. Via meta-regression and subgroup analyses, we assessed the following: different types of studies, patient selection method, sample pre-treatment method, sample conditions, and homogenization as potential sources of heterogeneity. At least four published studies were required to perform meta-analysis for predefined variable types. Stata version 15.0 (Stata Corp., College Station, TX, the USA) with midas module were used to generate the forest plots for the sensitivity and specificity with 95% CI in each study and to carry out meta-analyses and meta-regression analyses. midas is a command for idiot-proof implementation of the contemporary statistical methods for meta-analysis of diagnostic test accuracy [22].
Results
Characteristics of the studies
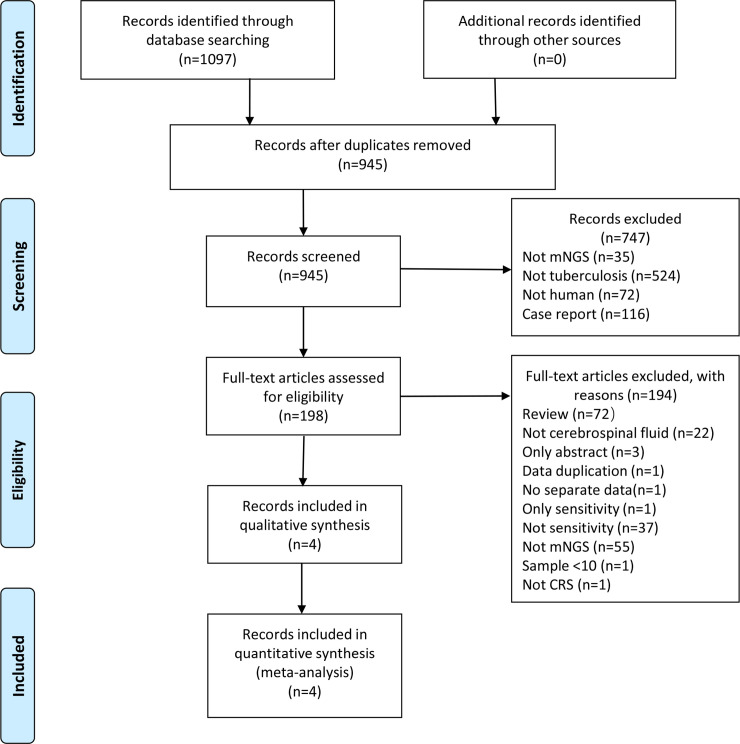
After searching relevant databases using our search strategy, we identified 1097 candidate articles. Of these articles, four, including two retrospective and two prospective, studies met the inclusion criteria [15–17, 23] (Fig 1). The kappa index of agreement for the selection of studies and data extraction was 0.665 (95% confidence interval [CI]: 0.355–0.975) between the two investigators. All studies were conducted in low-income areas with a high prevalence of TB. All articles have been published in English. CSF was used in all studies. The size of the specimen ranged from 29 to 213, with a median specimen size of 50 and a total specimen size of 342. All characteristics of the included study are depicted in Table 1. We excluded one article that reported sensitivity, but not specificity [24], and another one that presented data that were discussed in an article we already included [25].
Fig 1. Flow chart of literature retrieval.
In total, 507, 53, 0, 242, and 295 articles were found in Embase, PubMed, Cochrane Library, CNKI, and Wanfang Data, respectively.
Table 1. Characteristics of the included studies.
| Author | Year | County | sample type | reference | N | TP | FP | FN | TN | Type of research | Sample condition | Homogenisation | sample pre-treatment | patient selection method | patient population |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Wang S | 2019 | China | CSF | CRS | 29 | 18 | 0 | 5 | 6 | Retrospective | Frozen | Mechanical | With bead-beating | Convenience | Low |
| Zhou X | 2019 | China | CSF | CRS | 49 | 7 | 1 | 9 | 32 | Prospective | Fresh/Frozen | No | Without bead-beating | Consecutive | Low |
| Xing X | 2020 | China | CSF | CRS | 213 | 12 | 6 | 32 | 163 | Prospective | Frozen | No | With bead-beating | Convenience | Low |
| Yan L | 2020 | China | CSF | CRS | 51 | 38 | 0 | 7 | 6 | Retrospective | Fresh | Mechanical | With bead-beating | Convenience | Low |
CSF, cerebrospinal fluid; CRS, composite reference standard; TP, true-positive; FP, false-positive; FN, false-negative; TN, true-negative.
Study quality
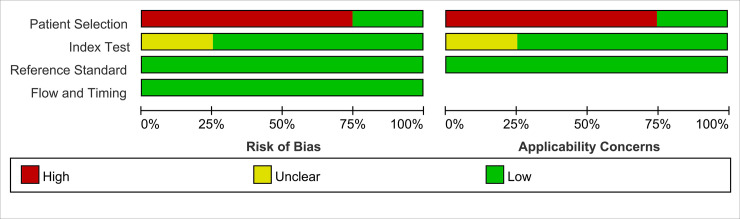
The overall methodological quality of the included studies, using a CRS as the gold reference, is presented in Fig 2. The risk of bias was primarily attributed to patient selection and index test. The risk of bias from the flow and timing was low, as was the risk of bias from the reference standard. An assessment of publication bias was not performed due to the limited number of studies included (<10).
Fig 2. Methodological quality graphs (risk of bias and applicability concerns) were presented as percentages across the included studies using a composite reference standard.
Diagnostic accuracy of mNGS for TBM
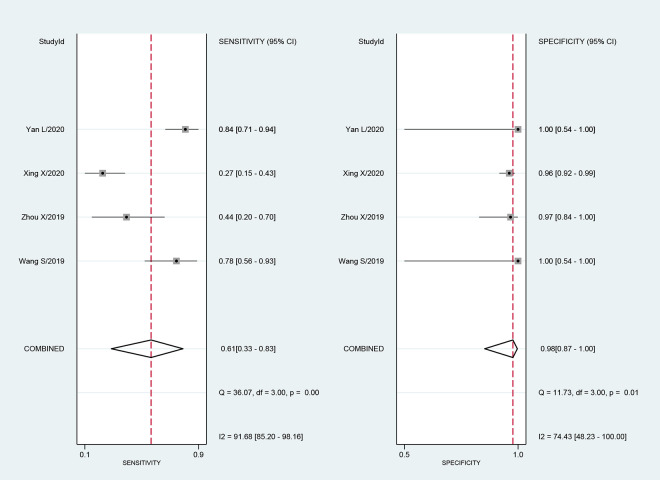
Four studies included 342 samples with a CRS. The sensitivity of mNGS for TBM ranged from 27% (95% CI: 15–43) to 84% (95% CI: 71–94). The combined sensitivity of mNGS for the diagnosis of TBM was 61% (95% CI: 33–83), and the I2 value was 92% (95% CI: 85–98). The specificity of mNGS ranged from 96% (95% CI: 92–99) to 100% (95% CI: 54–100). Furthermore, the combined specificity of mNGS was 98% (95% CI: 87–100), and the I2 value was 74% (95% CI: 48–100) (Fig 3). The heterogeneity between studies in terms of sensitivity and specificity was significant. The AUC of the SROC was 0.98 (95% CI: 0.96–0.99).
Fig 3. Forest plot for the sensitivity and specificity of mNGS for the diagnosis of tuberculosis meningitis using a composite reference standard.
We explored the heterogeneity among studies using subgroup, meta-regression, and sensitivity analyses. Subgroup and meta-regression analyses were performed on the predefined subgroups for the different types of studies, patient selection method, sample pre-treatment method, and homogenization used in the assay. Meta-regression analysis demonstrated that the different types of studies (prospective vs. retrospective) had effect on the sensitivity and specificity of mNGS for TBM compared with CRS (32% vs. 82%, meta-regression P < 0.01; 97% vs. 100%, meta-regression P < 0.01). The same results were also observed in studies using different homogenization methods (mechanical or not) (Table 2). With or without bead-beating for sample pre-treatment might have an effect on the specificity of mNGS (98% vs. 97%, meta-regression P = 0.03), but not sensitivity (66% vs. 43%, meta-regression P = 0.43). The patient selection methods (consecutive or convenience) had no effect on the sensitivity and specificity of mNGS (meta-regression P > 0.05). However, limited data from relevant studies were included to facilitate subgroup and sensitivity analysis. One study was unable to distinguish sample conditions [23], so meta-regression analysis for sample conditions could not be performed.
Table 2. Meta-regression analysis for different parameters.
| Parameter | Sensitivity (95% CI) | Specificity (95% CI) | |
|---|---|---|---|
| Type of study | Prospective (2 studies) | 32% (20–43%) | 97% (94–99%) |
| Retrospective (2 studies) | 82% (73–91%) | 100% (100–100%) | |
| Meta-regression P-value | <0.01 | <0.01 | |
| Homogenization | Mechanical (2 studies) | 82% (73–91%) | 100% (100–100%) |
| No (2 studies) | 32% (20–43%) | 97% (94–99%) | |
| Meta-regression P-vaule | <0.01 | <0.01 | |
| Sample pre-treatment | With bead-beating (3 studies) | 66% (38–93%) | 98% (94–100%) |
| Without bead-beating (1 study) | 43% (11–98%) | 97% (91–100%) | |
| Meta-regression P-vaule | 0.39 | 0.03 | |
| Patient selection | Consecutive (1 study) | 43% (11–98%) | 97% (91–100%) |
| Convenience (3 studies) | 66% (38–93%) | 98% (94–100%) | |
| Meta-regression P-vaule | 0.44 | 0.21 | |
CI: confidence interval
Discussion
Similar to other types of EPTB, the paucibacillary nature makes early diagnosis of TBM more difficult [26]. Delays in the diagnosis and treatment of TBM can lead to serious complications such as severe disability or death. This can significantly affect the patient’s prognosis and quality of life [27]. Lumbar puncture is a vital step in the diagnosis of patients. CSF obtained via lumbar puncture is the most common specimen used to diagnose TBM. Due to the low bacterial content in the sample, TBM is still challenging to diagnose via AFB and CSF culture [28]. By contrast, TBM is easily confused with cryptococcal meningitis or viral meningitis, and the diagnosis of the causative organism is quite difficult [16]. Hence, TBM is often missed or misdiagnosed. Therefore, to reduce mortality and disability from TBM and to facilitate early identification of the causative organisms, rapid and effective diagnostic methods should be developed to obtain a differential diagnosis of TBM.
NAATs, such as Xpert MTB/RIF assay and the loop-mediated isothermal amplification assay, are fast and efficient detection methods. Hence, they have been widely used in the early diagnosis of TB [29, 30]. Moreover, they have an extremely good diagnostic performance in paucibacillary EPTB [31]. However, these methods have limitations. That is, a single test can only detect MTB DNA and can only provide indirect assistance in the differential diagnosis of TBM.
The mNGS technology is a new detection method for all currently known pathogenic microorganisms, including MTB [12]. mNGS allows for an unbiased and detailed test of the total DNA or RNA content of the microbiome [32]. Moreover, it takes about 3 days to obtain the results, which is still relatively fast compared with culture. mNGS can detect a wide range of pathogenic microorganisms, and it is useful in obtaining the differential diagnosis of suspected TBM. The current findings on the use of mNGS for the diagnosis of TBM are controversial. Moreover, systematic review and meta-analysis of the use of mNGS for the diagnosis of TBM were not conducted. Therefore, the current study was conducted.
The current study included four studies that conducted a comparison between mNGS and CRS, showing that the combined sensitivity and specificity of mNGS for the diagnosis of TBM were 61% (95% CI: 33–83) and 98% (95% CI: 87–100), respectively. The sensitivity widely varied among the studies. The AUC of the SROC was 0.98 (95% CI: 0.96–0.99), thereby indicating that mNGS had an extremely good diagnostic performance for TBM. However, a substantial level of heterogeneity was observed, regardless of sensitivity and specificity. Therefore, the results should be treated with caution. Publication bias was not assessed because of the limited number of studies included. The type of research, patient selection method, sample pre-treatment method, sample conditions, and homogenization differed among the studies. The patient selection methods were non-consecutive for most of the included studies. However, most of the studies used bead-beating for sample pre-treatment. Meta-regression analysis demonstrated that the different types of studies and homogenization methods had effect on the sensitivity and specificity of mNGS for TBM compared with CRS. Sample pre-treatment methods might have effect on the specificity of mNGS, but not sensitivity. Different homogenization and sample pre-treatment methods might lead to different distribution of MTB in solution, which might affect the diagnostic performance of mNGS. However, whether these methods really affect the diagnostic performance of mNGS remains to be explored further, and the mechanisms need to be clarified further. The patient selection methods had no effect on the sensitivity and specificity of mNGS. However, the limited number of studies included did not allow for further subgroup. These factors might be a source of heterogeneity among studies. The impact of these factors on diagnostic validity needs to be further confirmed by larger clinical trials. Furthermore, sensitivity analysis could not be conducted due to the limited number of studies. For paucibacillary TBM, culture might not be a perfect reference standard. However, CRS, which uses a combination of factors, might be a more appropriate reference standard. In addition, the number of studies comparing the diagnostic efficacy between culture and mNGS for TBM is extremely limited. Therefore, in this study, CRS was used as the reference standard. The CRS used in the included studies was also inconsistent. For example, some studies had not included response to treatment, which might be a source of heterogeneity.
This meta-analysis had several limitations. Despite a comprehensive search, some articles might have been missed, and the number of studies included in this meta-analysis is limited. In addition, some studies cannot distinguish meningitis data, which might have led to some bias in the results. In addition, studies on the diagnosis of other pathogens via mNGS were not included in the current analysis. In addition, the CRS differed among the studies. Different CRS might lead to changes in patient classification and thus affect the sensitivity and specificity of the test. The heterogeneity among the studies was substantial, and the combined estimates must be treated with caution.
Conclusions
This study showed that the sensitivity of mNGS for the diagnosis of TBM was moderate. Moreover, the specificity was extremely high, and the AUC indicated a very good diagnostic efficacy. mNGS could be used as an early diagnostic method for TBM. However, the heterogeneity between studies was extremely significant. Hence, the results should be treated with caution.
Supporting information
(DOC)
(DOCX)
Acknowledgments
We thank the authors of the studies included in this meta-analysis.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
The author(s) received no specific funding for this work.
References
- 1.World Health Organization. Global tuberculosis report 2019. Geneva, Switzerland; 2019.
- 2.Donovan J, Thu DDA, Phu NH, Dung VTM, Quang TP, Nghia HDT, et al. Xpert MTB/RIF Ultra versus Xpert MTB/RIF for the diagnosis of tuberculous meningitis: a prospective, randomised, diagnostic accuracy study. Lancet Infect Dis. 2020;20(3):299–307. 10.1016/S1473-3099(19)30649-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu G, Zhong F, Ye B, Xu X, Chen D, Shen Y. Diagnostic Accuracy of the Xpert MTB/RIF Assay for Lymph Node Tuberculosis: A Systematic Review and Meta-Analysis. Biomed Res Int. 2019;2019:4878240 10.1155/2019/4878240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cresswell FV, Tugume L, Bahr NC, Kwizera R, Bangdiwala AS, Musubire AK, et al. Xpert MTB/RIF Ultra for the diagnosis of HIV-associated tuberculous meningitis: a prospective validation study. Lancet Infect Dis. 2020;20(3):308–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sheu JJ, Yuan RY, Yang CC. Predictors for outcome and treatment delay in patients with tuberculous meningitis. Am J Med Sci. 2009; 338: 134–139. 10.1097/MAJ.0b013e3181a590f1 [DOI] [PubMed] [Google Scholar]
- 6.Bahr NC, Boulware DR. Methods of rapid diagnosis for the etiology of meningitis in adults. Biomark Med. 2014; 8: 1085–1103. 10.2217/bmm.14.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bahr NC, Nuwagira E, Evans EE, Cresswell FV, Bystrom PV, Byamukama A, et al. Diagnostic accuracy of Xpert MTB/RIF Ultra for tuberculous meningitis in HIV-infected adults: a prospective cohort study. Lancet Infect Dis. 2018;18(1):68–75. 10.1016/S1473-3099(17)30474-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tyagi S, Sharma N, Tyagi JS, Haldar S. Challenges in pleural tuberculosis diagnosis: existing reference standards and nucleic acid tests. Future Microbiol. 2017;12:1201–1218. 10.2217/fmb-2017-0028 [DOI] [PubMed] [Google Scholar]
- 9.Kohli M, Schiller I, Dendukuri N, Dheda K, Denkinger CM, Schumacher SG, et al. Xpert® MTB/RIF assay for extrapulmonary tuberculosis and rifampicin resistance. Cochrane Database Syst Rev. 2018;8(8):CD012768 10.1002/14651858.CD012768.pub2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bahr NC, Marais S, Caws M, van Crevel R, Wilkinson RJ, Tyagi JS, et al. Tuberculous Meningitis International Research Consortium. GeneXpert MTB/Rif to Diagnose Tuberculous Meningitis: Perhaps the First Test but not the Last. Clin Infect Dis. 2016;62(9):1133–5. 10.1093/cid/ciw083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miao Q, Ma Y, Wang Q, Pan J, Zhang Y, Jin W, et al. Microbiological Diagnostic Performance of Metagenomic Next-generation Sequencing When Applied to Clinical Practice. Clin Infect Dis. 2018. Nov 13;67(suppl_2):S231–S240. 10.1093/cid/ciy693 [DOI] [PubMed] [Google Scholar]
- 12.Ji XC, Zhou LF, Li CY, Shi YJ, Wu ML, Zhang Y, et al. Reduction of Human DNA Contamination in Clinical Cerebrospinal Fluid Specimens Improves the Sensitivity of Metagenomic Next-Generation Sequencing. J Mol Neurosci. 2020;70(5):659–666. 10.1007/s12031-019-01472-z [DOI] [PubMed] [Google Scholar]
- 13.Ai JW, Li Y, Cheng Q, Cui P, Wu HL, Xu B, et al. Diagnosis of local hepatic tuberculosis through next-generation sequencing: Smarter, faster and better. Clin Res Hepatol Gastroenterol. 2018;42(3):178–181. 10.1016/j.clinre.2018.04.007 [DOI] [PubMed] [Google Scholar]
- 14.Parize P, Muth E, Richaud C, Gratigny M, Pilmis B, Lamamy A, et al. Untargeted next-generation sequencing-based first-line diagnosis of infection in immunocompromised adults: a multicentre, blinded, prospective study. Clin Microbiol Infect. 2017;23(8):574.e1–574.e6. 10.1016/j.cmi.2017.02.006 [DOI] [PubMed] [Google Scholar]
- 15.Wang S, Chen Y, Wang D, Wu Y, Zhao D, Zhang J, et al. The Feasibility of Metagenomic Next-Generation Sequencing to Identify Pathogens Causing Tuberculous Meningitis in Cerebrospinal Fluid. Front Microbiol. 2019;10:1993 10.3389/fmicb.2019.01993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xing XW, Zhang JT, Ma YB, He MW, Yao GE, Wang W, et al. Metagenomic Next-Generation Sequencing for Diagnosis of Infectious Encephalitis and Meningitis: A Large, Prospective Case Series of 213 Patients. Front Cell Infect Microbiol. 2020;10:88 10.3389/fcimb.2020.00088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan L, Sun W, Lu Z, Fan L. Metagenomic Next-Generation Sequencing (mNGS) in cerebrospinal fluid for rapid diagnosis of Tuberculosis meningitis in HIV-negative population. Int J Infect Dis. 2020;96:270–275. 10.1016/j.ijid.2020.04.048 [DOI] [PubMed] [Google Scholar]
- 18.Yu G, Zhao W, Shen Y, Zhu P, Zheng H. Metatagenomic NextGeneration Sequencing for diagnosis of tuberculosis meningitis: A protocol of systematic review and meta-analysis. Inplasy protocol 202070100. 10.37766/inplasy2020.7.0100 [DOI] [Google Scholar]
- 19.Moher D, Liberati A, Tetzlaff J, Altman DG; PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6(7):e1000097 10.1371/journal.pmed.1000097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155(8):529–36. 10.7326/0003-4819-155-8-201110180-00009 [DOI] [PubMed] [Google Scholar]
- 21.Shen Y, Yu G, Zhong F, Kong X. Diagnostic accuracy of the Xpert MTB/RIF assay for bone and joint tuberculosis: A meta-analysis. PLoS One. 2019;14(8):e0221427 10.1371/journal.pone.0221427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dwamena Ben, 2007. "MIDAS: Stata module for meta-analytical integration of diagnostic test accuracy studies," Statistical Software Components S456880, Boston: College Department of Economics, revised 05 Feb 2009. https://ideas.repec.org/c/boc/bocode/s456880.html [Google Scholar]
- 23.Zhou X, Wu H, Ruan Q, Jiang N, Chen X, Shen Y, et al. Clinical Evaluation of Diagnosis Efficacy of Active Mycobacterium tuberculosis Complex Infection via Metagenomic Next-Generation Sequencing of Direct Clinical Samples. Front Cell Infect Microbiol. 2019;9:351 10.3389/fcimb.2019.00351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma H, Wang X, Wang Y, Wang F, Qin L, Li W, et al. The significance of second-generation sequencing in the early diagnosis of tuberculous meningitis. J Med Forum. 2019; 40: 19–21,5. Chinese. [Google Scholar]
- 25.Xing X. Diagnostic value of high-throughput sequencing of metagenome in infectious diseases of central nervous system. Chinese People's Liberation Army medical college; 2019. Chinese. [Google Scholar]
- 26.Mezochow A, Thakur K, Vinnard C. Tuberculous Meningitis in Children and Adults: New Insights for an Ancient Foe. Curr Neurol Neurosci Rep. 2017;17(11):85 10.1007/s11910-017-0796-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thwaites GE, van Toorn R, Schoeman J. Tuberculous meningitis: more questions, still too few answers. Lancet Neurol. 2013; 12: 999–1010. 10.1016/S1474-4422(13)70168-6 [DOI] [PubMed] [Google Scholar]
- 28.Mai NT, Thwaites GE. Recent advances in the diagnosis and management of tuberculous meningitis. Curr Opin Infect Dis. 2017; 30: 123–8. 10.1097/QCO.0000000000000331 [DOI] [PubMed] [Google Scholar]
- 29.Yu G, Shen Y, Zhong F, Ye B, Yang J, Chen G. Diagnostic accuracy of the loop-mediated isothermal amplification assay for extrapulmonary tuberculosis: A meta-analysis. PLOS ONE. 2018; 13:e0199290 10.1371/journal.pone.0199290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen Y, Fang L, Xu X, Ye B, Yu G. CapitalBio Mycobacterium real-time polymerase chain reaction detection test: Rapid diagnosis of Mycobacterium tuberculosis and nontuberculous mycobacterial infection. Int J Infect Dis. 2020;98:1–5. 10.1016/j.ijid.2020.06.042 [DOI] [PubMed] [Google Scholar]
- 31.Daley P, Thomas S, Pai M. Nucleic acid amplification tests for the diagnosis of tuberculous lymphadenitis: a systematic review. Int J Tuberc Lung Dis. 2007;11:1166–1176. [PubMed] [Google Scholar]
- 32.Fei X, Li C, Zhang Y, Zhang H, Liu X, Ji X, et al. Next-generation sequencing of cerebrospinal fluid for the diagnosis of neurocysticercosis. Clin Neurol Neurosurg. 2020;193:105752 10.1016/j.clineuro.2020.105752 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
(DOCX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.